Abstract
Background
Interleukin-10 (IL-10) is a powerful modulator of anti-tumor immune responses. The IL-10 promoter region polymorphisms are known to regulate IL-10 production, and thus are thought to be implicated in tumorigenesis. Recently, the roles of these polymorphisms in urologic cancer have been extensively studied, with conflicting results. Therefore, we conducted the present meta-analysis to better elucidate the correlations between IL-10 polymorphisms and urologic cancer risk.
Methods
Eligible articles were searched in PubMed, Medline, Embase, Scopus and CNKI up to May 2016. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to detect any potential associations between IL-10 polymorphisms and the risk of urologic cancer.
Results
A total of 22 case–control studies including 8572 patients and 9843 controls were analyzed. The overall meta-analysis results showed that IL-10 −592C>A polymorphism was significantly associated with urologic cancer in CA versus AA (P = 0.04, OR 0.87, 95% CI 0.76–0.99) and AA versus CC+CA (P = 0.03, OR 1.15, 95% CI 1.02–1.31). Subgroup analyses by cancer types suggested there were significant associations between all the three investigated IL-10 polymorphisms and bladder cancer. However, subgroup analyses by ethnicity only detected a weak association between IL-10 −819C>T and Asian population.
Conclusions
Our findings suggests that IL-10 −592C>A polymorphism may implicate with urologic cancer risk. Besides, promoter region polymorphisms of IL-10 may serve as potential biological markers, especially for bladder cancer. Furthermore, IL-10 −819C>T polymorphism may contribute to urologic cancer susceptibility in Asians while all the three studied variants of IL-10 did not relate to Caucasian urologic cancer predisposition.
Similar content being viewed by others
Background
Commonly seen urologic cancers such as prostate cancer, renal cancer, and bladder cancer are leading causes of cancer-related morbidity and mortality globally (Siegel et al. 2014). Despite rapid advances in early diagnosis and surgical treatment over the past few decades, the numbers of new urologic cancer cases and associated deaths are continue to increase, making it becomes one of the major threats to public health worldwide (Ferlay et al. 2015).
To date, the exact cause of urologic cancer remains unclear. Certain environmental factors like smoking habit, heavy alcohol intake, high caloric diet and chemical dyes have been identified as potential etiological factors for urologic cancer. However, the fact that only a small portion of individuals exposed to these carcinogenic agents ultimately develop urologic cancer suggests that genetic susceptibility factors may play a crucial part in its pathogenesis (Jiang et al. 2014).
Interleukin-10 (IL-10), encoded by the IL-10 gene located on chromosome 1q31–32, is a potent regulator of anti-tumor immune responses (Eskdale et al. 1997; Mocellin et al. 2005). As a result, certain polymorphisms located in the promoter region of IL-10 gene (−592C>A, −819C>T and −1082A>G), which regulate the expression level of IL-10 protein (Turner et al. 1997; Kingo et al. 2005), were thought to be implicated in the pathogenesis of various kinds of cancers. Recently, many genetic association studies have been carried out to investigate the potential correlations between IL-10 promoter region polymorphisms and urologic cancer risk. However, results of these studies were controversial and the statistical power of individual studies was insufficient. Therefore, we conducted the present meta-analysis to better assess the potential associations of IL-10 genetic polymorphisms with the risk of urologic cancer.
Methods
Literature searching strategy
To retrieve all relevant articles, a systematic literature search of PubMed, Medline, Embase, Scopus and China National Knowledge Infrastructure (CNKI) was performed using the following keywords: “Interleukin-10”, “IL-10”, “Interleukin 10”, “IL 10”, “polymorphism”, “variant”, “genotype”, “allele”, “prostate”, “renal”, “bladder”, “urinary”, “urologic”, “cancer”, “tumor”, “carcinoma”, “neoplasm” and “malignancy”. The initial search was conducted in September 2015 and the latest update was performed in May 2016. In addition, the reference lists of all retrieved articles were reviewed manually for further identification of potentially relevant articles.
Inclusion criteria
The inclusion criteria for the present study were set prior to the literature search. Eligible studies met all the following conditions: (1) case–control study of unrelated urologic cancer patients and control subjects; (2) evaluation of the associations between IL-10 polymorphisms (−592C>A, −819C>T and −1082A>G) and the risk of urologic cancer; (3) presentation of sufficient data to calculate the odds ratios (ORs) and corresponding 95% confidence intervals (CIs); (4) full text in English or Chinese available. If the report was duplicated or identical patients were enrolled in two studies, only the most recent and complete article was included. Abstracts, family-based association studies, case reports, case series, reviews, editorials, expert opinions and conference presentations were intentionally excluded.
Data extraction and quality assessment
From each included studies, the following data were extracted: references, country of origin, ethnicity of study population, the number of cases and controls, types of urologic cancer, allelic and genotypic frequencies of IL-10 polymorphisms in urologic cancer patients and control subjects, and whether the distributions of IL-10 polymorphisms in the control group were in accordance with Hardy–Weinberg equilibrium (HWE). The Newcastle-ottawa quality assessment scale (NOS), a classical rating tool which evaluates the credibility of observational studies from three perspectives: selection, comparability and exposure, was used to assess the reliability of all case–control studies included (Zhang et al. 2014). This rating system has a score range of 0–9, and studies with a score of more than 7 were assumed to be of high quality. Two reviewers (Shi and Xie) conducted the data extraction and quality assessment independently. When necessary, the reviewers wrote to the corresponding authors for extra information or raw data. Any discrepancies between two reviewers were resolved by discussion until reaching a consensus. The final results were reviewed by a senior reviewer (Li).
Statistical analysis
All data analyses were performed using Review Manager Version 5.3.3 (The Cochrane Collaboration, Software Update, Oxford, United Kingdom). HWE was explored with the Chi square test. ORs and 95% CIs were employed to evaluate potential associations between IL-10 polymorphisms and the risk of urologic cancer. Heterogeneity between studies was assessed by using the Q test and I2 statistic. If probability value (P value) of Q test was less than 0.1 or I2 was greater than 50%, the random-effect model (REM) would be adopted for analyses due to the existence of significant between-study heterogeneity. Otherwise, the fixed-effect model (FEM) would be applied for analyses. Subgroup analyses were performed based on types of cancer and ethnicity of study population. Sensitivity analyses were carried out by omitting one individual study each time. Publication bias was further evaluated with funnel plots. And a P value of 0.05 or less was considered to be statistically significant for all analyses.
Results
Included studies
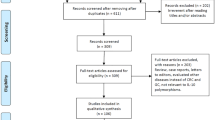
The literature search yielded 462 results. After exclusion of irrelevant or duplicate articles by reading titles and abstracts, 39 articles were selected for further evaluation. Among these, a total of 22 case–control studies including 8572 urologic cancer patients and 9843 control subjects met our inclusion criteria (see Fig. 1), 14/22 were about the IL-10 −592C>A polymorphism, 13/22 were about the IL-10 −819C>T polymorphism, and 20/22 were about the IL-10 −1082A>G polymorphism. All included studies were published between 2002 and 2016. Of these, there were 16 studies of prostate cancer, 4 studies of renal cancer, and 2 studies of bladder cancer. All articles were published in English except for two in Chinese. Characteristics of studies analyzing IL-10 −592C>A polymorphism were summarized in Table 1, characteristics of studies examining IL-10 −819C>T polymorphism were summarized in Table 2, and characteristics of studies investigating IL-10 −1082A>G polymorphism were summarized in Table 3.
Risk of bias in included studies
As shown in Tables 1, 2 and 3, the average NOS score of included studies was 7.59 (range from 7 to 8), suggesting that all enrolled articles were of relatively high quality. The improper selection of controls and mismatching baseline characteristics of urologic cancer cases and control subjects (age and/or ethnicity) were the major sources of biases.
IL-10 −592C>A polymorphism and urologic cancer risk
For IL-10 −592C>A polymorphism, a total of 14 studies including 5899 urologic cancer patients and 6181 control subjects were investigated (Dluzniewski et al. 2012; Dwivedi et al. 2015a, b; Eder et al. 2007; Faupel-Badger et al. 2008; Liu et al. 2010; VanCleave et al. 2010; Wang et al. 2009; Winchester et al. 2015; Xu et al. 2005; Zabaleta et al. 2008; Basturk et al. 2005; Cozar et al. 2007; Chang et al. 2016; Chen et al. 2013). HWE test for the control group of each included studies demonstrated that only 1 study deviated from HWE (see Table 1). In order to explore the association between IL-10 −592C>A polymorphism and urologic cancer risk, we compared distribution of genotypes and alleles in every genetic model. For CC versus AA, CA versus AA, CA versus CC+AA, and AA versus CC+CA, between-study heterogeneity was mild, and analyses were performed with FEMs. For CC versus CA, CC versus CA+AA, and C versus A, REMs were selected due to severe between-study heterogeneity. A significant association with urologic cancer was found for IL-10 −592C>A polymorphism in CA versus AA (P = 0.04, OR 0.87, 95% CI 0.76–0.99) and AA versus CC+CA (P = 0.03, OR 1.15, 95% CI 1.02–1.31) (see Figs. 2, 3).
Forest plots on association between IL-10 −592C>A polymorphism and urologic cancer risk. a Forest plot of CC versus CA for IL-10 −592C>A polymorphism and urologic cancer risk is shown. b Forest plot of CC versus AA for IL-10 −592C>A polymorphism and urologic cancer risk is shown. c Forest plot of CA versus AA for IL-10 −592C>A polymorphism and urologic cancer risk is shown. d Forest plot of CC versus CA+AA for IL-10 −592C>A polymorphism and urologic cancer risk is shown
Forest plots on association between IL-10 −592C>A polymorphism and urologic cancer risk. a Forest plot of CA versus CC+AA for IL-10 −592C>A polymorphism and urologic cancer risk is shown. b Forest plot of AA versus CC+CA for IL-10 −592C>A polymorphism and urologic cancer risk is shown. c Forest plot of C versus A for IL-10 −592C>A polymorphism and urologic cancer risk is shown
IL-10 −819C>T polymorphism and urologic cancer risk
A total of 13 studies with 4655 cancer cases and 6344 healthy controls were enrolled to evaluate the association between IL-10 −819C>T polymorphism and urologic cancer risk (Dwivedi et al. 2015a, b; Faupel-Badger et al. 2008; Liu et al. 2010; VanCleave et al. 2010; Winchester et al. 2015; Zabaleta et al. 2008; Basturk et al. 2005; Cozar et al. 2007; Chang et al. 2016; Chen et al. 2013; Kesarwani et al. 2009; Michaud et al. 2006; Ahirwar et al. 2009). HWE test for the control group of eligible studies revealed that 4 studies violated HWE (see Table 2). All genetic models were tested to detect any differences in genotypic and allelic frequencies of cases and controls. For CT versus TT, there was only trivial between-study heterogeneity, and FEM was employed for analysis. For CC versus CT, CC versus TT, CC versus CT+TT, CT versus CC+TT, TT versus CC+CT, and C versus T, between-study heterogeneity was obvious, and REMs were adopted for analyses. No significant association with urologic cancer was found for IL-10 −819C>T polymorphism in any genetic models (see Figs. 4, 5).
Forest plots on association between IL-10 −819C>T polymorphism and urologic cancer risk. a Forest plot of CC versus CT for IL-10 −819C>T polymorphism and urologic cancer risk is shown. b Forest plot of CC versus TT for IL-10 −819C>T polymorphism and urologic cancer risk is shown. c Forest plot of CT versus TT for IL-10 −819C>T polymorphism and urologic cancer risk is shown. d Forest plot of CC versus CT+TT for IL-10 −819C>T polymorphism and urologic cancer risk is shown
Forest plots on association between IL-10 −819C>T polymorphism and urologic cancer risk. a. Forest plot of CT versus CC+TT for IL-10 −819C>T polymorphism and urologic cancer risk is shown. b Forest plot of TT versus CC+CT for IL-10 −819C>T polymorphism and urologic cancer risk is shown. c Forest plot of C versus T for IL-10 −819C>T polymorphism and urologic cancer risk is shown
IL-10 −1082A>G polymorphism and urologic cancer risk
Of the 20 included studies for IL-10 −1082A>G polymorphism, there were 7401 urologic cancer patients and 8437 controls (Dluzniewski et al. 2012; Faupel-Badger et al. 2008; Liu et al. 2010; VanCleave et al. 2010; Wang et al. 2009; Winchester et al. 2015; Xu et al. 2005; Zabaleta et al. 2008; Basturk et al. 2005; Cozar et al. 2007; Chang et al. 2016; Chen et al. 2013; Kesarwani et al. 2009; Michaud et al. 2006; Ahirwar et al. 2009; Ianni et al. 2013; McCarron et al. 2002; Niu 2011; Omrani et al. 2009; Havranek et al. 2005). Deviations from HWE were found in 5 studies while the remaining 15 studies were in accordance with HWE (see Table 3). For evaluation of the association between IL-10 −1082A>G polymorphism and urologic cancer risk, frequencies of genotypes and alleles in cases and control subjects were compared in every genetic model. REMs were used for all analyses on account of striking between-study heterogeneity, and no significant association was detected between IL-10 −1082A>G polymorphism and urologic cancer risk (see Figs. 6, 7).
Forest plots on association between IL-10 −1082A>G polymorphism and urologic cancer risk. a Forest plot of AA versus AG for IL-10 −1082A>G polymorphism and urologic cancer risk is shown. b Forest plot of AA versus GG for IL-10 −1082A>G polymorphism and urologic cancer risk is shown. c Forest plot of AG versus GG for IL-10 −1082A>G polymorphism and urologic cancer risk is shown. d Forest plot of AA versus AG+GG for IL-10 −1082A>G polymorphism and urologic cancer risk is shown
Forest plots on association between IL-10 −1082A>G polymorphism and urologic cancer risk. a Forest plot of AG versus AA+GG for IL-10 −1082A>G polymorphism and urologic cancer risk is shown. b Forest plot of GG versus AA+AG for IL-10 −1082A>G polymorphism and urologic cancer risk is shown. c Forest plot of A versus G for IL-10 −1082A>G polymorphism and urologic cancer risk is shown
Subgroup analysis
For IL-10 polymorphisms (−592C>A, −819C>T, −1082A>G) and urologic cancer risk, subgroup analyses were performed by stratifying available data according to types of cancer and ethnicity of study population. When data were stratified by cancer types, we found that IL-10 −592C>A polymorphism was significantly associated with bladder cancer risk in CC versus AA (P = 0.002, OR 0.51, 95% CI 0.33–0.78), CA versus AA (P = 0.004, OR 0.64, 95% CI 0.48–0.87), CC versus CA+AA (P = 0.02, OR 0.62, 95% CI 0.41–0.93), CA versus CC+AA (P = 0.04, OR 0.74, 95% CI 0.56–0.99), AA versus CC+CA (P = 0.0004, OR 1.65, 95% CI 1.25–2.19), and C versus A (P = 0.00001, OR 0.66, 95% CI 0.54–0.82). Besides, IL-10 −819C>T polymorphism was significantly correlated with bladder cancer risk in CC versus CT (P = 0.03, OR 0.71, 95% CI 0.52–0.96), CC versus TT (P = 0.0005, OR 0.57, 95% CI 0.41–0.78), CC versus CT+TT (P = 0.002, OR 0.63, 95% CI 0.47–0.84), and C versus T (P < 0.00001, OR 0.72, 95% CI 0.61–0.84). Additionally, IL-10 −1082A>G polymorphism was also significantly associated with the risk of bladder cancer in AA versus GG (P = 0.02, OR 2.00, 95% CI 1.13–3.55), AG versus GG (P = 0.01, OR 2.03, 95% CI 1.16–3.55), and GG versus AA+AG (P = 0.009, OR 0.49, 95% CI 0.28–0.84). When data were subsequently stratified by ethnicity, we observed a significant association with urologic cancer risk for IL-10 −819C>T polymorphism in CT versus TT (P = 0.0009, OR 0.81, 95% CI 0.69–0.95). No any other associations were found in subgroup analyses (see Tables 4, 5, 6).
Sensitivity analysis
Sensitivity analyses were carried out through removing one individual study each time. For IL-10 −519C>A polymorphism, when the study performed by Dluzniewski et al. (2012), Faupel-Badger et al. (2008) or Chen et al. (2013) was excluded, the significant association with urologic cancer was no longer observed in CA versus AA, and AA versus CC+CA. For IL-10 −819C>T polymorphism, when the study of Liu et al. (2010) or Michaud et al. (2006) was removed, the null association with urologic cancer in CT versus TT was altered. For IL-10 −1082A>G polymorphism, however, removing any study did not impact the overall results.
Publication bias
Potential publication bias was evaluated with funnel plots. Visual inspection of funnel plots revealed no apparent asymmetry for IL-10 −592C>A, −819C>T, and −1082A>G polymorphisms. And these results indicated that significant publication bias was unlikely.
Discussion
Urologic cancer is a major public health problem. According to a recent survey, prostate cancer, renal cancer and bladder cancer altogether accounted for 13.3% (1879,000/14090,000) new cancer cases and 7.5% (616,000/8201,000) cancer-related deaths worldwide in 2012, making the urologic cancer ranked as the second most common group of malignancies in terms of morbidity, and the third most common group of malignancies in terms of mortality (Ferlay et al. 2015).
To date, the etiologies of urologic cancer are still largely unknown in spite of extensive studies. However, it has become evident recently that multiple immunomodulatory cytokines are implicated in the process of tumor genesis (Kurzrock 2001; Smyth et al. 2004). Among these cytokines, IL-10 is a multifunctional immunological regulator mainly produced by B cells, T cells and activated monocytes/marcophages. As an important modulator of immune responses, IL-10 can be both tumor-promoting and tumor-inhibiting since it has both immunosuppressive and anti-angiogenic functions (Mocellin et al. 2005). On the one hand, the immunosuppressive property of IL-10 may suppress anti-tumor immune responses and promote tumor development. On the other hand, the anti-angiogenic property of IL-10 may inhibit microvasculature formation and tumor growth. Previous studies have found that serum level of IL-10 was significantly elevated in urologic cancer, and it was closely correlated with tumor progression and metastasis (Stearns et al. 1999; Uwatoko et al. 2002; Dwivedi et al. 2015a, b), which suggested that IL-10 may play a vital role in the development of urologic cancer.
IL-10 gene is located on chromosome 1q31–32. Common promoter region polymorphisms of IL-10 gene, −592C>A (rs1800872), −819C>T (rs1800871) and −1082A>G (rs1800896) were found to influence the production of IL-10 (Turner et al. 1997; Kingo et al. 2005). Consequently, it is biologically plausible that these polymorphisms may be associated with susceptibility to urologic cancer.
Recently, numerous studies have tried to explore the potential associations between IL-10 polymorphisms and the risk of urologic cancer, but the results were contradicted. Thus, we conducted the present meta-analysis to solve the conflict and obtain a more conclusive result. And our overall analyses suggested that IL-10 −592C>A polymorphism was significantly associated with the risk of urologic cancer in CA versus AA, and AA versus CC+CA. However, we failed to detect any significant associations with urologic cancer for IL-10 −819C>T and −1082A>G polymorphisms in overall analyses. Considering the differences of carcinogenic mechanisms for each type of cancer and the importance of ethnic background in genetic investigations, stratified analyses were subsequently performed by categorizing included studies into different subgroups on the basis of types of cancer and ethnicity of study population. When data were stratified by types of cancer, we found that IL-10 −592C>A, −819C>T and −1082A>G polymorphisms were all significantly associated with the risk of bladder cancer in certain genetic models. In addition, the A allele of −592C>A polymorphism and T allele of −819C>T polymorphism conferred an increased susceptibility to bladder cancer. When data were stratified by ethnicity of study population, a significant association with urologic cancer risk in Asians was detected for IL-10 −819C>T polymorphism in CT versus TT. No any other significant associations between IL-10 polymorphisms and urologic cancer risk were observed in subgroup analyses. For the evaluation of the heterogeneity, we found that the between-study heterogeneity remained significant in several subgroup comparisons, suggesting that differences in cancer type and ethnicity could not fully elucidate the observed inconsistent results, and other unmeasured characteristics of study participants may partially attribute to the heterogeneity between studies. Moreover, we noticed a substantial decrease of heterogeneity for IL-10 −592C>A polymorphism when the study performed by Zabaleta et al. (2008) was omitted, and that for IL-10 −819C>T polymorphism when the study conducted by Chen et al. (2013) was removed or that for IL-10 1082A>G polymorphism when the studies of Ianni et al. (2013) and Niu (2011) were excluded, which suggested that these studies were the major sources of the observed heterogeneity.
This study is certainly not without limitations. Firstly, the number of studies investigating the associations of certain IL-10 polymorphisms with renal cancer or bladder cancer is still limited, and sample size of several included studies were obviously not sufficient, which precluded us from drawing definite conclusions. Secondly, our results were based on unadjusted estimates since the majority of included studies failed to report baseline characteristics of individuals, such as age, sex, smoking status and eating habits. And lack of analyses adjusted for these potential confounding factors may affect the reliability of our results. Thirdly, although funnel plots revealed no apparent publication bias, we still could not eliminate the possibility of publication bias since only published studies were included. Fourthly, all included studies were published in English or Chinese, therefore, maybe some qualified articles in other languages were missed. Fifthly, genetic associations of IL-10 polymorphisms with urologic cancer may also be influenced by gene–gene and gene-environmental interactions. It is possible that one certain polymorphism may be associated with the risk of urologic cancer, but due to interactions with multiple genes and environmental factors, the association would no longer be observed.
Conclusions
In conclusion, the current meta-analysis suggests that IL-10 −592C>A polymorphism may implicate with urologic cancer risk. Besides, promoter region polymorphisms of IL-10 may serve as potential biological markers, especially for bladder cancer. Furthermore, IL-10 −819C>T polymorphism may contribute to urologic cancer susceptibility in Asians while all the three studied variants of IL-10 did not relate to Caucasian urologic cancer predisposition. However, it should be pointed out that the present results concerning renal cancer and bladder cancer were based on limited number of case–control studies, and further multi-center studies with larger sample size from different populations are warranted to confirm our results. Besides, given that immunomodulating cytokines play a crucial role in regulating anti-tumor immune responses, future investigations are needed to explore the potential roles of other polymorphisms of these cytokine genes in the occurrence and development of urologic cancer.
References
Ahirwar D, Mandhani A, Mittal RD (2009) Interleukin-10 G-1082A and C-819T polymorphisms as possible molecular markers of urothelial bladder cancer. Arch Med Res 40:97–102
Basturk B, Yavascaoglu I, Vuruskan H et al (2005) Cytokine gene polymorphisms as potential risk and protective factors in renal cell carcinoma. Cytokine 30:41–45
Chang WS, Liao CH, Tsai CW et al (2016) The role of IL-10 promoter polymorphisms in renal cell carcinoma. Anticancer Res 36:2205–2209
Chen ZG, Zhou W, Dai MJ, Wu ZG, Jin R (2013) Association between the interaction polymorphisms of interleukin-10 and smoking on patients with bladder cancer risk from a case–control study. Zhonghua Liu Xing Bing Xue Za Zhi 34:183–186
Cozar JM, Romero JM, Aptsiauri N et al (2007) High incidence of CTLA-4 AA (CT60) polymorphism in renal cell cancer. Hum Immunol 68:698–704
Dluzniewski PJ, Wang MH, Zheng SL et al (2012) Variation in IL10 and other genes involved in the immune response and in oxidation and prostate cancer recurrence. Cancer Epidemiol Biomark Prev 21:1774–1782
Dwivedi S, Goel A, Khattri S et al (2015a) Genetic variability at promoters of IL-18 (pro-) and IL-10 (anti-) inflammatory gene affects susceptibility and their circulating serum levels: an explorative study of prostate cancer patients in North Indian populations. Cytokine 74:117–122
Dwivedi S, Goel A, Mandhani A et al (2015b) Functional genetic variability at promoters of pro-(IL-18) and anti-(IL-10) inflammatory affects their mRNA expression and survival in prostate carcinoma patients: five year follow-up study. Prostate 75:1737–1746
Eder T, Mayer R, Langsenlehner U et al (2007) Interleukin-10 [ATA] promoter haplotype and prostate cancer risk: a population-based study. Eur J Cancer 43:472–475
Eskdale J, Kube D, Tesch H, Gallagher G (1997) Mapping of the human IL10 gene and further characterization of the 5′ flanking sequence. Immunogenetics 46:120–128
Faupel-Badger JM, Kidd LC, Albanes D, Virtamo J, Woodson K, Tangrea JA (2008) Association of IL-10 polymorphisms with prostate cancer risk and grade of disease. World J Urol 19:119–124
Ferlay J, Soerjomataram I, Dikshit R et al (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136:E359–E386
Havranek E, Howell WM, Fussell HM, Whelan JA, Whelan MA, Pandha HS (2005) An interleukin-10 promoter polymorphism may influence tumor development in renal cell carcinoma. J Urol 173:709–712
Ianni M, Porcellini E, Carbone I et al (2013) Genetic factors regulating inflammation and DNA methylation associated with prostate cancer. Prostate Cancer Prostatic Dis 16:56–60
Jiang W, Sun G, Xiong J, Xi X, Shi Z (2014) Association of CYP1B1 L432V polymorphism with urinary cancer susceptibility: a meta-analysis. Diagn Pathol 9:113
Kesarwani P, Ahirwar DK, Mandhani A et al (2009) IL-10 1082 G>A: a risk for prostate cancer but may be protective against progression of prostate cancer in North Indian cohort. World J Urol 27:389–396
Kingo K, Ratsep R, Koks S, Karelson M, Silm H, Vasar E (2005) Influence of genetic polymorphisms on interleukin-10 mRNA expression and psoriasis susceptibility. J Dermatol Sci 37:111–113
Kurzrock R (2001) Cytokine deregulation in cancer. Biomed Pharmacother 55:543–547
Liu J, Song B, Bai X et al (2010) Association of genetic polymorphisms in the interleukin-10 promoter with risk of prostate cancer in Chinese. BMC Cancer 10:456
McCarron SL, Edwards S, Evans PR et al (2002) Influence of cytokine gene polymorphisms on the development of prostate cancer. Cancer Res 62:3369–3372
Michaud DS, Daugherty SE, Berndt SI et al (2006) Genetic polymorphisms of interleukin-1B (IL-1B), IL-6, IL-8, and IL-10 and risk of prostate cancer. Cancer Res 66:4525–4530
Mocellin S, Marincola FM, Young HA (2005) Interleukin-10 and the immune response against cancer: a counterpoint. J Leukoc Biol 78:1043–1051
Niu WQ (2011) The study on the association between the IL-10 promoter genetic polymorphisms and prostate cancer in Hubei Hans population. Int J Lab Med 32:41–43
Omrani MD, Bazargani S, Bageri M (2009) Interlukin-10, interferon-g and tumor necrosis factor—a genes variation in prostate cancer and benign prostatic hyperplasia. Curr Urol 2:175–180
Siegel R, Ma J, Zou Z, Jemal A (2014) Cancer statistics, 2014. CA Cancer J Clin 64:9–29
Smyth MJ, Cretney E, Kershaw MH, Hayakawa Y (2004) Cytokines in cancer immunity and immunotherapy. Immunol Rev 202:275–293
Stearns ME, Rhim J, Wang M (1999) Interleukin 10 (IL-10) inhibition of primary human prostate cell-induced angiogenesis: IL-10 stimulation of tissue inhibitor of metalloproteinase-1 and inhibition of matrix metalloproteinase (MMP)-2/MMP-9 secretion. Clin Cancer Res 5:189–196
Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV (1997) An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet 24:1–8
Uwatoko N, Tokunaga T, Hatanaka H et al (2002) Expression of interleukin-10 is inversely correlated with distant metastasis of renal cell carcinoma. Int J Oncol 20:729–733
VanCleave TT, Moore JH, Benford ML et al (2010) Interaction among variant vascular endothelial growth factor (VEGF) and its receptor in relation to prostate cancer risk. Prostate 70:341–352
Wang MH, Helzlsouer KJ, Smith MW et al (2009) Association of IL10 and other immune response-and obesity-related genes with prostate cancer in CLUE II. Prostate 69:874–885
Winchester DA, Till C, Goodman PJ et al (2015) Variation in genes involved in the immune response and prostate cancer risk in the placebo arm of the prostate cancer prevention trial. Prostate 75:1403–1418
Xu J, Lowey J, Wiklund F et al (2005) The interaction of four genes in the inflammation pathway significantly predicts prostate cancer risk. Cancer Epidemiol Biomark Prev 14:2563–2568
Zabaleta J, Lin HY, Sierra RA et al (2008) Interactions of cytokine gene polymorphisms in prostate cancer risk. Carcinogenesis 29:573–578
Zhang R, Wang J, Yang R et al (2014) Effects of Pro12Ala polymorphism in peroxisome proliferator-activated receptor-γ2 gene on metabolic syndrome risk: a meta-analysis. Gene 535:79–87
Authors’ contributions
XHS and SWL conceived of the study, participated in its design. XHS and XCX conducted the systematic literature review. XSX and YXJ performed data analyses. XHS, XCX and SWL drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors have no support or funding to report.
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Shi, X., Xie, X., Xun, X. et al. Associations of IL-10 genetic polymorphisms with the risk of urologic cancer: a meta-analysis based on 18,415 subjects. SpringerPlus 5, 2034 (2016). https://doi.org/10.1186/s40064-016-3705-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40064-016-3705-0