Abstract
Background
Many cases of acute lymphoblastic leukemia (ALL) carry visible acquired chromosomal changes of pathogenetic, diagnostic, and prognostic importance. Nevertheless, from one-fourth to half of newly diagnosed ALL patients have no visible chromosomal changes detectable by G-banding analysis at diagnosis. The introduction of powerful molecular methodologies has shown that many karyotypically normal ALLs carry clinically important submicroscopic aberrations.
Case presentation
We used fluorescence in situ hybridization (FISH), array comparative genomic hybridization (aCGH), RNA sequencing, reverse transcription (RT) and genomic polymerase chain reaction (PCR), as well as Sanger sequencing to investigate a case of pediatric ALL with a normal karyotype. FISH with a commercial PDGFRB breakapart probe showed loss of the distal part of the probe suggesting a breakpoint within the PDGFRB locus. aCGH revealed submicroscopic deletions in chromosome bands 5q32q35.3 (about 30 Mb long, starting within PDGFRB and finishing in the CANX locus), 7q34 (within TCRB), 9p13 (PAX5), 10q26.13 (DMBT1), 14q11.2 (TRAC), and 14q32.33 (within the IGH locus). RNA sequencing detected an in-frame GTF2I–PDGFRB and an out-of-frame IKZF1–TYW1 fusion transcript. Both fusion transcripts were verified by RT-PCR together with Sanger sequencing and interphase FISH. The GTF2I–PDGFRB fusion was also verified by genomic PCR and FISH. The corresponding GTF2I–PDGFRB fusion protein would consist of almost the entire GTF2I and that part of PDGFRB which harbors the catalytic domain of the tyrosine kinase. It would therefore seem to lead to abnormal tyrosine kinase activity in a manner similar to what has been seen for other PDGFRB fusion proteins.
Conclusions
The examined pediatric leukemia is a Ph-like ALL which carries novel GTF2I–PDGFRB and IKZF1–TYW1 fusion genes together with additional submicroscopic deletions. Because hematologic neoplasms with PDGFRB-fusion genes can be treated with tyrosine kinase inhibitors, the detection of such novel fusions may be clinically important. Since the GTF2I–PDGFRB could be detected only after molecular studies of the leukemic cells, further investigations of ALL-cases, perhaps especially but not exclusively with a normal karyotype, are needed in order to determine the frequency of GTF2I–PDGFRB in leukemia, and also to find out which clinical impact the fusion may have.
Similar content being viewed by others
Background
Most cases of acute lymphoblastic leukemia (ALL) carry visible acquired chromosomal changes of pathogenetic, diagnostic, and prognostic importance [1]. However, up to 43% of newly diagnosed ALL patients are reported to carry normal G-banded karyotypes at diagnosis [2,3,4]. The introduction of powerful molecular methodologies such as fluorescence in situ hybridization (FISH), array based gene expression analysis of mRNA and miRNA, array comparative genomic hybridization (aCGH), single nucleotide polymorphism (SNP) arrays, and, very recently, whole genome sequencing, whole-exome sequencing, and RNA sequencing, has shown that many of the karyotypically normal ALLs nevertheless carry clinically important submicroscopic genetic aberrations [2, 5,6,7,8,9]. Using SNP methodology, Paulsson et al. [6] showed that almost all 45 examined adult and adolescent ALL cases carried cryptic genetic changes. Similar results were obtained by Okamoto et al. [7] who studied with SNP methodology 75 adult ALLs and compared them with 399 pediatric ALLs, and by Othman et al. [8] who studied 61 karyotypically normal ALL cases. A common theme in the published studies is that deletions of genes involved in B lymphopoiesis and cell-cycle regulation, such as CDKN2A, EBF1, ETV6, IKZF1, PAX5, and RB1, occur with a high frequency [6,7,8].
In the present study, we used FISH, aCGH, and RNA sequencing to further examine a karyotypically normal case of pediatric ALL finding novel fusion genes and submicroscopic deletions.
Case presentation
Case report
The patient was a 3 years old girl who for 2 months had symptoms and signs of disease with fever and reduced general condition. The last week she had nose bleedings, petechiae, and pain from the throat and abdomen. The initial blood tests showed hemoglobin 4.3 g/dL (normal 11–14 g/dL), leukocytes 72 × 109/L (normal 4 × 109/L–15 × 109/L), and thrombocytes 19 × 109/L (normal 150 × 109/L–450 × 109/L). The blood and bone marrow smears revealed that the patient had acute lymphoblastic leukemia. Immunophenotyping of peripheral blood showed 80% pre-B-lymphoblasts, in the bone marrow 88%. The patient was treated according to the NOPHO ALL 2008 protocol, high-risk group [10]. She started with induction treatment, but the bone marrow on day 15 showed 90% lymphoblasts. The treatment was therefore changed to block treatment according to the protocol, and on day 34 minimal residual disease (MRD) was less than 0.01%. Also repeated later measurements have shown MRD of less than 0.01%.
Genetic analyses
The G-banding analysis at diagnosis of bone marrow and blood metaphase cells revealed a normal karyotype, 46,XX, in all 25 examined metaphases (Fig. 1a). Interphase FISH analyses with the Cytocell (Cytocell, Banbury, Oxfordshire, UK) multiprobe ALL panel did not detect aberrations of MYC, CDKN2A, TCF3, MLL, and IGH, no ETV6-RUNX1 or BCR-ABL1 fusions, nor was hyperdiploidy seen in 200 examined nuclei (data not shown). FISH with the PDGFRB breakapart probe (Cytocell) showed loss of the distal part of the probe in 176 out of 201 examined interphase nuclei from white blood cells and 42 out of 100 examined interphase nuclei from bone marrow cells, suggesting a genetic breakpoint in the PDGFRB locus in 5q32 (Fig. 1b).
G-banding, FISH, and aCGH analyses. a The G-banding analysis showed a normal karyotype, 46,XX. b Interphase FISH with the PDGFRB breakapart probe on a normal nucleus and on a nucleus with loss of the distal probe suggesting a genetic breakpoint in the PDGFRB locus. c aCGH showing deletions on chromosome bands 5q32q35.3, 7q34, 9p13, 10q26.13, 14q11.2, and 14q32.33
aCGH was performed with genomic DNA extracted from the patientʼs peripheral blood cells using the Maxwell 16 Instrument System and the Maxwell 16 Cell DNA Purification Kit (Promega, Madison, USA). Promegaʼs human genomic female DNA (Promega, Madison, USA) was used as reference DNA. For aCGH, the CytoSure array products were used (Oxford Gene Technology, Begbroke, Oxfordshire, UK) following the company’s protocols. The CytoSure Genomic DNA Labelling Kit was used for labelling of one μg of patient’s and reference DNA, the CytoSure Cancer +SNP array was used for hybridization, and the CytoSure Interpret analysis software was used to analyse the results.
aCGH revealed submicroscopic deletions in chromosome bands 5q32q35.3, 7q34 (within TCRB), 9p13 (PAX5), 10q26.13 (DMBT1), 14q11.2 (TRAC), and 14q32.33 (within the IGH locus) (Fig. 1c, Table 1). The deletion on 5q was 30 Mb long, started between exons 8 and 9 of PDGFRB (5q32), and finished in the CANX locus (5q35.3). The result was in agreement with the FISH data obtained with the PDGFRB breakapart probe (Fig. 1b, c). Because both FISH and aCGH findings indicated a possible PDGFRB-fusion gene, one µg of the total RNA, extracted from the patient´s bone marrow at the time of diagnosis using miRNeasy Mini Kit (Qiagen Nordic, Oslo, Norway), was sent to the Genomics Core Facility at the Norwegian Radium Hospital, Oslo University Hospital (http://genomics.no/oslo/) for high-throughput paired-end RNA-sequencing. For library preparation from total RNA the Illumina TruSeq RNA Access Library Prep kit was used according to Illuminaʼs protocol (Illumina, San Diego, CA, USA; https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_truseq/truseqrnaaccess/truseq-rna-access-library-prep-guide-15049525-b.pdf). Sequencing was performed on NextSeq 550 System (Illumina) and 16 million reads were generated.
Because the raw fastq RNA sequencing data were in the text-based format, we used the “grep” command-line utility to search for sequences which contained part of the ninth exon of PDGFRB [11]. Using the search term “TCCCTGTCCGAGTGCTGG”, which corresponds to 1713–1730 nt in the PDGFRB reference sequence with accession number NM_002609.3, only one 76 bp long sequence was extracted (Fig. 2a). BLAT of this sequence on the human genome browser-hg19 assembly (http://genome-euro.ucsc.edu/cgi-bin/hgGateway) showed that the sequence between nucleotides 26–76 mapped on chromosome 5 at position 149510177–149510227 and was part of exon 9 of PDGFRB. The sequence between nucleotides 1–27 (GCCAGTTGGAAGTTCCAGCCACAGAAG) mapped on chromosome 7 at three different positions: (a) chr7:74172307–74172333 (exon 32 of general transcription factor Iii, GTF2I, reference sequence: NM_032999.3), (b) chr7: 74603796–74603822 (exon 22 of general transcription factor IIi pseudogene 1, GTF2IP1, reference sequence: NR_002206.3), and (c) chr7:72618618–72618644 (exon 22 of general transcription factor IIi pseudogene 4, GTF2IP4, reference sequence: NR_003580.2). These data were verified when we used the BLAST algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to compare the sequence with the reference sequences NM_002609.3 (PDGFRB), NM_032999.3 (GTF2I), NR_002206.3 (GTF2IP1), and NR_003580.2 (GTF2IP4).
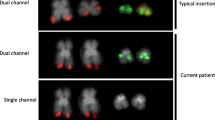
Molecular genetic and FISH analyses for identification of the GTF2I–PDGFRB fusion gene. a The 76 bp sequence obtained from the raw data of RNA sequencing using the command “grep”. The search term “TCCCTGTCCGAGTGCTGG” is underlined. b Partial sequence chromatogram of the PCR amplified cDNA fragment showing the fusion (arrow) of GTF2I and PDGFRB. c Partial sequence chromatogram of the PCR amplified genomic DNA fragment showing the fusion (arrow) of GTF2I and PDGFRB. The identification of the GTF2I gene as the PDGFRB-fusion partner was possible only after FISH experiments using appropriate commercial BAC probes for PDGFRB and GTF2I. d Ideogram of chromosome 5 showing the mapping position of the PDGFRB gene (vertical red line). e Diagram showing the FISH probe RP11-21I20 for PDGFRB. Additional genes in this region are also shown. f Ideogram of chromosome 7 showing the mapping position of the GTF2I gene (vertical green line). g Diagram showing the FISH probe RP11-137E8 for GTF2I. Additional genes in this region are also shown. h FISH on interphase nuclei with the PDGFRB (red signal) and GTF2I (green signal) probes showing a red signal, a green signal and one yellow-fusion signal (arrow)
In order to confirm the existence of the GTF2I–PDGFRB fusion gene, reverse transcription (RT) and genomic PCR analyses were performed as previously described [12]. The primers used for PCR amplifications and Sanger sequencing analyses are shown in Table 2. RT-PCR with the primers GTF2I-3306F1/PDGFRB-1732R1 amplified an 84 bp long cDNA fragment. Sanger sequencing of the PCR products verified the fusion which was found upon searching the RNA sequencing data using the “grep” command (Fig. 2b). Thus, the leukemic cells carried either the fusion transcript GTF2IP1-PDGFRB, or GTF2I–PDGFRB, or GTF2IP4-PDGFRB. Genomic PCR with the primers GTF2I-3317F1 and PDGFRB-1737R1 amplified a single 1200 bp fragment which by Sanger sequencing was shown to be a hybrid genomic DNA fragment in which intron 8 of PDGFRB is fused with either intron 22 of GTF2IP1, intron 32 of GTF2I, or intron 22 of GTF2IP4 (Fig. 2c). Additional interphase FISH experiments were performed to detect the GTF2I–PDGFRB fusion gene (Fig. 2d–h). BACs RP11-21I20 and RP11-137E8 were retrieved from the Human “32K” BAC Re-Array library (BACPAC Resources, https://bacpacresources.org/home.htm). RP11-21I20, the probe for the PDGFRB gene, mapped to band 5q32 (Position: chr5: 149,320,375–149,496,703; UCSC Genome Browser on Human February 2009 GRCh37/hg19 Assembly) and was labeled red (Fig. 2d, e). RP11-137E8, the probe for the GTF2I gene, mapped to band 7q11.23 (Position: chr7: 73,944,720–74,129,587) and was labeled green (Fig. 2f, g). Detailed information about the FISH procedure was given previously [13]. Fluorescent signals were captured and analyzed using the CytoVision system (Leica Biosystems, Newcastle, UK). FISH analysis showed a fusion signal in 44 out of 100 examined interphase nuclei from bone marrow cells suggesting a GTF2I–PDGFRB fusion gene (Fig. 2h). Thus, FISH with specific probes for PDGFRB and GTF2I was crucial to show that a novel GTF2I–PDGFRB fusion gene had been formed (Fig. 2d–h).
Using the FusionCatcher software [14] with the fastq files of the RNA sequencing data, an out-of-frame IKZF1–TYW1 fusion transcript was found (Fig. 3a).
Molecular genetic and FISH analyses for identification of the IKZF1–TYW1 fusion gene. a The sequence which was obtained using the FusionCatcher software with the fastq files of the RNA sequencing data. b Partial sequence chromatogram of the PCR amplified cDNA fragment showing the fusion (arrow) of IKZF1 and TYW1. c Ideogram of chromosome 7 showing the mapping position of IKZF1 (vertical red line) and TYW1 (vertical green line). d Diagram showing the FISH probe (RP11-813K3 and RP11-95E2) for IKZF1. Additional genes in this region are also shown. e Diagram showing the FISH probe (RP11-458F8 and RP11-166O4) for TYW1. Additional genes in this region are also shown. f FISH on interphase nuclei with the IKZF1 (red signal) and TYW1 (green signal) probes showing a red signal, a green signal, and two yellow-fusion signals (arrow)
RT-PCR with the primers IKZF1-469F1/TYW1-1282R1 (Table 2) amplified a 319 bp long cDNA fragment which by Sanger sequencing was shown to contain IKZF1–TYW1 (Fig. 3b). The fusion point thus detected was identical to that found by analysis of the RNA sequencing data using the FusionCatcher software (Fig. 3a, b). In the IKZF1–TYW1 transcript, exon 4 of IKZF1 (nt 642 in sequence with accession number NM_006060 version 6) was fused out-of-frame to exon 8 of TYW1 (nt 1131 in NM_018264 version 4) (Fig. 3b).
The IKZF1–TYW1 fusion gene would encode a putative truncated 159 aa IKZF1 protein containing the first 140 aa of IKZF1 (NP_006051) and 19 aa from the fused TYW1. This protein would not contain the functional domains of the normal IKZF1 protein. Alterations of IKZF1 (often deletions) are strongly associated with BCR-ABL1-positive as well as Ph-like ALL [15, 16].
Additional interphase FISH experiments were performed to detect the IKZF1–TYW1 fusion gene using a home-made dual color dual fusion probe. The probes were made from commercial BACs which were purchased from BACPAC Resources Center (https://bacpacresources.org). The probe for the IKZF1 gene was constructed from a pool of the clones RP11-813K3 (Accesion number AC020743; Position: chr7: 50157413–50339940) and RP11-95E2 (Accesion number AC018705; chr7: 50475184–50648153) and was labeled red (Fig. 3c, d). The probe for the TYW1 gene was constructed from a pool of the clones RP11-458F8 (Accesion number AC073335; Position: chr7: 66297269–66454983) and RP11-166O4 (Accesion number AC006480; Position: Chr7: 66699524–66859231) and was labeled green (Fig. 3c, e).
FISH analysis showed double fusion signals in 91 out of 100 examined interphase nuclei from white blood cells suggesting an IKZF1–TYW1 fusion gene (Fig. 3f).
Discussion
We present here a case of pediatric leukemia with a normal karyotype, 46,XX, but with multiple hidden aberrations identified using a combination of aCGH, RNA sequencing, and FISH methodologies. The initial FISH analysis with a PDGFRB breakapart probe showed loss of the distal part of the probe suggesting a genetic rearrangement of the PDGFRB gene. Because loss of the distal part of the probe was seen in interphase nuclei and not on metaphase spreads, we concluded that the normal karyotype was from non-leukemic cells whereas the leukemic cells did not divide in vitro. Subsequent analyses by aCGH revealed submicroscopic deletions within the TCRB (7q34), PAX5 (9p13), DMBT1 (10q26.13), 14q11.2 (TRAC), and IGH (14q32) loci, as well as loss of a 30 Mb stretch which started with PDGFRB (5q32) and finished in the CANX locus (5q35.3). The aCGH results were in agreement with previous findings of submicroscopic deletions found at high frequencies in ALL [2, 6,7,8]. Many PAX5 deletions were reported in BCR-ABL1-positive and Ph-like ALL [6, 7, 17]. DMBT1 deletions were reported in tumors of the central nervous system but this gene’s role, if any, in ALL development remains unknown [18, 19]. Finally, concomitant deletions within the immune loci TCR (alpha/delta and gamma) and IGH, which are often accompanied by deletions within IKZF1, were shown to be associated with lymphoid blast transformation of chronic myeloid leukemia [20]. The deletion of 30 Mb in 5q32q35.3, found by aCGH and FISH using a PDGFRB breakapart probe, started within PDGFRB (5q32), indicating the presence of a PDGFRB-fusion gene which we set out to find using RNA sequencing. Combining RNA sequencing, PCR/Sanger sequencing, and FISH methodologies we found and verified two fusion transcripts: The first was a novel in-frame GTF2I–PDGFRB fusion transcript. Due to the extensive homology of the GTF2I gene with two pseudogenes, GTF2IP1 and GTF2IP4, neither RNA sequencing nor the subsequent PCR/Sanger sequencing analyses could verify the precise fusion. FISH analyses based on hybridization with specific probes for the GTF2I and PDGFRB genes helped to identify the fusion as a GTF2I–PDGFRB, however. Similar to the FISH experiments with the PDGFRB breakapart probe, the fusion signal suggesting a GTF2I–PDGFRB hybrid gene was seen on interphase nuclei but not on the examined metaphase spreads. In all likelihood, the leukemic cells did not divide in vitro.
Based on the reference sequences NM_032999.3/NP_127492.1 (GTF2I) and NM_002609.3/NP_002600.1 (PDGFRB), the GTF2I–PDGFRB fusion gene would code for a putative 1671 amino-acid-residues (aa) chimeric protein containing almost the entire GTF2I protein (979 out of 998 aa) and the part of PDGFRB (aa 415 to 1106 in NP_002600.1) which contains the catalytic domain of the protein tyrosine kinase. The putative GTF2I–PDGFRB fusion protein seems to be an abnormal protein tyrosine kinase in a similar way to what has been seen with other PDGFRB fusion proteins [21].
Hematologic neoplasms with PDGFRB-fusion genes can be treated with tyrosine kinase inhibitors [22,23,24]. Whether this applies also to the present case, remains unknown since complete remission was obtained using standard high-risk ALL treatment.
The second novel fusion transcript was an out-of-frame IKZF1–TYW1 which would code for a truncated 159 aa IKZF1 protein containing the first 140 aa of IKZF1 (NP_006051) and 19 aa resulting from the fusion with TYW1. The IKZF1 and TYW1 genes were mapped on chromosome subbands 7p12.2 and 7q11.21. Thus, IKZF1–TYW1 was most probably the product of a 7p12.2/7q11.21 recombination event: maybe an inv(7)(p12q11) or a t(7;7)(p12;q11) rearrangement. Fusions of IKZF1 with various partner genes have been reported before in ALL [25, 26]. However, to the best of our knowledge, this is the first time that an IKZF1–TYW1 fusion transcript is reported. Alterations of IKZF1 (often deletions) are known to be strongly associated with BCR-ABL1-positive and Ph-like ALL [15, 16].
Conclusions
The examined pediatric leukemia was a Ph-like ALL which carried a novel GTF2I–PDGFRB fusion gene, a novel IKZF1–TYW1 fusion gene, and submicroscopic deletions on chromosome bands 5q32q35.3, 7q34 (within TCRB), 9p13 (PAX5), 10q26.13 (DMBT1), and 14q32.33 (within the IGH locus).
Availability of data and materials
All available data are included in the manuscript and its figures.
Abbreviations
- aCGH:
-
array comparative genomic hybridization
- ALL:
-
acute lymphoblastic leukemia
- BAC:
-
bacterial artificial chromosome
- FISH:
-
fluorescence in situ hybridization
- RT-PCR:
-
reverse transcription-polymerase chain reaction
- SNP:
-
single nucleotide polymorphism
References
Heim S, Mitelman F. Cancer cytogenetics: chromosomal and molecular genetic abberations of tumor cells. 4th ed. New York: Wiley-Blackwell; 2015.
Forero-Castro M, Robledo C, Benito R, Abaigar M, Africa Martin A, Arefi M, et al. Genome-wide DNA copy number analysis of acute lymphoblastic leukemia identifies new genetic markers associated with clinical outcome. PLoS ONE. 2016;11:e0148972.
Coccé MC, Alonso CN, Rossi JG, Bernasconi AR, Rampazzi MA, Felice MS, et al. Cytogenetic and molecular findings in children with acute lymphoblastic leukemia: experience of a single institution in argentina. Mol Syndromol. 2015;6:193–203.
Moorman AV, Chilton L, Wilkinson J, Ensor HM, Bown N, Proctor SJ. A population-based cytogenetic study of adults with acute lymphoblastic leukemia. Blood. 2010;115:206–14.
Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102:2951–9.
Paulsson K, Cazier JB, Macdougall F, Stevens J, Stasevich I, Vrcelj N, et al. Microdeletions are a general feature of adult and adolescent acute lymphoblastic leukemia: unexpected similarities with pediatric disease. Proc Natl Acad Sci USA. 2008;105:6708–13.
Okamoto R, Ogawa S, Nowak D, Kawamata N, Akagi T, Kato M, et al. Genomic profiling of adult acute lymphoblastic leukemia by single nucleotide polymorphism oligonucleotide microarray and comparison to pediatric acute lymphoblastic leukemia. Haematologica. 2010;95:1481–8.
Othman MA, Melo JB, Carreira IM, Rincic M, Glaser A, Grygalewicz B, et al. High rates of submicroscopic aberrations in karyotypically normal acute lymphoblastic leukemia. Mol Cytogenet. 2015;8:45.
Liu YF, Wang BY, Zhang WN, Huang JY, Li BS, Zhang M, et al. Genomic profiling of adult and pediatric b-cell acute lymphoblastic leukemia. EBioMedicine. 2016;8:173–83.
Toft N, Birgens H, Abrahamsson J, Griskevicius L, Hallbook H, Heyman M, et al. Results of NOPHO ALL2008 treatment for patients aged 1–45 years with acute lymphoblastic leukemia. Leukemia. 2018;32:606–15.
Panagopoulos I, Gorunova L, Bjerkehagen B, Heim S. The, “grep” command but not FusionMap, FusionFinder or ChimeraScan captures the CIC-DUX4 fusion gene from whole transcriptome sequencing data on a small round cell tumor with t(4;19)(q35;q13). PLoS ONE. 2014;9:e99439.
Panagopoulos I, Gorunova L, Zeller B, Tierens A, Heim S. Cryptic FUS-ERG fusion identified by RNA-sequencing in childhood acute myeloid leukemia. Oncol Rep. 2013;30:2587–92.
Panagopoulos I, Gorunova L, Torkildsen S, Tjonnfjord GE, Micci F, Heim S. DEK-NUP214-fusion identified by RNA-sequencing of an acute myeloid leukemia with t(9;12)(q34;q15). Cancer Genomics Proteomics. 2017;14:437–43.
Nicorici D, Satalan H, Edgren H, Kangaspeska S, Murumagi A, Kallioniemi O, et al. Fusioncatcher—a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv. 2014. https://doi.org/10.1101/011650.
Iacobucci I, Mullighan CG. Genetic basis of acute lymphoblastic leukemia. J Clin Oncol. 2017;35:975–83.
Marke R, van Leeuwen FN, Scheijen B. The many faces of IKZF1 in B-cell precursor acute lymphoblastic leukemia. Haematologica. 2018;103:565–74.
Schwab CJ, Chilton L, Morrison H, Jones L, Al-Shehhi H, Erhorn A, et al. Genes commonly deleted in childhood B-cell precursor acute lymphoblastic leukemia: association with cytogenetics and clinical features. Haematologica. 2013;98:1081–8.
Pang JC, Dong Z, Zhang R, Liu Y, Zhou LF, Chan BW, et al. Mutation analysis of DMBT1 in glioblastoma, medulloblastoma and oligodendroglial tumors. Int J Cancer. 2003;105:76–81.
Motomura K, Mittelbronn M, Paulus W, Brokinkel B, Keyvani K, Sure U, et al. DMBT1 homozygous deletion in diffuse astrocytomas is associated with unfavorable clinical outcome. J Neuropathol Exp Neurol. 2012;71:702–7.
Nacheva EP, Brazma D, Virgili A, Howard-Reeves J, Chanalaris A, Gancheva K, et al. Deletions of immunoglobulin heavy chain and T cell receptor gene regions are uniquely associated with lymphoid blast transformation of chronic myeloid leukemia. BMC Genomics. 2010;11:41.
Vega F, Medeiros LJ, Bueso-Ramos CE, Arboleda P, Miranda RN. Hematolymphoid neoplasms associated with rearrangements of PDGFRA, PDGFRB, and FGFR1. Am J Clin Pathol. 2015;144:377–92.
David M, Cross NC, Burgstaller S, Chase A, Curtis C, Dang R, et al. Durable responses to imatinib in patients with PDGFRB fusion gene-positive and BCR-ABL-negative chronic myeloproliferative disorders. Blood. 2007;109:61–4.
Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371:1005–15.
Walz C, Metzgeroth G, Haferlach C, Schmitt-Graeff A, Fabarius A, Hagen V, et al. Characterization of three new imatinib-responsive fusion genes in chronic myeloproliferative disorders generated by disruption of the platelet-derived growth factor receptor beta gene. Haematologica. 2007;92:163–9.
Gu Z, Churchman M, Roberts K, Li Y, Liu Y, Harvey RC, et al. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun. 2016;7:13331.
Lilljebjorn H, Henningsson R, Hyrenius-Wittsten A, Olsson L, Orsmark-Pietras C, von Palffy S, et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7:11790.
Acknowledgements
Not applicable.
Funding
This work was supported by Grants from Radiumhospitalets Legater.
Author information
Authors and Affiliations
Contributions
IP designed and supervised the research, performed molecular genetic experiments, and wrote the manuscript. MB performed molecular genetic experiments. LG interpreted the cytogenetics and FISH data. MS performed aCGH experiments and interpreted the data. RAUS performed aCGH experiments and interpreted the data. JS performed molecular genetic experiments. KA performed cytogenetics and FISH experiments and interpreted the data. IK performed the bioinformatic analyses. TSH performed the bioinformatic analyses. SL performed RNA sequencing. TF made hematologic evaluations and treated the patient. TAN made the hematologic evaluations and treated the patient. FM interpreted the cytogenetic and FISH data. SH evaluated the cytogenetics and FISH data, and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Regional Committee for Medical and Health Research Ethics, South-East Norway (REK Sør-Øst; http://helseforskning.etikkom.no) and written informed consent was obtained from the patient’s parents to publication of the case details. The ethics committee’s approval included a review of the consent procedure. All patient information has been de-identified.
Consent for publication
Written informed consent was obtained from the patient’s parents to publication of the case details. A copy of the consent form is available on request for review by the Editor of this journal.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Panagopoulos, I., Brunetti, M., Stoltenberg, M. et al. Novel GTF2I–PDGFRB and IKZF1–TYW1 fusions in pediatric leukemia with normal karyotype. Exp Hematol Oncol 8, 12 (2019). https://doi.org/10.1186/s40164-019-0136-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40164-019-0136-y