Abstract
Background
Bovine Respiratory Disease (BRD) is a multifactorial and economically important illness of cattle. The current study was designed to characterize the major bacterial pathogens associated with BRD and determine the antibiotic susceptibility patterns of isolates. Samples were collected from 400 pneumonic cases of cattle.
Results
Laboratory assay revealed isolation of 376 (94.0%) bacterial pathogens. The most prevalent bacterial pathogens recovered were Mannheimia haemolytica (M. haemolytica) followed by Pasteurella multocida (P. multocida), Histophilus somni (H. somni), and Bibersteinia trehalosi (B. trehalosi) from 191 (50.80%), 81 (21.54%), 56 (14.89%), and 48 (12.77%) samples, respectively. M. haemolytica strains were confirmed using multiplex PCR assay through the amplification of PHSSA (~ 325 bp) and Rpt2 (~ 1022 bp) genes. Capsular typing of P. multocida revealed amplification of serogroup A (hyaD-hyaC) gene (~ 1044 bp) and serogroup D (dcbF) gene (~ 657 bp). B. trehalosi isolates displayed amplification of the sodA gene (~ 144 bp). Besides, serotyping of M. haemolytica showed the distribution of serotype A:1 (82.20%), A:2 (10.47%), and A:6 (7.33%). Whereas, biotyping of P. multocida revealed a higher prevalence of biotype A:3 (83.95%), then A:1 (8.64%), A:2 (4.94%), and A:12 (2.47%). The majority of the retrieved isolates showed remarkable susceptibility to enrofloxacin, ciprofloxacin, sulfamethoxazole-trimethoprim, florfenicol, and ceftiofur (100%). Besides, varying degree of antimicrobial resistance was observed against streptomycin, gentamicin, penicillin-G, and ampicillin.
Conclusions
The current findings confirmed that M. haemolytica (A:1) strain is the most common bacterial pathogen identified from BRD cases in the study areas of Ethiopia. Hence, continuous outbreak monitoring and evaluation of antibiotics susceptibility patterns of bacterial pathogens associated with BRD are indispensable to reduce the impact of BRD in the study areas. Further investigation of bacterial pathogens and genotypic analysis of pathogens from a wider area of the country is essential to design a cost-efficient control strategy.
Similar content being viewed by others
Introduction
Ethiopia is a home for various livestock species and is assumed to be among the leading countries in livestock population in Africa [1]. The total cattle population is estimated to be 65.3 million [2]. Despite the huge cattle population, the current productivity and commercialization of cattle remain very low due to diseases, inadequate feed, genetics of local breed, inefficient production system, and poor infrastructure along the value chain [3, 4]. The persistence of animal diseases such as Bovine respiratory disease (BRD) has continued to be a major constraint to the cattle population. It causes huge economic losses and reduced performance during and after the illness [5, 6]. Stresses, viral infections, nutritional, and environmental conditions are the predisposing factors that enhance the vulnerability of cattle to respiratory illness [7]. The bacterial pathogens that cause BRD include M. haemolytica, P. multocida, H. somni, Mycoplasma species, and Trueperella Pyogens (T. pyogens) [8, 9].
M. haemolytica and B. trehalosi strains were initially classified as Pasteurella haemolytica under the genus Pasteurella. Currently, these two pathogens are classified in two different genera of Bibersteinia and Mannheimia using DNA-DNA hybridization and 16S RNA [10]. The two biotypes are further classified into 17 serotypes. Thirteen serotypes classified to biotype A include (1, 2, 5, 6, 7, 8, 9, 11, 12, 13, 14, 16, and 17) and reclassified as M. haemolytica. On the other hand, four serotypes (3, 4, 10, and 15) are worth mentioned as B. trehalosi that belong to biotype T [11]. Serotype A:11 was later reclassified as M. glucosidal [12]. P. multocida strains are currently classified into five capsular types or serogroups (A, B, D, E, and F) based on capsular polysaccharide and into 16 Heddleston lipopolysaccharides (LPS) serovars using the gel diffusion precipitation assay [13, 14]. Besides, Polymerase chain reaction (PCR) assay [15] and the development of multiplex PCR assay used to determine each capsular serogroup [16]. Moreover, serogroups were classified into eight LPS (L1 – L8) genotypes [17].
Regardless of the considerable studies conducted over the past several years on BRD, it is still a serious concern to exert a huge economic impact on the cattle population [18]. Effective control of BRD likely requires a combination of more definitive diagnosis, efficacious vaccines, therapeutic intervention, and improved management practices [19]. Thus, BRD is one of the diseases that demand efficient control strategies. Previous reports showed the extent of respiratory disease losses, estimated to be higher in Ethiopia [20,21,22]. Furthermore, the emergence of multidrug-resistant bacterial pathogens associated with BRD is considered a potential threat to the cattle population [23]. Hence, these call for continuous outbreak monitoring, identification of bacterial pathogens diversity, and surveillance of antibiotic susceptibility. Therefore, the current study was designed to characterize and determine the antimicrobial susceptibility pattern of the major bacterial pathogens associated with BRD in Ethiopia.
Results
Clinical and pneumonic lung examination
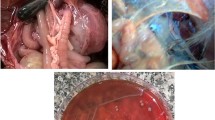
Cattle suspected of respiratory infection were exhibited marked depression, loss of appetite (anorexia), severe respiratory distress, and pyrexia (high fever > 40.0 °C) which is commonly known as DART. Besides, coughing, salivation, lacrimation, and respiratory grunts were observed in advanced cases of the diseases. Cattle slaughtered at the abattoir were inspected for typical gross pathological lesions. Examined lung showed firm, friable, irregularity in shape, consolidation, and dark red color. In advanced cases, pulmonary parenchymal consolidation and interstitial edema were observed (Fig. 1).
Bacterial isolation and distribution
A total of 400 cattle were examined in the present study. Bacteriological and PCR assay revealed the identification of 376 (94.0%) bacterial pathogens. Isolates were identified from 182 (91.0%) nasopharyngeal swab and 194 (97.0%) pneumonic lung tissue samples. The distribution of bacterial pathogens revealed a higher prevalence in adult cattle 220 (95.65%) than calves 156 (91.76%). Prevalence was slightly higher in female cattle 120 (94.49%) compared to male cattle 256 (93.77%). Besides, the prevalence in cross breeds 34 (94.44%) was higher than 342 (93.96%) local breeds. The highest prevalence was observed in poor body condition cattle 83 (97.65%) than moderate 109 (93.16%) and good body conditions 184 (92.93%). Table 1 shows the prevalence of the major bacterial pathogens associated with BRD. The current finding revealed that there was a significant difference (P < 0.05) between the prevalence of bacterial pathogens and age, sex, and body condition. However, there was no significant difference (P > 0.05) between the prevalence of pathogens and breed.
The major bacterial pathogens encountered from suspected cases of BRD in the current study were shown in Table 2. The commonest bacterial pathogens encountered in the current study were M. haemolytica, P. multocida, H. somni, and B. trehalosi with the prevalence of 191 (50.80%), 81 (21.54%), 56 (14.89%), and 48 (12.77%), respectively. Out of these isolates, 111 (88.8%) were isolated from the nasopharyngeal swabs of calves and 71 (94.67%) from adult cattle. Besides, 45 (100%) and 149 (96.13%) were identified from pneumonic lung tissue of calves and adult cattle, respectively. There was no significant difference (P > 0.05) between the prevalence of pathogens and isolates type identified.
PCR assay
Multiplex PCR assay of M. haemolytica targeting PHSSA and Rpt2 gene showed the desired amplification of band size at ~ 325 bp and ~ 1022 bp, respectively. While B. trehalosi isolates were found positive for the presence of the sodA gene by conventional PCR assay (Fig. 2A and B). B. trehalosi also revealed the desired band size of the sodA gene at ~ 144 bp. PCR assay of P. multocida revealed amplification of ~ 460 bp size for species-specific detection. P. multocida capsular typing confirmed by the presence of the hyaD-hyaC gene of serogroup A specific and amplified product showed band size of ~ 1044 bp (Fig. 3A and B).
Serotypes and biovars
M. haemolytica serotyping revealed the identification of 157 (82.20%), 20 (10.47%), and 14 (7.33%) serotype A:1, A:2, and A:6, respectively. Isolates from nasopharyngeal swab samples showed serotype A:1 (73.79%), A:2 (18.48%), and A:6 (8.74%). Whereas, isolates from pneumonic lung tissue samples revealed serotype A:1 (92.05%), A:2 (2.27%), and A:6 (5.68%) as shown in Table 3. Further, characterization of P. multocida revealed the identification of P. multocida subspecies multocida. Thus, Ornithine decarboxylase (ODC) producing isolates belonged to biovar A:3, A:1, A:2, and A:12 from 78.69, 11.47, 6.56, and 3.28%, respectively.
Antimicrobial susceptibility test (AST)
Tables 4 and 5 showed the antimicrobial susceptibility and multidrug-resistance patterns of the major bacterial pathogens against antimicrobial agents. Isolates were 100% susceptible to enrofloxacin, ciprofloxacin, ceftiofur, and florfenicol. Besides, susceptibility was observed in tetracycline (> 75%), and Oxytetracycline (> 80%). M. haemolytica showed varying degrees of multidrug-resistance against streptomycin, gentamicin, penicillin-G, and ampicillin 78.5, 72.8, 43.2, and 38.7%, respectively. P. multocida revealed multidrug resistance against streptomycin (84.0%), gentamicin (80.2%), Penicillin-G (48.1%), and Ampicillin (43.3%). H. somni exhibited multidrug resistance against streptomycin (71.4%), gentamicin (66.1%), Penicillin-G (51.8%), and Ampicillin (42.9%).
Discussion
Differentiation of BRD based on visual and clinical examination is difficult. Hence, diagnosis has to be supported with the identification of the exact pathogens and AST to overcome the growing global concern of antimicrobial resistance. The finding in the present study revealed an overall incidence of 376 (94.0%) bacterial pathogens associated with BRD. Isolates were recovered from 182 (91.0%) nasopharyngeal swabs and 194 (97.0%) pneumonic lung tissue samples. The most prevalent bacterial pathogen recovered in this study was M. haemolytica strain 191 (50.80%) followed by P. multocida 81 (21.54%). Besides, H. somni and B. trehalosi were isolated from 56 (14.89%) and 48 (12.77%) pneumonic samples, respectively.
The current finding showed the potential impact of M. haemolytica and P. multocida in the study areas. The incidence of M. haemolytica was higher than previous reports of 29.2% [21], 10.13% [24], 10.67% [20], and 46.4% [22] which were identified from different parts of the country. The present study proves a higher incidence of M. haemolytica as compared to the other pathogens associated with BRD. Besides, P. multocida recovery from 81 (21.54%) pneumonic cases of cattle was higher than the previous reports of 3.34% [22] and 13.29% [20] but lower than 39.2% [21]. The occurrence of B. trehalosi from 48 (12.77%) cases was in agreement with 14.3% [22] and 12.67% [24]. Moreover, this study proves the presence of H. somni with an incidence rate of 56 (14.89%) in the study areas, which has been described only recently from cases of BRD in Ethiopia.
Isolation and differentiation of M. haemolytica and B. trehalosi isolates is difficult due to their phenotypic relatedness. Thus, the molecular assay was used as a confirmatory method in the current study. M. haemolytica virulence-associated genes are ideal targets for rapid molecular characterization due to M. haemolytica genomic fragments homologous to PHSSA have been identified from many strains of the isolate [25]. PHSSA represents M. haemolytica virulence-associated (species-specific) genes [26] and Rpt2 (species-specific) locus in M. haemolytica has a possible role in modulation of type III restriction-modification system [27]. Multiplex PCR assay of M. haemolytica revealed simultaneous amplification of the two gene fragments (PHSSA and Rpt2). Hence, PCR assay targeting PHSSA and Rpt2 genes become an appropriate molecular diagnostic tool with a high degree of discriminating efficiency. In the present study majority of M. haemolytica strains displayed the desired amplification band size of PHSSA (~ 325 bp) and Rpt2 (~ 1022 bp) genes. Moreover, presumptively identified B. trehalosi PCR products showed amplification of the sodA gene (~ 144 bp) coding manganese-dependent superoxide dismutase.
Capsular typing of P. multocida targeting hyaD-hyaC gene (~ 1044 bp) confirmed the preponderance of serogroup A strains in the present study. Capsular type A was recovered from 76 (93.83%) isolates and capsular type D was identified from 5 (6.17%) isolates. The current finding is in agreement with the report of 93.7% capsular type A and 6.3% capsular type D [28] Moreover, previous study reported that serogroup A is the most prevalent isolate from cattle [29]. Further serotyping analysis of 191 M. haemolytica isolates revealed that 157 (82.20%), 20 (10.47%), and 14 (7.33%) isolates were classified to serotype A:1, A:2, and A:6, respectively. Thus, serotype A:1 was considered as the prevalent pathogen to cause BRD in the study areas. Likewise, Ornithine decarboxylase (ODC) producing P. multocida isolates categorized to biotype A:3 (78.69%), followed by biotype A:1 (11.47%), A:2 (6.56%), and A:12 (3.28%). Thus, P. multocida A:3 strain was considered among the principal respiratory pathogens in cattle.
Antimicrobial resistance is a growing global threat that calls for appropriate use and antimicrobial choice during treatment. In this study, the antibiotics susceptibility pattern of M. haemolytica, P. multocida, and H. somni strains were exhibited remarkable susceptibility to enrofloxacin, ciprofloxacin, ceftiofur, and florfenicol (100% for each), tetracycline (> 75%), and Oxytetracycline (> 80%). However, M. haemolytica showed varying degrees of multidrug resistance against streptomycin (78.5%), gentamicin (72.8%), penicillin-G (43.2%), and ampicillin (38.7%). P. multocida revealed multidrug resistance against streptomycin (84.0%), gentamicin (80.2%), Penicillin-G (48.1%), and Ampicillin (43.3%). In addition, H. somni exhibited multidrug resistance against streptomycin (71.4%), gentamicin (66.1%), Penicillin-G (51.8%), and Ampicillin (42.9%). The current antibiotics susceptibility pattern-finding was supported by a few studies in Ethiopia [22]. However, the finding suggests further studies to investigate the contributing factors associated with multidrug-resistance and measure the association between antimicrobials use and exposure to BRD in the country.
In this study, the findings showed remarkable evidence of the major bacterial pathogens associated with BRD and their antibiotic susceptibility pattern. M. haemolytica (A:1) strain is the most predominant bacterial pathogen followed by P. multocida (A:3) strain to cause BRD. These two pathogens were considered as the principal bacterial pathogens associated with BRD infection in the study areas of Ethiopia. Besides, B. trehalosi and H. somni were associated with few pneumonic cases and perhaps considered as potential pathogens to cause significant impact in the study areas. However, the current finding lacks to show the potential impact of mycoplasma species and other emerging bacterial pathogens associated with BRD. Hence, these bacterial pathogens could be further investigated to know more about the current epidemiological scenario in Ethiopia. Such studies could help in designing efficient prevention and control strategies.
Conclusion
The current finding described the major bacterial pathogens prevalence, serotypes, and antibiotics susceptibility pattern. The phenotypic and molecular assay confirmed that M. haemolytica (A:1) is the most common bacterial pathogen identified from BRD cases in the study areas of Ethiopia. Besides, continuous outbreak monitoring and surveillance of antimicrobial susceptibility is indispensable to decide on the drug of choice attributable to the development of multidrug-resistant strains. Therefore, the current findings suggest further comprehensive studies to investigate strain distribution, the antigenic relationship among strains to understand the molecular epidemiology, and other bacterial pathogens associated with BRD at the national level to design a cost-efficient control strategy.
Methods
Study area and animal
Samples were collected from different agro-ecological zones of Ethiopia (Bale-Robe located at 7°7′N, 40°0′E, Asosa situated at 10°04′N, 34°31′E, Bishoftu found at 8°45′N, 38°59′E), Yabelo located 4°53′N, 38°5′E), and Mekele set at 13°29′N, 39°28′E). These areas were located from 550 to 2492 m above sea level (m.a.s.l). Samples were collected from a total of 400 BRD suspected cases. Laboratory analysis was carried out at the National Veterinary Institute (NVI), Ethiopia.
Samples were collected from study animals based on respiratory clinical signs and postmortem examination associated with age, sex, breed, and body conditions. Clinical signs of bacterial pneumonia in active cases were evaluated for combination of signs including depression and fever (39 °C - 40 °C), serous to mucopurulent nasal discharge, moist cough, and a rapid and shallow respiratory rate are the classic components of a case definition for early BRD cases. Sever cases characterized for pleurisy, irregular breathing pattern, grunting, and unthrift appearance associated with pulmonary abscesses.
Sample collection and sample size
A cross-sectional study with a purposive sampling method was employed to collect samples. Samples were collected from suspected BRD cases brought to veterinary clinics and an abattoir survey was conducted to collect pneumonic lung tissue samples. Since there is no similar research study in the area, the expected prevalence was assumed 50%. The sample size for the study was calculated using a 95% confidence level and required 5% precision [30].
Where N is the required sample size,
Pexp is expected prevalence, and
d is required precision.
The required sample size was calculated to be 384, but 5% samples were considered for the precision of sampling from the study areas and this makes the total samples to be 400.
Nasopharyngeal swab
Respiratory cases of cattle were examined using the Wisconsin clinical respiratory scoring method from 0 (normal) to 3 (severe) cases. The scoring was made by assessing the five clinical signs (fever, lacrimation, nasal discharge, coughing, and ear position). Cattle with higher clinical respiratory scoring (≥ 5) were considered for sampling. Nasopharyngeal swab samples from the nasopharynx were collected from suspected cattle. The sample was collected using a laryngeal swab (MWE dry swab, England). Briefly, the external nares was cleaned with a dry paper towel for any frank discharge or detritus from the nares. A sterile plastic swab was directed via the ventral nasal meatus into the nasopharynx, rotated vigorously against the pharyngeal mucosa for 30 – 45 s at the contra-lateral side. The swab was retracted by taking care not to touch the nares and samples were placed into a sterile screw-capped test tube with a modified Cary-Blair Medium (Park Scientific, UK).
Pneumonic lung
Pneumonic lung tissue samples were inspected and evaluated grossly. A small portion of the lung tissue (~ 3 × 3 mm) sample was aseptically taken from the edge of the lesion. Samples were collected immediately after slaughter and kept in a sterile screw-capped universal bottle. Samples were transported and maintained in a cold chain.
Bacteriological assay
Nasopharyngeal samples were inoculated comparably onto MacConkey and blood agar base (HiMedia, India) supplemented with 5% sheep blood. Lung samples were processed (minced, vortex, centrifuged (3200 x g, 3 min), supernatant discarded, sediment reconstituted) and the suspension was cultured onto MacConkey and blood agar. The remaining lung tissue suspension was stored in 20% glycerol at − 80 °C and later processed to isolate H. somni. Ten microliter of the frozen stock was inoculated onto blood agar (37 °C for 48 h in 5-10% CO2). Presumptive colonies of the major bacterial pathogens were identified based on the standard morphological, cultural, and biochemical assay.
Biochemical assay
Isolates were further analyzed for Grams staining, oxidase, catalase, ornithine decarboxylase (ODC) reaction, indole production, urease, and nitrate reduction. Identification of bacteria pathogens to species level was carried out based on sugar fermentation reaction (glucose, sucrose, lactose, arabinose, trehalose, dulcitol, mannitol, sorbitol, and D-xylose).
Serotyping of M. haemolytica
Rapid plate agglutination assay was employed to characterize the serotypes of M. haemolytica strains as described previously [23]. Rabbit antisera were prepared against reference strains of M. haemolytica which were kindly provided by the NVI, Ethiopia.
Molecular Characterization
DNA extraction
Genomic DNA was extracted using DNeasy® Blood and Tissue kit (QIAGEN GmbH, Germany) following the manufacturer’s instructions.
Multiplex PCR assay of M. haemolytica
M. haemolytica serotype-specific virulence-associated (PHSSA) genes and methyltransferase coding (Rpt2) gene were used in multiplex PCR assay as described in prior studies [25]. The oligonucleotide sequences used in this study was illustrated in Table 6. PCR assay was conducted in a final volume of 25 μl reaction mixture containing IQ supermix (10 μl of Bio-Rad, USA), primer pair (2 μl of 5 pm/μl), RNase free water (3 μl), and template DNA (3 μl). Alpha thermal cycler (PCR max, Ac 296, UK) was used for amplification (Table 7).
PCR assay of P. multocida
Species-specific primers were used in P. multocida PCR assay based on a previous report [26]. A reaction mix of 20 μl containing IQ supermix (10 μl), primer pair (2 μl of 5 pmol), RNase free water (3 μl), and DNA template (3 μl) was used in the PCR assay. Capsular typing was assayed using serogroup-specific (A, B, D, and E) primers. PCR assay was conducted in a final volume of 40 μl reaction mixture containing IQ supermix (20 μl), primer pair (6 μl of 5 pm/μl), RNase free water (2 μl), and template DNA (6 μl).
PCR detection of B. trehalosi
Specific primer targeting sodA gene coding for manganese-dependent superoxide dismutase was used to detect B. trehalosi as described in a previous study [31].
Electrophoresis
Electrophoresis was carried out in agarose gel (2%). PCR product (10 μl) was mixed with a 6x loading buffer. One hundred bp or 1 kb plus DNA molecular marker (10 μl) was added into the first and last lane and run at 120 V for 60 min. The expected band size of PCR products was visualized under a gel documentation system (Uvitec, UK).
AST
AST was conducted using the Kirby-Bauer disk diffusion method to evaluate the sensitivity pattern of the commonly used antimicrobials in the treatment of BRD. Antibiogram of the major bacterial pathogens was carried out against Enrofloxacin (ENR 5 μg), Ciprofloxacin (CIP 5 μg), Penicillin-G (P 10 units), Ampicillin (AMP 10 μg), Streptomycin (S 25 μg), Gentamicin (CN 10 μg), Tetracycline (TE 30 μg), Oxytetracycline (OT 30 μg), Sulfamethoxazole-trimethoprim (SXT 25 μg), Florfenicol (FFC 30 μg), and Ceftiofur (EFT 30 μg). The result was interpreted as described by the Clinical and Laboratory Standards Institute (CLSI) [32] and European committee on antimicrobial susceptibility testing EUvet-CAST [33].
Analysis
Data were coded and stored in an excel spreadsheet. Descriptive statistics and Chi-square test was used for the analysis using STATA (STATA software version 11.0). Statistical significant level was considered at P < .05.
Availability of data and materials
All data supporting the findings of this study can be obtained from the corresponding author upon formal request.
Abbreviations
- AST:
-
Antimicrobial susceptibility test
- BRD:
-
Bovine respiratory disease
- DNA:
-
Deoxyribose nucleic acid
- NVI:
-
National Veterinary Institute
- PCR:
-
Polymerase chain reaction
References
Shapiro BI, Gebru G, Desta S, Negassa A, Nigussie K, Aboset G, et al. Ethiopia livestock sector analysis: A 15 year livestock sector strategy; 2017. https://cgspace.cgiar.org/bitstream/handle/10568/92057/LSA_Ethiopia.pdf
Central Statistical Agency, Agricultural Sample Survey, 2019/20. Volume II: Report on livestock and livestock characteristics (Private peasant holdings). Statistical Bulletin 587. Addis Ababa: Central Statistical Agency (CSA), Federal Democratic Republic of Ethiopia; 2019.
Gizaw S, Desta H, Alemu B, Tegegne A, Wieland B. Importance of livestock diseases identified using participatory epidemiology in the highlands of Ethiopia. Trop Anim Health Prod. 2020;52(4):1745–57. https://doi.org/10.1007/s11250-019-02187-4.
Getabalew M, Alemneh T, Akeberegn D. Dairy Production in Ethiopia-Existing Scenario and Constraints. Biomed J sci Tech. 2019;16(5):12304–9. https://doi.org/10.26717/BJS-TR.2019.16.002903.
Murray GM, More SJ, Clegg TA, Earley B, O’Neill RG, Johnston D, et al. Risk factors associated with exposure to bovine respiratory disease pathogens during the peri-weaning period in dairy bull calves. BMC Vet Res. 2018;14(1):1–11. https://doi.org/10.1186/s12-917018-1372-9.
Delabouglise A, James A, Valarcher JF, Hagglünd S, Raboisson D, Rushton J. Linking disease epidemiology and livestock productivity: The case of bovine respiratory disease in France. PLoS One. 2017;12(12). https://doi.org/10.1371/journal.pone.0189090.
Taylor JD, Fulton RW, Lehenbauer TW, Step DL, Confer AW. The epidemiology of bovine respiratory disease: What is the evidence for predisposing factors? Can Vet J. 2010;51(10):1095–102.
Griffin D, Chengappa MM, Kuszak J, McVey DS. Bacterial pathogens of the bovine respiratory disease complex. Vet Clin Food Anim Pract. 2010;26(2):381–94. https://doi.org/10.1016/j.cvfa.20-10.04.004.
Peek SF, Ollivett TL, Divers TJ. Respiratory diseases. Rebhun's Dis Dairy Cattle. 2018;94. https://doi.org/10.1016/B978-0-323-39055-2.00004-8.
Murugananthan A, Shanthalingam S, Batra SA, Alahan S, Srikumaran S. Leukotoxin of Bibersteinia trehalosi contains a unique neutralizing epitope, and a non-neutralizing epitope shared with Mannheimia haemolytica leukotoxin. Toxins. 2018;10(6):220. https://doi.org/10.3390/toxins10060220.
Haig SJ. Adherence of Mannheimia haemolytica to ovine bronchial epithelial cells. Biosci Horiz. 2011;4(1):50–60. https://doi.org/10.1093/biohorizons/hzr007.
Abed AH, El-Seedy FR, Hassan HM, Nabih AM, Khalifa E, Salem SE, et al. Serotyping, genotyping and virulence genes characterization of Pasteurella multocida and Mannheimia haemolytica Isolates Recovered from Pneumonic Cattle Calves in North Upper Egypt. Vet Sci. 2020;7(4):174. https://doi.org/10.3390/vetsci7040174.
Aski HS, Tabatabaei M. Occurrence of virulence-associated genes in Pasteurella multocida isolates obtained from different hosts. Microb Pathog. 2016;96:52–7. https://doi.org/10.1016/j.micpath.20-16.04.008.
Heddleston KL, Gallagher JE, Rebers PA. Fowl cholera: gel diffusion precipitin test for serotyping Pasteurella multocida from avian species. Avian Dis. 1972;925-936. https://doi.org/10.2307/1588773.
Townsend KM, Frost AJ, Lee CW, Papadimitriou JM, Dawkins HJ. Development of PCR assays for species-and type-specific identification of Pasteurella multocida isolates. J Clin Microbiol. 1998;36(4):1096–100. https://doi.org/10.1128/JCM.36.4.1096-1100.1998.
Townsend KM, Boyce JD, Chung JY, Frost AJ, Adler B. Genetic organization of Pasteurella multocida cap loci and development of a multiplex capsular PCR typing system. J Clin Microbiol. 2001;39(3):924–9. https://doi.org/10.1128/JCM.39.3.924-929.2001.
Harper M, John M, Turni C, Edmunds M, St. Michael F, Adler B, et al. Development of a rapid multiplex PCR assay to genotype Pasteurella multocida strains by use of the lipopolysaccharide outer core biosynthesis locus. J Clin Microbiol. 2015;53(2):477–85. https://doi.org/10.1128/JCM.02824-14.
Fernández M, Ferreras MD, Giráldez FJ, Benavides J, Pérez V. Production significance of bovine respiratory disease lesions in slaughtered beef cattle. Animals. 2020;10(10):1770. https://doi.org/10.3390/ani10101770.
Confer AW, Ayalew S. Mannheimia haemolytica in bovine respiratory disease: immunogens, potential immunogens, and vaccines. Anim Health Res Rev. 2018;19(2):79–99. https://doi.org/10.1017/S14662523-18000142.
Gebremeskel AK, Tesema TS, Yegoraw AA, Birhanu BT, Mekuria SA. Isolation and Characterization of Bacterial Species from Respiratory Tracts of Cattle Slaughtered in Addis Ababa City, Central Ethiopia. World Vet J. 2017;7(1):14–20. https://doi.org/10.5455/wvj.20170289.
Musteria M, Getnet F, Anteneh W, Yehualashet B. Isolation of Pasteurella multocida and Mannheimia hemolytica from Pneumonic Calves and Their Antibiotic Susceptibility in Harar and Haramaya, Eastern Ethiopia. Glob Vet. 2017;19:627–32. https://doi.org/10.5829/idosi.gv.2017.627.632.
Abera D, Sisay T, Birhanu T. Isolation and identification of Mannhemia and Pasturella species from pneumonic and apparently healthy cattle and their antibiogram susceptibility pattern in Bedelle District, Western Ethiopia. Afr J Microbiol Res. 2014;6(5):32–41. https://doi.org/10.5897/JBR2014.0143.
Klima CL, Zaheer R, Briggs RE, McAllister TA. A multiplex PCR assay for molecular capsular serotyping of Mannheimia haemolytica serotypes 1, 2, and 6. J Microbiol Methods. 2017;139:155–60. https://doi.org/10.1016/j.mimet.2017.05.010.
Mebratu A, Habtamu B, Melkamu B. Isolation and characterization of respiratory tract bacterial species from domestic animals with pneumonic lungs from Elphora abattoir, Ethiopia. Int J Microbiol Res. 2015;6(1):13–9. https://doi.org/10.5829/idosi.ijmr.20-15.6.1.91162.
Legesse A, Abayneh T, Mamo G, Gelaye E, Tesfaw L, Yami M, et al. Molecular characterization of Mannheimia haemolytica isolates associated with pneumonic cases of sheep in selected areas of Central Ethiopia. BMC Microbiol. 2018;18(1):1–10. https://doi.org/10.1186/s12866-018-1338-x.
El-Jakee JK, Ali SS, El-Shafii SA, Hessain AM, Al-Arfaj AA, Mohamed MI. Comparative studies for serodiagnosis of haemorrhagic septicaemia in cattle sera. Saudi J Biol Sci. 2016;23(1):48–53. https://doi.org/10.1016/j.sjbs.2015.06.011.
Ryan KA, Lo RY. Characterization of a CACAG pentanucleotide repeat in Pasteurella haemolytica and its possible role in modulation of a novel type III restriction-modification system. Nucleic Acids Res. 1999;27(6):1505–11. https://doi.org/10.1093/nar/27.6.1505.
Katsuda K, Hoshinoo K, Ueno Y, Kohmoto M, Mikami O. Virulence genes and antimicrobial susceptibility in Pasteurella multocida isolates from calves. Vet Microbiol. 2013;167(3-4):737–41. https://doi.org/10.1016/j.vetmic.2013.09.029.
Kong LC, Wang Z, Wang YM, Dong WL, Jia BY, Gao D, et al. Antimicrobial susceptibility and molecular typing of Pasteurella multocida isolated from six provinces in China. Trop Anim Health Prod. 2019;51(4):987–92. https://doi.org/10.1007/s11250-018-1754-9.
Thrufield M. Veterinary Epidemiology. 3rd ed. Berlin: Blackwell Wissenschafts verla; 2005.
Dassanayake RP, Call DR, Sawant AA, Casavant NC, Weiser GC, Knowles DP, et al. Bibersteinia trehalosi inhibits the growth of Mannheimia haemolytica by a proximity-dependent mechanism. Appl Environ Microbiol. 2010;76(4):1008–13. https://doi.org/10.1128/AEM.02086-09.
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. CLSI document VET01S. 5th ed; 2020. https://clsi.org/standards/products/new-products/documents/vet01s/
The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 11.0; 2021. http://www.eucast.org
Acknowledgments
The authors highly acknowledge the Research and Development laboratories of the National Veterinary Institute of Ethiopia for providing the facility to conduct this research. Great appreciation was forwarded to Veterinary clinics located in the study areas, animal owners, and abattoir staff for their cooperation during sample collection. The authors also thank the Department of Biotechnology, Koneru Lakshmaiah Education Foundation (KLEF) for supporting the study.
Funding
This study was supported by the NVI, Ethiopia and KLEF, India for laboratory resources and technical assistance. However, financial aid was not granted for this particular study from any other sources.
Author information
Authors and Affiliations
Contributions
All authors participated in the conception and design of the study; MA conducted all laboratory works and drafted the manuscript. BV, TA, and EG analyzed the data and revised the paper. TD and GD participated in laboratory work. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Samples collection followed scientific procedures and animal handling employed with basic animal welfare protocols. Laboratory assay was performed following the standard bacteriological and molecular methods. Animal owners gave their consent for sample collection and use of data.
Consent for publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Akalu, M., Vemulapati, B., Abayneh, T. et al. Serotyping, antibiogram, and detection of bacterial pathogens associated with bovine respiratory disease in selected areas of Ethiopia. Ir Vet J 75, 3 (2022). https://doi.org/10.1186/s13620-022-00210-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13620-022-00210-z





