Abstract
The inhibition of tyrosinase is the most effective method to decrease melanin synthesis during the process of pigmentation. We aimed to find compounds from traditional Chinese medicines (TCM) that are more effective than the most commonly used tyrosinase inhibitor, arbutin. First, we employed homology modeling to construct a tyrosinase-modeled structure, and structure-based virtual screening to screen from 61,000 TCM compounds. We also adopted the following quantitative structure-activity relationship (QSAR) models for ligand-based validation: support vector machine, multiple linear regression, and Bayesian network. Molecular dynamics (MD) simulation was used to confirm the stability of ligand binding. We found that merresectine C might more effectively bind and inhibit the activity of tyrosinase than arbutin. This study provides useful evidence for the potential development of a novel non-toxic bleaching or whitening ingredient.
Similar content being viewed by others
Introduction
Hyperpigmentation, hypermelanosis, skin darkening, or tanning can be caused by ultraviolet(UV) exposure, drugs, or post-inflammatory conditions (Praetorius et al. 2013). UV radiation stimulates melanin synthesis in the epidermal melanocytes (Johnson et al. 1972). Drug-induced hyperpigmentation can be caused by many compounds including minocycline, amiodarone, oral contraceptives, and anticancer drugs (Holm and Nelson 2006; Rappersberger et al. 1989; Kim et al. 2012; Ibrahimi and Anderson 2010; Kew et al. 1977). Post-inflammatory hyperpigmentation can be caused by dermatological inflammatory diseases, or as a side effect of laser treatment (Ortonne and Bissett 2008; Fisher and James 2010). Tyrosinase function has been studied in the context of clinically significant diseases such as albinism or vitiligo (Chian and Wilgram 1967; Hertz et al. 1977; Betterle et al. 1984; Bowcock and Fernandez-Vina 2012), and tyrosinase dysfunction is responsible for these depigmentation diseases (Spritz et al. 1990; Song et al. 1994; Robert et al. 2003).
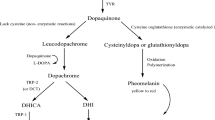
Studies on the mechanism of pigmentation and melanogenesis have been previously reported (Diffey et al. 1995; Bagnara et al. 1979). Melanocytes produce melanin, which determines differences in skin or hair color (Schallreuter et al. 1994). Melanogenesis, which is the synthesis and distribution of melanin in the epidermis, begins with the transcription of proteins required for melanin synthesis. Then, melanosomes are produced and transported to the melanocyte dendrites, and then to adjacent keratinocytes (McGuire and Moellmann 1972; Lin and Fisher 2007). Tyrosinase is a copper-binding enzyme that is produced only by melanocytic cells (Setty et al. 2008). The first biochemical survey of pigmentation was carried out on the mushroom because of its color and since then, the enzyme has been found widely distributed from bacteria to mammals (Fitzpatrick et al. 1950; Kukita and Fitzpatrick 1955; Wood and Ingraham 1965). Tyrosinase catalyzes the first two important reactions of melanin synthesis: L-tyrosinase to L-DOPA through hydroxylation and L-DOPA to dopaquinone through oxidation (Wykes et al. 1971; Pomerantz 1969; Korner and Pawelek 1982; Mirica et al. 2005). Tyrosine hydroxylase, the other DOPA-related enzyme in the nervous system, is not expressed in usual melanocytes. Tyrosinase is the central enzyme involved in eumelanin and pheomelanin synthesis via activation of melanocortin 1 receptor (MC1R), then expression of microphthalmia-associated transcription factor (MITF) (Sealy et al. 1982; Yaar 2013). Other enzymes involved in melanin synthesis include tyrosinase-related protein 1 (Trp 1) and Trp 2 (Sendoel et al. 2010). Inhibition of tyrosinase is the most effective method to decrease melanin synthesis (Bulengo-Ransby et al. 1993; Stern 2004).
By definition, true tyrosinase inhibitors are different from melanin inhibitors, which interfere with melanin formation by blocking its upstream signal transduction or downstream transportation, regardless of direct enzyme interaction. Blocking upstream signal transduction of tyrosinase includes down-regulation of MC1R activity and MITF expression. Blocking downstream transportation includes involvement in melanosomal transfer or epidermal abrasion leading to melanin loss. Tyrosinase inhibitors can chelate copper to prevent substrate binding (Bae-Harboe and Park 2012). Well-known whitening agents, kojic acid and hydroquinone, may induce adverse reactions such as skin irritation, dermatitis, depigmentation, and even cancer. Kojic acid may cause liver toxicity by increased glutathione S-transferase levels, and promote hepatocarcinogenesis (Chusiri et al. 2011; Ota et al. 2009). Hydroquinone may disturb immune response by affecting the function of endotoxin-activated neutrophils or microvascular endothelial cells (Hebeda et al. 2012; Hebeda et al. 2011). Herbs used for cosmetic whitening are widely used among Asians who practice traditional Chinese medicines (TCM). These agents are often composed of hundreds or thousands of compounds. It is difficult to distinguish which types of compounds that whiten skin effectively are safe for routine use (Ernst 2002; Chan 2011).
Searching for pure, safe, and effective ingredients that can achieve skin lightening would be beneficial. Fortunately, computational techniques have rapidly emerged in small molecular drug design (Tang and Chen 2015). TCMs used to lighten skin are often gentle, and have therapeutic advantage in some diseases. The use of a TCM database makes it possible to find new molecules that could be used as future drugs. We aimed to find potent compounds that can inhibit the activity of tyrosinase using computational simulation and the TCM Database@Taiwan (http://tcm.cmu.edu.tw/).
Methods
Compound database
We used the TCM Database@Taiwan (http://tcm.cmu.edu.tw/) to perform potential tyrosinase inhibitor screening. The TCM Database@Taiwan is a large database of TCM compounds and includes 61,000 small molecules. All the small molecules in the database were passed through Lipinski’s rule of five, absorption, distribution, metabolism, excretion, and toxicity (ADMET) to rule out potential toxic compounds in Discovery Studio (DS) (Chen 2011).
Homology modeling
We acquired the human tyrosinase sequence from the Uniprot Knowledgebase (P14679). The 3D structure of tyrosinase from Bacillus megaterium was acquired from the Protein Data Bank (PDB ID: 3NM8). We aligned the sequence of human tyrosinase (P14679) and homologous Bacillus megaterium protein (3NM8) by using the “Modeler protocol" in Accelrys Discovery Studio (DS, San Diego, CA, USA). Based on the results of the sequence alignment, the percentage of identity and similarity was estimated. We used the Build Homology Models module in DS to perform homology modeling of tyrosinase. We confirmed the tyrosinase-modeled structure by Ramachandran plot with Rampage mode in DS.
Disorder prediction
We used the PONDR-FIT protocol in the DisProt website to exclude the disordered residues of the tyrosinase 3D structure.
Structure-based virtual screening
A docking protocol was performed with tyrosinase for all small compounds from the TCM Database@Taiwan and the control (arbutin) by LigandFit mode in DS. The protocol included hydrogen bonds (H-bond), pi bonds, and charge interactions. All docking poses between the ligand and tyrosinase were restricted by the force field of Chemistry at HARvard Molecular Mechanics (CHARMm). We also used the LIGPLOT protocol to display H-bonds and hydrophobic contact between the ligand and tyrosinase.
Quantitative structure-activity relationship (QSAR) models
We used the support vector machine (SVM) and multiple linear regression (MLR) models and Bayesian network to predict the activities of selected TCM compounds. We obtained 24 compounds and pIC50 data of tyrosinase inhibitors from two previous studies: Lee et al. (2009) and Bandgar et al. (2012) (Lee et al. 2009; Bandgar et al. 2012). We transformed these compounds to 2D and 3D structures with ChemBioDraw software. Then, we used the Calculate Molecular Property module and Genetic Function Approximation module in DS to find and estimate the appropriate molecular descriptor for every ligand. We selected ten optimum descriptors for predicting activity. These descriptors, which constructed the SVM and MLR models, were verified by libSVM and Matlab Statistics Toolbox, respectively. We normalized the description between [−1,+1] with the SVM training model. The value of the square correlation coefficient (R2) was used to validate the model. We used the data from these compounds to predict the selected candidates and the control. The Bayes Net Toolbox (BNT), which is a Matlab package for Bayesian network modeling, predicted the pIC50 values. The predicted models used five-fold cross validation. We chose the highest R2of the SVM, MLR, and Bayesian network to be the predicted activity models.
Molecular dynamics (MD) simulation
The trajectories of MD simulations were illustrated by the GROningen MAchine for Chemical Simulations (GROMACS) program (Stockholm, Sweden). Every ligand-tyrosinase complex passed through minimization, heating, equilibration, and production phases. We demonstrated the trajectories of root mean square deviation (RMSD), gyrate, mean square deviation (MSD), total energy, root mean square fluctuation (RMSF), and the central distance between ligand and protein. Cluster analysis, database of secondary structure assignment (DSSP), matrices of smallest distance of residues, and principal component analysis were also calculated.
Ligand pathway
We used the CAVER software (Brno, Czech Republic) to find all possible ligand pathways while the ligand is bound with tyrosinase. The ligand pathway was also found to compute the possible tunnels inside tyrosinase to which the ligand bound. The most important parameters were set as the following description. Shell_radius, which defined the shell probe, was set at a radius of 4. Shell_depth, which specified the maximal depth of the surface region, was set at 5. Probe_radius, which identified the minimum tunnel radius, was set at 0.9 (Chovancova et al. 2012).
Results
Homology modeling
The sequence alignment between P14679_Human and the template (3NM8) had an overall identity of 31.8% and similarity was 50.7% (Figure 1). The Ramachandran plot of the tyrosinase-modeled structure demonstrates that 91.3% of residues were in the favored region, 4.7% were in the allowed region, and 4% were in the outlier region (Figure 2).
Disorder prediction
The residues of binding sites for the tyrosinase-modeled structure did not fall in the disordered area, so there was not any influence on the shape of the binding domains (Figure 3).
Structure-based virtual screening
Tyrosinase catalyzes two important reactions of hydroxylation and oxidation in the presence of copper atoms. His180, His202, His211, His363, His367, and His390 were the key residues that cooperated with the copper atoms to achieve enzyme activity. The binding sites were set around the six key residues. Only 46,583 compounds could dock with the tyrosinase protein. There were 8581 compounds better than the control (arbutin) based on the Dock score. Table 1 lists the Dock score, LigScore, binding energy, H-bond quantity, pi bond quantity, and predicted activity of the top ten TCM compounds ranked by Dock score. We selected 5-hydroxy-L-tryptophan, merresectine C, and bufotenine as the candidates for further survey. Arbutin, the most commonly used tyrosinase inhibitor, was chosen as the control (Figure 4).
The binding amino acids between the ligand and tyrosinase protein were investigated. 5-hydroxy-L-tryptophan formed an H-bond with Glu203, Lys334, and Asp356. Merresectine C formed an H-bond with Glu203 and Lys334. Bufotenine formed an H-bond with Glu203, Lys334, Ala355, and Asp356. The control formed an H-bond with Glu203, Arg308, Lys334, and Asn364. The H-bond, pi bond, and charge interaction are also important binding forces between the ligand and tyrosinase. 5-hydroxy-L-tryptophan formed a pi bond with His202 and Lys334. It also had a charge interaction with Glu203 and Lys334. Merresectine C formed a pi bond with His202 and Lys334. It also had a charge interaction with Asp197 and Glu203. Bufotenine formed a pi bond with His202. It also had a charge interaction with Asp199 and Glu203 (Figure 5). Hydrophobic contact is another essential force between the ligand and tyrosinase. 5-hydroxy-L-tryptophan had hydrophobic contact with Ile253. Merresectine C had hydrophobic contact with Asp82, Asp84, Lys191, His248, Asn249, and Ile253. Bufotenine had hydrophobic contact with Asp84, Phe232, Asn249, and Val262. The control formed hydrophobic contact with Phe232 and Ile239 (Figure 6).
Quantitative structure-activity relationship (QSAR) models
We chose the following ten optimum descriptors for constructing the ligand-based drug design models: Molecular_Solubility, Num_AromaticBonds, Num_AromaticRings, Num_AtomClasses, Dipole_X, Dipole_Y, Jurs_RASA, Strain_Energy, Shadow_XZfrac, and Shadow_YZfrac. We constructed SVM and MLR models with these descriptors. The predictive models were generated by using these descriptors:
p(IC50) = 7.636 + 0.444 × Molecular_Solubility + 0.689 × Num_AromaticBonds − 2.922 × Num_AromaticRings − 0.004 × Num_AtomClasses + 0.050 × Dipole_X − 0.107 × Dipole_Y + 0.405 × Jurs_RASA − 0.00008 × Strain_Energy − 3.092 × Shadow_XZfrac − 1.310 × Shadow_YZfrac (1)
The 24 identified compounds were randomly divided into 18 training sets and six test sets for validation. The R2 values of the predicted activity for SVM, MLR, and Bayesian network were 0.8419, 0.934, and 0.6538, respectively (Figure 7).
Molecular dynamics (MD) simulation
The trajectories of protein and ligand RMSD were drawn to compare the degree of deviation of the top three compounds and the control. The merresectine C protein-ligand complex had the lowest average protein RMSD value. The control protein-ligand complex had the largest average protein RMSD value. Conversely, merresectine C had the largest average ligand RMSD value (Figure 8). We demonstrated the trajectory of protein gyrate to investigate the average atoms’ distance to the center of every corresponding protein, which demonstrates the compact degree of every corresponding protein. The merresectine C protein-ligand complex had the lowest average protein gyrate value. The bufotenine protein-ligand complex had the largest average protein gyrate value. To calculate the deviation of each ligand-protein complex, the trajectory of protein MSD was found. The MSD trajectory of 5-hydroxy-L-tryptophan was exceeded by that of the control at the end of MD (Figure 9).
The trajectory of total energy was found to assess the stability of the ligand-protein complex. To compare the binding stability of the ligand-protein complex, we calculated the average potential and kinetic energies needed for the ligand to bind to tyrosinase (Table 2). The average total energy for 5-hydroxy-L-tryptophan, merresectine C, bufotenine and the control were −640977, −640214, −640627, and −640355 KJ/mol, respectively (Figure 10). RMSF was calculated to survey the fluctuation of every amino acid of the ligand-protein complex. The largest fluctuations of 5-hydroxy-L-tryptophan, merresectine C, bufotenine, and the control were near residues 240, 150, 175, and 80, respectively. There were no prominent influences from the key binding residues, Glu203 and Lys334 (Figure 11).
We used cluster analysis to investigate the representative structure of the ligand-protein complex. The representative structure of the 5-hydroxy-L-tryptophan ligand-protein complex was cluster 10 from 12.5 to 20 ns. The representative structure of the merresectine C ligand-protein complex was cluster 5 from 1.5 to 19.5 ns. The representative structure of the bufotenine ligand-protein complex was cluster 3 from 0.5 to 10.5 ns. The representative structure of the control ligand-protein complex was cluster 2 from 0.5 to 20 ns (Figure 12). The distance of the gravity center between the ligand and tyrosinase was found to compare the top three candidates and the control. The control and 5-hydroxy-L-tryptophan had increased distance between the ligand and protein after 12 and 14 ns (Figure 13).
Secondary structure changes were investigated to survey the structural component changes of the ligand-protein complex. There were large changes from residue 50 to 150 for the top three candidates and the control (Figure 14). Matrices of the smallest distance of residues were created to find the variation of smallest distance for any given residue. There were not any apparent differences between 5-hydroxy-L-tryptophan, merresectine C, bufotenine, or the control (Figure 15).
We performed principle component analysis to find the two eigenvectors (PC1 and PC2) based on the backbone of 5-hydroxy-L-tryptophan, merresectine C, bufotenine, and the control ligand-protein complex. There were similar eigenvectors among 5-hydroxy-L-tryptophan, bufotenine, and the control (Figure 16). The eigenvalues (PC1 and PC2) were comparable with those of principle component analysis. There were similar eigenvalues of PC1 and PC2 among 5-hydroxy-L-tryptophan, bufotenine, and the control (Figure 17).
Ligand pathway
A 3D simulation of the ligand pathway was created to estimate all possible pathways for the ligand to bind with tyrosinase. All candidates and the control had different estimated binding pathways. There were 9, 3, 7, and 4 possible pathways for 5-hydroxy-L-tryptophan, merresectine C, bufotenine, and the control, respectively (Figure 18). Aside from the binding forces of the three candidates and the control, the number and pathway of tunnels were also different.
Discussion
Homology modeling
We chose the human tyrosinase sequence (P14679)and the Bacillus megaterium (3NM8) template for homology modeling to simulate the human tyrosinase structure. 3NM8 was the most approximate crystal structure to human tyrosinase. The high percentage of identity (31.8%) and similarity (50.7%) of sequence alignment, and high percentage of residues in the favored (91.3%) and allowed (4.7%) region implied that the tyrosinase-modeled structure was reliable.
Structure-based virtual screening
Based on the docking score, binding energy, and the quantity of important binding forces, we concluded that 5-Hydroxy-L-tryptophan, merresectine C, and bufotenine had better binding capacity than that of the control. All of the top three candidates and the control formed H-bonds with Glu203 and Lys334. Aside from the H-bonds, all of the top three candidates formed pi bonds with His202 and charge interactions with Glu203. These inhibitors occupied the original space of the copper atoms. Therefore, Glu203 and Lys334arethe key residues of the ligand-protein complex. 5-hydroxy-L-tryptophan, merresectine C, and bufotenine had more stable binding energy and binding forces than that of the control.
Quantitative structure-activity relationship (QSAR) models
The high R2 values of predicted activity for SVM, MLR, and Bayesian network indicate that the predicted activity of any chosen compound is probably similar to its observed activity. The SVM values for 5-hydroxy-L-tryptophan, merresectine C, bufotenine, and the control were 4.629, 4.694, 4.783 and 4.871, respectively. The MLR values were 5.837, 4.194, 4.812, and 5.710, respectively. The BNT values were 3.990, 5.742, 4.090, and 4.331, respectively. Integrating the results of these predictive models, the MLR value of 5-hydroxy-L-tryptophan was higher than that of the control. The BNT value of merresectine C was higher than that of the control. Therefore, 5-hydroxy-L-tryptophan and merresectine C might have better biological activities than that of the control.
Molecular dynamics (MD) simulation
Merresectine C and bufotenine had lower values for protein RMSD, MSD, and the distance of the gravity center compared with the control. The total energy result was consistent with that of protein RMSD, gyrate, and MSD. Therefore, the binding stability of merresectine C and bufotenine are probably better than that of the control, and they could bind with tyrosinase successfully and stably.
There were no similar RMSF values among 5-hydroxy-L-tryptophan, merresectine C, bufotenine, and the control. This finding was consistent with that of the cluster analysis, which showed different groups of representative structure for the top three candidates and the control. This finding implies that the dynamic condition of tyrosinase bound with each ligand was different. The principle component analysis yielded a similar finding. However, all ligands could induce changes in structure of tyrosinase. Binding of each of the top three candidates and the control resulted in large changes from residue 50 to 150 in DSSP figures. There were no apparent differences between the top three candidates and the control in the matrices of smallest residue distances. To find the individual residue or conformational changes, we conducted RMSF, cluster analysis, database of secondary structure assignment (DSSP), and the matrices of smallest distance of residues. Although the RMSF and cluster analysis patterns were different, the appearance of DSSP and the smallest distance of residues were similar. Therefore, the three candidates could induce similar changes in tyrosinase structure, similar to the control, despite the different changes in individual residues. All the ligands could therefore potentially inhibit tyrosinase activity.
According to the docking results, QSAR models, and MD simulation, merresectine C is the best potential lead compound for future development of a novel tyrosinase inhibitor (Figure 19).
References
Bae-Harboe YS, Park HY (2012) Tyrosinase: a central regulatory protein for cutaneous pigmentation. J Invest Dermatol 132(12):2678–2680, doi:10.1038/jid.2012.324
Bagnara JT, Matsumoto J, Ferris W, Frost SK, Turner WA Jr, Tchen TT, Taylor JD (1979) Common origin of pigment cells. Science 203(4379):410–415
Bandgar BP, Adsul LK, Chavan HV, Shringare SN, Korbad BL, Jalde SS, Lonikar SV, Nile SH, Shirfule AL. (2012) Synthesis, biological evaluation, and molecular docking of N-{3-[3-(9-methyl-9H-carbazol-3-yl)-acryloyl]-phenyl}-benzamide/amide derivatives as xanthine oxidase and tyrosinase inhibitors. Bioorg Med Chem 20(18):5649–5657, doi:10.1016/j.bmc.2012.07.001. S0968-0896(12)00540-8
Betterle C, Mirakian R, Doniach D, Bottazzo GF, Riley W, Maclaren NK (1984) Antibodies to melanocytes in vitiligo. Lancet 1(8369):159
Bowcock AM, Fernandez-Vina M (2012) Targeting skin: vitiligo and autoimmunity. J Invest Dermatol 132(1):13–15, doi:10.1038/jid.2011.353.jid2011353
Bulengo-Ransby SM, Griffiths CE, Kimbrough-Green CK, Finkel LJ, Hamilton TA, Ellis CN, Voorhees JJ (1993) Topical tretinoin (retinoic acid) therapy for hyperpigmented lesions caused by inflammation of the skin in black patients. N Engl J Med 328(20):1438–1443, doi:10.1056/NEJM199305203282002
Chan TY (2011) Inorganic mercury poisoning associated with skin-lightening cosmetic products. Clin Toxicol (Phila) 49(10):886–891, doi:10.3109/15563650.2011.626425
Chen CY (2011) TCM Database@Taiwan: the world's largest traditional Chinese medicine database for drug screening in silico. PLoS One 6(1), e15939, doi:10.1371/journal.pone.0015939
Chian LT, Wilgram GF (1967) Tyrosinase inhibition: its role in suntanning and in albinism. Science 155(3759):198–200
Chovancova E, Pavelka A, Benes P, Strnad O, Brezovsky J, Kozlikova B, Gora A, Sustr V, Klvana M, Medek P, Biedermannova L, Sochor J, Damborsky J. (2012) CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput Biol 8(10), e1002708, doi:10.1371/journal.pcbi.1002708. PCOMPBIOL-D-12-00584
Chusiri Y, Wongpoomchai R, Kakehashi A, Wei M, Wanibuchi H, Vinitketkumnuan U, Fukushima S (2011) Non-genotoxic mode of action and possible threshold for hepatocarcinogenicity of Kojic acid in F344 rats. Food Chem Toxicol 49(2):471–476, doi:10.1016/j.fct.2010.11.027.S0278-6915(10)00685-X
Diffey BL, Healy E, Thody AJ, Rees JL (1995) Melanin, melanocytes, and melanoma. Lancet 346(8991–8992):1713
Ernst E (2002) Toxic heavy metals and undeclared drugs in Asian herbal medicines. Trends Pharmacol Sci 23(3):136–139, doi:S0165-6147(00)01972-6 10.1016/S0165-6147(00)01972-6
Fisher DE, James WD (2010) Indoor tanning–science, behavior, and policy. N Engl J Med 363(10):901–903, doi:10.1056/NEJMp1005999
Fitzpatrick TB, Becker SW Jr, Lerner AB, Montgomery H (1950) Tyrosinase in human skin: demonstration of its presence and of its role in human melanin formation. Science 112(2904):223–225
Hebeda CB, Pinedo FJ, Bolonheis SM, Ferreira ZF, Muscara MN, Teixeira SA, Farsky SH (2012) Intracellular mechanisms of hydroquinone toxicity on endotoxin-activated neutrophils. Arch Toxicol 86(11):1773–1781, doi:10.1007/s00204-012-0886-3
Hebeda CB, Pinedo FJ, Vinolo MA, Curi R, Farsky SH (2011) Hydroquinone stimulates inflammatory functions in microvascular endothelial cells via NF-kappaB nuclear activation. Basic Clin Pharmacol Toxicol 109(5):372–380, doi:10.1111/j.1742-7843.2011.00739.x
Hertz KC, Gazze LA, Kirkpatrick CH, Katz SI (1977) Autoimmune vitiligo: detection of antibodies to melanin-producing cells. N Engl J Med 297(12):634–637, doi:10.1056/NEJM197709222971204
Holm AN, Nelson WK (2006) Images in clinical medicine. Minocycline-induced hyperpigmentation. N Engl J Med 355(20):e23, doi:355/20/e23 10.1056/NEJMicm053666
Ibrahimi OA, Anderson RR (2010) Images in clinical medicine. Bleomycin-induced flagellate hyperpigmentation. N Engl J Med 363(24), e36, doi:10.1056/NEJMicm1002334
Johnson BE, Mandell G, Daniels F Jr (1972) Melanin and cellular reactions to ultraviolet radiation. Nat New Biol 235(57):147–149
Kew MC, Mzamane D, Smith AG, Shuster S (1977) Melanocyte-stimulating-hormone levels in doxorubicin-induced hyperpigmentation. Lancet 1(8015):811
Kim NH, Cheong KA, Lee TR, Lee AY (2012) PDZK1 upregulation in estrogen-related hyperpigmentation in melasma. J Invest Dermatol 132(11):2622–2631, doi:10.1038/jid.2012.175. jid2012175
Korner A, Pawelek J (1982) Mammalian tyrosinase catalyzes three reactions in the biosynthesis of melanin. Science 217(4565):1163–1165
Kukita A, Fitzpatrick TB (1955) Demonstration of tyrosinase in melanocytes of the human hair matrix by autoradiography. Science 121(3156):893–894
Lee KH, Ab Aziz FH, Syahida A, Abas F, Shaari K, Israf DA, Lajis NH. (2009) Synthesis and biological evaluation of curcumin-like diarylpentanoid analogues for anti-inflammatory, antioxidant and anti-tyrosinase activities. Eur J Med Chem 44(8):3195–3200, doi:10.1016/j.ejmech.2009.03.020. S0223-5234(09)00157-3
Lin JY, Fisher DE (2007) Melanocyte biology and skin pigmentation. Nature 445(7130):843–850, doi:nature05660 10.1038/nature05660
McGuire J, Moellmann G (1972) Cytochalasin B: effects on microfilaments and movement of melanin granules within melanocytes. Science 175(4022):642–644
Mirica LM, Vance M, Rudd DJ, Hedman B, Hodgson KO, Solomon EI, Stack TD. (2005) Tyrosinase reactivity in a model complex: an alternative hydroxylation mechanism. Science 308(5730):1890–1892, doi:308/5730/1890 10.1126/science.1112081
Ortonne JP, Bissett DL (2008) Latest insights into skin hyperpigmentation. J Investig Dermatol Symp Proc 13(1):10–14, doi:10.1038/jidsymp.2008.7.jidsymp20087
Ota Y, Imai T, Onose J, Takami S, Cho YM, Hirose M, Nishikawa A. (2009) A 55-week chronic toxicity study of dietary administered kojic acid (KA) in male F344 rats. J Toxicol Sci 34(3):305–313, doi:JST.JSTAGE/jts/34.305
Pomerantz SH (1969) L-tyrosine-3,5-3H assay for tyrosinase development in skin of newborn hamsters. Science 164(3881):838–839
Praetorius C, Grill C, Stacey SN, Metcalf AM, Gorkin DU, Robinson KC, Van Otterloo E, Kim RS, Bergsteinsdottir K, Ogmundsdottir MH, Magnusdottir E, Mishra PJ, Davis SR, Guo T, Zaidi MR, Helgason AS, Sigurdsson MI, Meltzer PS, Merlino G, Petit V, Larue L, Loftus SK, Adams DR, Sobhiafshar U, Emre NC, Pavan WJ, Cornell R, Smith AG, McCallion AS, Fisher DE et al (2013) A Polymorphism in IRF4 Affects Human Pigmentation through a Tyrosinase-Dependent MITF/TFAP2A Pathway. Cell 155(5):1022–1033
Rappersberger K, Honigsmann H, Ortel B, Tanew A, Konrad K, Wolff K (1989) Photosensitivity and hyperpigmentation in amiodarone-treated patients: incidence, time course, and recovery. J Invest Dermatol 93(2):201–209
Robert C, Spatz A, Faivre S, Armand JP, Raymond E (2003) Tyrosine kinase inhibition and grey hair. Lancet 361(9362):1056, doi:S0140-6736(03)12805-X
Schallreuter KU, Wood JM, Pittelkow MR, Gutlich M, Lemke KR, Rodl W, Swanson NN, Hitzemann K, Ziegler I (1994) Regulation of melanin biosynthesis in the human epidermis by tetrahydrobiopterin. Science 263(5152):1444–1446
Sealy RC, Hyde JS, Felix CC, Menon IA, Prota G (1982) Eumelanins and pheomelanins: characterization by electron spin resonance spectroscopy. Science 217(4559):545–547
Sendoel A, Kohler I, Fellmann C, Lowe SW, Hengartner MO (2010) HIF-1 antagonizes p53-mediated apoptosis through a secreted neuronal tyrosinase. Nature 465(7298):577–583, doi:10.1038/nature09141. nature09141
Setty SR, Tenza D, Sviderskaya EV, Bennett DC, Raposo G, Marks MS (2008) Cell-specific ATP7A transport sustains copper-dependent tyrosinase activity in melanosomes. Nature 454(7208):1142–1146, doi:10.1038/nature07163. nature07163
Song YH, Connor E, Li Y, Zorovich B, Balducci P, Maclaren N (1994) The role of tyrosinase in autoimmune vitiligo. Lancet 344(8929):1049–1052, doi:S0140-6736(94)91709-4
Spritz RA, Strunk KM, Giebel LB, King RA (1990) Detection of mutations in the tyrosinase gene in a patient with type IA oculocutaneous albinism. N Engl J Med 322(24):1724–1728, doi:10.1056/NEJM199006143222407
Stern RS (2004) Clinical practice. Treatment of photoaging. N Engl J Med 350(15):1526–1534, doi:10.1056/NEJMcp023168. 350/15/1526
Tang H-C, Chen Y-C (2015) Molecular insight and resolution for tumors harboring the H-ras (G12V) mutation. RSC Adv 5(27):20623–20633
Wood BJ, Ingraham LL (1965) Labelled Tyrosinase from Labelled Substrate. Nature 205:291–292
Wykes JR, Dunnill P, Lilly MD (1971) Conversion of tyrosine to L-dihydroxyphenylalanine using immobilized tyrosinase. Nat New Biol 230(14):187
Yaar M (2013) Cutaneous pigmentation in health and disease: novel and well-established players. J Invest Dermatol 133(1):11–13, doi:10.1038/jid.2012.442. jid2012442
Acknowledgements
This research was supported by grants from China Medical University Hospital and Asia University (DMR-104-118, DMR-104-084, DMR-102-001, Asia102-CMU-1 Asia102-CMU-2, and Asia102-CMU-3) and CMU under the Aim for Top University Plan of the Ministry of Education, Taiwan. This research was supported partly by the Taiwan Ministry of Health and Welfare Clinical Trial and Research Center of Excellence (MOHW104-TDU-B-212-113002).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HCT conceived of and designed the experiments. YCC performed the experiments. HCT and YCC analyzed the data and wrote the paper. YCC contributed reagents, materials, and analysis tools. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Tang, HC., Chen, YC. Identification of tyrosinase inhibitors from traditional Chinese medicines for the management of hyperpigmentation. SpringerPlus 4, 184 (2015). https://doi.org/10.1186/s40064-015-0956-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40064-015-0956-0