Abstract
Background
Inherited primary arrhythmias, such as long QT (LQT) syndromes, are electrical abnormalities of the heart mainly due to variants in 3 genes. We herein describe a novel stop-gain pathogenic variant in the KCNQ1 gene in an Iranian child with LQT syndrome 1.
Methods
The patient and his family underwent clinical evaluation, electrocardiographic Holter monitoring, and whole-exome sequencing. Sanger sequencing and segregation analysis were used to confirm the variant in the patient and his family, respectively. The pathogenicity of the variant was checked via an in silico analysis.
Results
The proband suffered from bradycardia and had experienced syncope without stress. The corrected QT interval was 470 ms (the Schwartz score ≥ 3.5), and the Holter monitoring showed sinus rhythm, infrequent premature atrial contractions, and a prolonged QT interval in some leads. Whole-exome and Sanger sequencing showed c.968G > A in 3 affected family members. According to the American College of Medical Genetics and Genomics criteria, c.968G > A was classified as a pathogenic variant.
Conclusions
The KCNQ1 gene is the main cause of LQT syndromes in our population. The common genes of LQT syndromes should be studied in our country’s different ethnicities to determine the exact role of these genes in these subpopulations.
Similar content being viewed by others
Introduction
Long QT (LQT) syndromes are inherited primary arrhythmias affecting 1 in every 5000 to 20000 neonates worldwide [13]. They are characterized by the prolongation of the corrected QT interval on the electrocardiogram (ECG), and they propagate the risk of ventricular arrhythmias, resulting in torsade de pointes [16]. LQT syndromes occur due to alterations in the expression and/or function of repolarizing ionic channels. The main clinical features of LQT syndromes usually include palpitations, syncope, seizures, and ventricular arrhythmias, increasing the risk of sudden cardiac death [12].
The congenital form of LQT syndromes is mostly caused by autosomal dominant pathogenic variants in K + channel proteins, namely Kv7.1 and Kv11.1, in cardiac cells. Kv7.1 and Kv11.1 channels play a significant role in delayed-rectifier K + currents, required for normal ventricular repolarization. Kv7.1 is encoded by the KCNQ1 gene (LQT syndromes type 1 or LQT1) [1]. Pathogenic variants in KCNQ1 are the most common cause of congenital LQT syndromes. Without medical treatment, the mortality rate is high within 1 year after the first syncope episode, [11] but the rate decreases significantly during a 15-year follow-up [13]. LQT syndromes can be diagnosed by clinical features, QT-interval prolongation on 12‐lead ECGs, provocative tests such as epinephrine infusion and exercise stress, family history, and genetic testing.
We have previously described the genetic spectrum of the common types of LQT syndromes in our population. [5, 8, 17]. Here, we report a novel stop-gain pathogenic variant in KCNQ1 causing LQT1 in an Iranian child.
Methods
Clinical evaluation of family recruitment and ethics approval
An Iranian family recruited in this study, had been referred to Rajaie Cardiovascular Medical and Research Center, Tehran, Iran, for the experience of recurrent syncope of its children. The proband was a 10-year-old boy with a 3-year-old brother (Fig. 1A-, III-1 & III-2, respectively). The parents were unrelated and healthy: the father was 41 years old, and the mother was 34. Routine cardiovascular examinations, including echocardiography, 12-lead ECGs, exercise tests, and 24-h Holter monitoring, were carried out for all the family members, affected or healthy. The patients (Fig. 1A, III-1 & III-2) underwent an electrophysiological examination with a 2-electrode voltage−clamp amplifier (TEC10CD, NPI Electronics, Tamm, Germany) featuring KCl-filled electrodes of about 0.8 MU resistance [18]. The currents were measured at room temperature by applying bath solutions, including 10 mM of HEPES (pH 7.2), 105 mM of NaCl, 1.8 mM of CaCl2, and 10 mM of KCl. The Bazett formula was used for calculating the heart rate QT intervals. The study was performed due to the Declaration of Helsinki and was approved by the Ethics Committee of Rajaie Cardiovascular Medical and Research Center (IR.RHC.REC.1401.054). Informed consent was achieved from all participants and from the parents of participants below 16-year-old.
The image presents the pedigree of the family with long QT (LQT) syndromes, as well as the results of the electrocardiogram (ECG) and sequencing chromatograms of the mutated nucleotide in the KCNQ1 gene. A The pedigree of the family with LQT syndromes is presented herein. The proband is indicated with the arrow. B The Sanger sequencing results of the KCNQ1 gene in the patient and his family members are shown here. The patients carried a heterozygous nonsense variant: c.G968A. C The image demonstrates the poor region of the Kv11.1 schematic structure with the W323X variant. D This region includes amino acids conserved among humans, mice, rats, rabbits, and horses
Whole-Exome sequencing and in Silico analysis
Blood samples of family members were obtained. Genomic DNA extraction was performed using the DNSol Midi Kit (Roche: Product No. 50072012). Whole-exome sequencing was done on the proband (Fig. 1A, III-I) at Macrogen (Amsterdam, the Netherlands). Enrichment and capture of all exones were carried out using SureSelect Exon V7 Library Prep Kit. Exome was sequenced on an Illumina HiSeq 6000 machine based on the manufacturer’s protocol. A read quality value of greater than 20 and a depth of greater than 7 was used for the next steps. The quality of the reads was surveyed with FastQC. The clean reads were aligned to the reference genome (UCSC Genome Browser, hg19) applying the Burrows–Wheeler Aligner (BWA-MEM v.07.17) [3]. Insertion and/or deletion and single-nucleotide polymorphism were called with the aid of the Genome Analysis Toolkit (GATK, v.4.1.4.1) [6]. Annotation of determined variants was done by ANNOVAR [15] and filtered according to the Exome Aggregation Consortium (ExAC), the 1000 Genomes Project, Exome-Sequencing Project ESP6500, and the Genome Aggregation Database (gnomAD) minor allele frequency (MAF) of 0.001. The candidate variants were investigated with bioinformatics tools, consisting of MutationTaster (www.mutationtaster.org), CADD (cadd.gs.washington.edu), PROVEAN (provean.jcvi.org), SIFT (https://sift.bii.a-star.edu.sg), and PolyPhen-2 (genetics.bwh.harvard.edu/pph2) according to the 2015 guidelines of the American College of Medical Genetics and Genomics (ACMG) [10]. Moreover, the conservation of the variants regions were analyzed using the GERP + + score and CLUSTALW Web Server (https://www.genome.jp/tools-bin/clustalw) by comparing the amino acid sequences of different species.
Variant validation and segregation analysis
The identified candidate variant, KCNQ1 c.968G > A: p.Trp323, was confirmed and segregated using polymerase chain reaction (PCR) and Sanger sequencing to assess the family members (healthy/patient).
Primer pair was designed using the Primer3 v.04.0 (http://bioinfo.ut.ee/primer3-0.4.0/) with the sequences: forward: 5ʹ-TGCTCTTTGTTGACGACCA-3′ and reverse: 5′-AGCGTGGAAGTGCCCTCT-3′. PCR was carried out on a SimpliAmp Thermal Cycler (Thermo Fisher Scientific) with 1.5 mmol/L of MgCl2, 10 pmol/L of the primers, 200 mmol/L of dNTP, 100 ng DNA, and 1 U of Taq DNA polymerase (Amplicon, UK). Thereafter, incubation at 95 °C for 5 min and 35 amplification cycles (30 s at 95 °C, 30 s at 62 °C, and 30 s at 72 °C) was applied. The products of the PCR were sequenced on an ABI Sequencer 3500XL PE (Applied Biosystems), and the sequences were surveyed with CodonCode Aligner (v.7.1.2) (Fig. 1B).
Results
Clinical findings
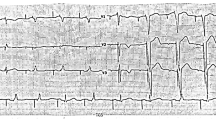
The proband (Fig. 1A, III-1) had suffered 2 syncope episodes without stress in the past year. Sinus rhythm, infrequent premature atrial contractions, and a prolonged QT interval in some leads were detected in his Holter recording (Fig. 2A, C). The minimum Schwartz score [2] was 3.5, indicating the high probability of LQT syndromes.
A The baseline 12-lead standard electrocardiogram indicates a normal sinus rhythm, a normal QRS frontal axis, and a normal corrected QT interval. B Four minutes after exercise cessation, the corrected QT interval is about 480 ms, deemed prolonged for this situation. C The image presents the leads of ambulatory monitoring. The corrected QT interval is normal in most of the leads but is prolonged in the right lower panel
Concerning the family history, the younger brother (Fig. 1A, III-2) also had suffered syncope and exhibited similar symptoms. The mother (Fig. 1A, II-4) had experienced chest pain and dyspnea in the preceding 5 months (no notable ECG manifestations). The father (Fig. 1A, II-3) had no history of arrhythmia or syncope, with no signs of heart disease or aberrant ECGs in his clinical examinations.
Molecular findings
Whole-exome sequencing was performed on the proband (Fig. 1A, III-1) to discover the causative variant. A novel stop-gain pathogenic variant, c.968G > A, was found in the seventh exon of KCNQ1. This variant substituted tryptophan at site 323 for a stop codon, proposed to cause a premature KCNQ1 truncated protein and/or nonsense-mediated KCNQ1 mRNA decay. Notably, the variant has not yet been reported either in the 1000 Genomes Project, ExAc, gnomAD, HGMD, and ClinVar or in publications. According to the ACMG, c.968G > A was determined as a pathogenic variant (criteria: PVS1, PM2, PP1, and PP4). This nonsense variant was considered the cause of the disease by SIFT, PolyPhen-2, PROVEAN, FATHMM, and GERP + + . The CADD Phred score was 41. The variant was confirmed in the proband (Fig. 1A, III-2) by PCR and Sanger sequencing in the heterozygous state. It was also detected in the proband’s affected brother (Fig. 1A, III-2) and the suspected mother (Fig. 1A, II-4) as a heterozygote. The father (Fig. 1A, II-3) had a normal sequence at this position (Fig. 1B). DNA from the other pedigree members was unavailable. A schematic secondary structure of the KCNQ1 protein is presented in Fig. 1C. In addition, based on the CLUSTALW results, tryptophan323 was located in the conserved part of the KCNQ1 protein (Fig. 1D).
Discussion
In many populations, inherited LQT syndromes (~ 75% of LQT syndromes) occur due to variants of 3 genes: KCNQ1 (~ 35%), KCNH2 (~ 30%), and SCN5A (~ 10%), which encode Kv7.1, Kv11.1, and Nav1.5, respectively [14]. However, other major genes might exist in some populations. For instance, in our previous study, we demonstrated that the common genes of LQT syndromes were responsible for only 43% of patients in a sample of the Iranian population [5]. Here, we describe an Iranian child suffering from syncope without stress and with a minimum Schwartz score of 3.5 due to a novel nonsense variant in KCNQ1.
Nonsense variants account for a lower percentage of point mutations in KCNQ1, with approximately 10% of KCNQ1 variants being nonsense [5]. In the present study, we found a novel nonsense variant, c.968G > A, leading to p.Trp323Ter in KCNQ1 in an Iranian proband whose brother and mother were also symptomatic. Notably, p.Trp323 is a hotspot in this gene because p.Trp323Ter due to c.969G > A (not c.968G > A) was reported in a patient by [4]. Our patients had bradycardia, a prolonged corrected QT interval (470 ms), sinus rhythm, infrequent premature atrial contractions, and a prolonged QT interval in some leads.
The pathogenic variant introduced herein may lead to more severe phenotypes, although a genotype–phenotype correlation with a significant number of affected individuals is required to elucidate the effects of this variant on clinical presentations and arrhythmic events. Our in silico analyses showed that this variant is pathogenic, and p.Trp323 is a conserved position among different species. Further, p.Trp323 is located in a region of poor channel condition, so any change in this position could have a potential pathogenic effect on the function of the channel such that the IKs (K slowed delayed rectifier) current is severely affected. As shown, any dysfunction in this current may lead to a prolonged cardiac action potential and U-wave and T-wave changes and trigger torsade de pointes [9].
Chain termination variants, such as nonsense variants, lead to truncated proteins and severe symptoms, whereas missense variants result in amino acid substitutions. Depending on their location and effects on messenger RNAs and proteins, splicing variants may have variable phenotypes in patients with such variants. We recommend further research on the effects of different KCNQ1 variants on symptoms and signs among patients with chain termination, missense, and splicing variants. In this regard, we have previously reported the effects of the splicing variant in the MYO15A gene [7].
Our findings suggest that KCNQ1 might be the principal cause of LQT in our population. There are many ethnicities in our country; the common genes of LQT syndromes should, therefore, be studied in these different ethnicities to determine the exact role of these genes in these subpopulations.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the ClinVar repository [https://www.ncbi.nlm.nih.gov/clinvar/variation/1723453/?new_evidence=false]. The accession number of the variant in ClinVar is as follows: NM_000218.3 (KCNQ1):c.968G > A (p.Trp323Ter): VCV001723453.1.
References
Adler A, Novelli V, Amin AS, Abiusi E, Care M, Nannenberg EA, Feilotter H, Amenta S, Mazza D, Bikker H, Sturm AC, Garcia J, Ackerman MJ, Hershberger RE, Perez MV, Zareba W, Ware JS, Wilde AAM, Gollob MH. An international, multicentered, evidence-based reappraisal of genes reported to cause congenital long QT syndrome. Circulation. 2020;141(6):418–28.
Alders M, Christiaans I. Long QT syndrome 2003 GeneReviews®. Seattle: University of Washington; 2017.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–9.
Lieve KV, Williams L, Daly A, Richard G, Bale S, Macaya D, Chung WK. Results of genetic testing in 855 consecutive unrelated patients referred for long QT syndrome in a clinical laboratory. Genet Test Mol Biomarkers. 2013;17(7):553–61.
Mahdieh N, Khorgami M, Soveizi M, Seyed Aliakbar S, Dalili M, Rabbani B. Genetic homozygosity in a diverse population: an experience of long QT syndrome. Int J Cardiol. 2020;316:117–24.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303.
Motavaf M, Soveizi M, Maleki M, Mahdieh N. MYO15A splicing mutations in hearing loss: a review literature and report of a novel mutation. Int J Pediatr Otorhinolaryngol. 2017;96:35–8.
Rabbani B, Khorgami M, Dalili M, Zamani N, Mahdieh N, Gollob MH. Novel cases of pediatric sudden cardiac death secondary to TRDN mutations presenting as long QT syndrome at rest and catecholaminergic polymorphic ventricular tachycardia during exercise: the TRDN arrhythmia syndrome. Am J Med Genet A. 2021;185(11):3433–45.
Ravens U, Cerbai E. Role of potassium currents in cardiac arrhythmias. Europace. 2008;10(10):1133–7.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17(5):405–23.
Schwartz PJ. Idiopathic long QT syndrome: progress and questions. Am Heart J. 1985;109(2):399–411.
Schwartz PJ, Ackerman MJ, George AL Jr, Wilde AAM. Impact of genetics on the clinical management of channelopathies. J Am Coll Cardiol. 2013;62(3):169–80.
Schwartz PJ, Stramba-Badiale M, Crotti L, Pedrazzini M, Besana A, Bosi G, Gabbarini F, Goulene K, Insolia R, Mannarino S, Mosca F, Nespoli L, Rimini A, Rosati E, Salice P, Spazzolini C. Prevalence of the congenital long-QT syndrome. Circulation. 2009;120(18):1761–7.
Tester DJ, Will ML, Haglund CM, Ackerman MJ. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2005;2(5):507–17.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164–e164.
Weiss JN, Garfinkel A, Karagueuzian HS, Chen PS, Qu Z. Early afterdepolarizations and cardiac arrhythmias. Heart Rhythm. 2010;7(12):1891–9.
Zafari Z, Dalili M, Zeinali S, Saber S, Far AFF, Akbari MT. Identification and characterization of a novel recessive KCNQ1 mutation associated with Romano-Ward Long-QT syndrome in two Iranian families. J Electrocardiol. 2017;50(6):912–8.
Zehelein J, Zhang W, Koenen M, Graf M, Heinemann SH, Katus HA. Molecular cloning and expression of cERG, the ether a go-go-related gene from canine myocardium. Pflugers Arch. 2001;442(2):188–91.
Acknowledgements
The authors wish to acknowledge the kind contribution of the family described herein. This research was funded by the Cardiogenetics Research Center, Rajaie Cardiovascular Medical and Research Center, Tehran, Iran.
Funding
The authors have received no specific funding for this research.
Author information
Authors and Affiliations
Contributions
SK and NM drafted the work. SK performed WES. MP contributed to the wet lab. MD and MM surveyed the patients clinically. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The current study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of Rajaie Cardiovascular Medical and Research Center (IR.RHC.REC.1401.054). Informed consent was obtained from all participants and from the parents of participants below 16yrs.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kalayinia, S., Dalili, M., Pourirahim, M. et al. A novel stop-gain pathogenic variant in the KCNQ1 gene causing long QT syndrome 1. Eur J Med Res 28, 23 (2023). https://doi.org/10.1186/s40001-023-00984-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40001-023-00984-0