Abstract
Background
A notable increase in severe cases of COVID-19, with significant hospitalizations due to the emergence and spread of JN.1 was observed worldwide in late 2023 and early 2024. However, no clinical data are available regarding critically-ill JN.1 COVID-19 infected patients.
Methods
The current study is a substudy of the SEVARVIR prospective multicenter observational cohort study. Patients admitted to any of the 40 participating ICUs between November 17, 2022, and January 22, 2024, were eligible for inclusion in the SEVARVIR cohort study (NCT05162508) if they met the following inclusion criteria: age ≥ 18 years, SARS-CoV-2 infection confirmed by a positive reverse transcriptase-polymerase chain reaction (RT-PCR) in nasopharyngeal swab samples, ICU admission for acute respiratory failure. The primary clinical endpoint of the study was day-28 mortality. Evaluation of the association between day-28 mortality and sublineage group was conducted by performing an exploratory multivariable logistic regression model, after systematically adjusting for predefined prognostic factors previously shown to be important confounders (i.e. obesity, immunosuppression, age and SOFA score) computing odds ratios (OR) along with their corresponding 95% confidence intervals (95% CI).
Results
During the study period (November 2022–January 2024) 56 JN.1- and 126 XBB-infected patients were prospectively enrolled in 40 French intensive care units. JN.1-infected patients were more likely to be obese (35.7% vs 20.8%; p = 0.033) and less frequently immunosuppressed than others (20.4% vs 41.4%; p = 0.010). JN.1-infected patients required invasive mechanical ventilation support in 29.1%, 87.5% of them received dexamethasone, 14.5% tocilizumab and none received monoclonal antibodies. Only one JN-1 infected patient (1.8%) required extracorporeal membrane oxygenation support during ICU stay (vs 0/126 in the XBB group; p = 0.30). Day-28 mortality of JN.1-infected patients was 14.6%, not significantly different from that of XBB-infected patients (22.0%; p = 0.28). In univariable logistic regression analysis and in multivariable analysis adjusting for confounders defined a priori, we found no statistically significant association between JN.1 infection and day-28 mortality (adjusted OR 1.06 95% CI (0.17;1.42); p = 0.19). There was no significant between group difference regarding duration of stay in the ICU (6.0 [3.5;11.0] vs 7.0 [4.0;14.0] days; p = 0.21).
Conclusions
Critically-ill patients with Omicron JN.1 infection showed a different clinical phenotype than patients infected with the earlier XBB sublineage, including more frequent obesity and less immunosuppression. Compared with XBB, JN.1 infection was not associated with higher day-28 mortality.
Similar content being viewed by others
Background
Following the emergence of the Omicron variant of SARS-CoV-2, several sublineages have co-circulated until the dominance of XBB recombinant variants in early 2023, which were subsequently replaced by a distinct branch of BA.2 named BA.2.86. Compared to XBB and the parental BA.2, the spike protein of BA.2.86 has more than 30 mutations [1]. Initially, BA.2.86 did not dominate other coexisting subvariants until it acquired an additional mutation (i.e., L455S), causing its progeny JN.1 to rapidly increase and become the dominant SARS-CoV-2 sublineage in several parts of the world. Subsequently, the WHO designated JN.1 as a variant of interest due to its increased transmissibility.
Several in vitro studies have shown that JN.1 has phenotypic characteristics that confer enhanced in vitro fitness. The L455S substitution in the spike protein enhances the ability of the virus to bind to the angiotensin-converting enzyme 2 receptor. JN.1 also appears to be one of the most immune-evading SARS-CoV-2 variants to date, contributing to its increased transmissibility compared to other Omicron sublineages [2].
Clinical reports from medical institutions indicate that the risk of serious illness due to JN.1 variant infection is low [3]. However, there has been a notable increase in severe cases of COVID-19, with significant hospitalizations due to COVID-19 in late 2023. Importantly, a certain proportion of patients is still admitted to intensive care units (ICUs) for COVID-19-associated acute respiratory failure, but their clinical phenotype and outcomes have changed since the early waves of the pandemic [4, 5], and those of patients admitted with severe COVID-19 due to the JN.1 subvariant are currently unknown. This information is critical as it could improve our ability to target individuals who may benefit from more personalized preventive measures, such as frequent vaccination and/or active immunoprophylaxis, as well as tailored therapeutic interventions, including early administration of antivirals in the event of infection.
As part of the SEVARVIR study, we have established a prospective French national multicenter cohort focused on patients admitted to ICUs with COVID-19-associated acute respiratory failure. In this specific substudy, our aims are (1) to assess day-28 mortality and (2) to comprehensively characterize the clinical presentation of patients infected with the emerging JN.1 variant and compare them with those infected with sublineages derived from XBB.
Methods
Study design and patients
The current study is a substudy of the SEVARVIR prospective multicenter observational cohort study. Patients admitted to any of the 40 participating ICUs between November 17, 2022, and January 22, 2024, were eligible for inclusion in the SEVARVIR cohort study (NCT05162508, see Supplementary Table 1 for the list of participating centers) if they met the following inclusion criteria: age ≥ 18 years, SARS-CoV-2 infection confirmed by a positive reverse transcriptase-polymerase chain reaction (RT-PCR) in nasopharyngeal swab samples, ICU admission for acute respiratory failure (i.e., peripheral oxygen saturation ≤ 90% and need for supplemental oxygen or any type of ventilatory support). Patients with SARS-CoV-2 infection but no acute respiratory failure or with a RT-PCR cycle threshold (Ct) value > 32 in nasopharyngeal swabs were not included. The study was approved by the Comité de Protection des Personnes Sud-Méditerranée I (N° EudraCT/ID-RCB: 2021-A02914-37). Informed consent was obtained from all patients or their relatives.
Demographics, clinical and laboratory variables were recorded upon ICU admission and during ICU stay. Patients’ frailty was assessed using the Clinical Frailty Scale [6]. The severity of the disease upon ICU admission was assessed using the World Health Organization (WHO) 10-point ordinal scale [7], the sequential organ failure assessment (SOFA [8]) score, and the simplified acute physiology score (SAPS [9]) II score. Acute respiratory distress syndrome (ARDS) was defined according to the Berlin definition [10]. Immunosuppression was defined as solid-organ transplant, active onco-hematological malignancy (within the past three years), HIV infection, long-term corticosteroid treatment (i.e., more than three months of > 0.5 mg/kg/day prednisone equivalent), and exposure to any other immunosuppressive treatment. Obesity was defined as a body mass index greater than 30 kg/m2. The primary clinical endpoint of the study was day-28 mortality.
SARS-CoV-2 variant determination
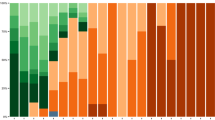
Full-length SARS-CoV-2 genomes from all included patients were sequenced by means of next-generation sequencing. For mutational pattern analysis at the amino acid level, only high-quality sequences, i.e., sequences covering ≥ 90% of the viral genome and 95% of the spike gene, were considered. Full-length viral genome sequence analysis yielding high coverage will be deposited in Genbank.
Statistical analysis
Descriptive results are presented as mean ± standard deviation [SD] or median (1st-3rd quartiles) for continuous variables, and as numbers with percentages for categorical variables. Exploratory unadjusted comparisons between patients infected with two groups of Omicron sublineages (including XBB sublineages, referred to as the “XBB group”, and emerging BA.2.86 sublineages, [parental BA.2.86, JN.1, and JN.3], referred to as the “JN.1 group”) were performed using Chi-squared or Fisher’s exact tests for categorical variables, and ANOVA or Kruskal–Wallis tests for continuous variables, as appropriate. Evaluation of the association between day-28 mortality and sublineage group was conducted by performing an exploratory multivariable logistic regression model, after systematically adjusting for predefined prognostic factors previously shown to be important confounders, i.e. body mass index, immunosuppression, age and SOFA score, computing odds ratios (OR) along with their corresponding 95% confidence intervals (95% CI).
The overall sample size of the SEVARVIR study was a priori defined (n = 2000). The sample size of this substudy was not predefined. Indeed, we had anticipated that data could be sequentially extracted from the prospective database based on epidemiological surges. Results have been reported according to the STROBE guidelines for cohort studies (Supplementary Table 2).
Two-sided p-values < 0.05 were considered statistically significant. No missing data imputation was performed and analyses were performed on complete cases. Analyses were performed with Stata V16.1 statistical software (StataCorp, College Station, TX, USA) and R 4.2.0 (R Foundation for Statistical Computing, Vienna, Austria).
Results
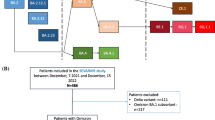
Between November 17, 2022, and January 22, 2024, 233 patients were admitted to one of the 40 participating ICUs and enrolled in the SEVARVIR cohort study. Of these, 126 patients in the “XBB group” and 56 patients in the “JN.1 group” were included in the analysis (Fig. 1).
The clinical phenotype of JN.1-infected patients differed from that of XBB-infected patients
No statistically significant differences were observed between patients infected with JN.1 and XBB sublineages with respect to age, gender and frequency of comorbidities. However, patients in the JN.1group were more likely to be obese (n = 20/56, 35.7% vs n = 26/125, 20.8%; p = 0.033), and had a statistically significant higher median body mass index (26.4 [22.4–33.4] vs 25.0 [21.2–28.7] kg/m2; p = 0.019). There were also significantly fewer immunosuppressed patients in the JN.1 group than in the XBB group (n = 10/49, 20.4% vs n = 48/116, 41.4%; p = 0.010) (Table 1).
The proportion of patients who had received at least one dose of SARS-CoV-2 vaccine did not show significant statistical differences between groups, although the median number of doses received was significantly higher in patients from the XBB group (3 [3–4] vs 3 [3–3]; p = 0.019). The median time from onset of first symptoms to ICU admission was significantly shorter in the JN.1 group than in the XBB group (3 [1–6] vs 5 [3–9] days; p = 0.006). Other variables related to SARS-CoV-2 virological characteristics, including median viral level in the upper respiratory tract measured by cycle threshold in RT-PCR and prevalence of positive SARS-CoV-2 anti-S antibodies at ICU admission, did not differ statistically significantly between groups (Table 1).
There was no statistically significant difference between the two groups in the severity of illness at ICU admission, as reflected by the SOFA and SAPS II scores and the WHO 10-point ordinal scale (Table 1). Invasive mechanical ventilation support was required in 22.0% (n = 40/182) of patients within 24 h of ICU admission, with no statistically significant difference between groups. No patient required extracorporeal membrane oxygenation (ECMO) support on ICU admission. Respiratory failure was eventually attributed to SARS-CoV-2 pneumonia without bacterial co-infection in about half of cases in both JN.1 and XBB groups (Table 2).
Day-28 mortality in JN.1-infected patients did not differ from that of XBB-infected patients
Day-28 mortality was not statistically significantly different between JN.1- and XBB-infected patients (14.6%, n = 7/48 vs 22.0%, n = 27/124; p = 0.28) (Table 3). In univariable logistic regression analysis and in multivariable analysis adjusting for confounders defined a priori, we found no statistically significant association between the infecting sublineage and day-28 mortality (Table 4). Age and a body mass index < 18 kg/cm2 showed a statistically significant association with day-28 mortality.
During the ICU stay, 32.6% (n = 59/181) of patients required invasive mechanical ventilation, with no statistical significant differences between the subgroups. There was also no statistical significant difference between groups regarding the need for other organ support (Table 3). Regarding COVID-19 management, JN.1-infected patients were treated with dexamethasone significantly more often than their XBB counterparts (n = 42/48, 87.5% vs n = 66/111, 59.5%; p < 0.001). No statistically significant differences between groups were observed in the use of other treatments, including anti-IL-6 antagonists, convalescent plasma and antivirals (Table 3).
Discussion
The current study is the first to describe the in-hospital mortality and the clinical phenotype associated with the newly emerging Omicron sublineage JN.1 in patients with severe COVID-19 requiring ICU admission. Our data provide reassuring evidence that this emerging sublineage does not cause more severe outcomes than XBB variants that emerged and spread earlier in the population. We observed unexpected phenotypic differences, with more frequent obesity and less frequent immunosuppression in patients infected with JN.1, as compared to those infected with XBB sublineages.
The main result of our study is that day-28 mortality of JN.1-infected patients did not significantly differ from that of XBB-infected patients. Recent epidemiologic data have confirmed the increased transmissibility of JN.1. Its proportion of the circulating variants in the US had increased to more than 90% according to nowcast estimates from the US Centers for Disease Control and Prevention (CDC) [11]. In France, JN.1 represented more than 90% of circulating variants, according to the Santé Publique France report of January 31st, 2024 [12]. In this context, and given the surge in COVID-19 cases during the winter of 2024 [3], obtaining clinical data reporting the clinical phenotype and lethality of patients infected with this subvariant as compared with the previous ones is crucial to inform public health authorities and clinicians managing these patients. Our data provide reassuring evidence regarding the severity of disease associated with JN.1 infection, showing not only a non-significant difference in day 28 mortality compared to patients infected with XBB, but also no significant differences in other outcomes, including the need for invasive mechanical ventilation and length of stay in the ICU. Consistently, there was also no significant association between sublineage and day-28 mortality in uni- and multivariable logistic regression analysis.
Patients infected with sublineage JN.1 were more likely to be obese and less likely to be immunosuppressed than those infected with XBB in our study. Such a finding was unexpected because immunosuppression has been reported to be the most common comorbidity in COVID-19 patients infected with the Omicron variant since the “ancestral” BA.1 Omicron sublineage [4, 13], occurring in almost 50% of cases, and may reflect an inherently less pathogenic variant as reported in a hamster model [14]. On the other hand, the higher prevalence of obesity, a previously reported risk factor for severity with previous SARS-CoV-2 variants, including the ancestral variant, is consistent with previous data reporting obesity as a risk factor for severity [15]. These findings may have important implications for the updated use of pre-exposure monoclonal antibodies use [16] as well as COVID-19 vaccination recommendations. Initial estimates of the updated XBB.1.5 COVID-19 vaccine showed sustained vaccine efficacy against symptomatic JN.1 lineage infection [17].
The day-28 mortality rate measured in the current cohort (JN.1 group: 14.6%; XBB group: 22.0%) of critically ill COVID-19 patients was numerically lower than that of previously published cohorts involving other variants (i.e., Wuhan: 31% [18]; Alpha: 26% [19]; Delta: 29% [4]) or Omicron sublineages (i.e., BA.1: 35%; BQ.1.1: 22% [4, 5]), suggesting a milder severity of JN.1 infection in the ICU. Such finding is corroborated by the fact that only one third of JN.1-infected patients required invasive mechanical ventilation and extra-corporeal membrane oxygenation support was almost never required, despite the fact that the majority of patients were categorized as having SARS-CoV-2 pneumonia at cause for acute respiratory failure (as opposed to SARS-CoV-2-associated cardiogenic pulmonary edema or decompensated chronic respiratory failure). The higher prevalence of obesity in JN.1- than in XBB-infected patients might in part account for the numerically better outcome in the former patients, as previously demonstrated [20]. In contrast, patients with a lower body mass index had a higher risk of day-28 mortality. In terms of ICU management, patients in the JN.1 group received dexamethasone more frequently than their counterparts in the XBB group, possibly because they were less likely to be immunosuppressed. Other aspects of treatment did not differ.
Our study certainly has limitations, including a limited sample size in the JN.1 group, which limits our statistical power to perform subgroup analyses and adjust for confounding variables. The generalizability of our findings is also restricted to the population studied. Indeed, we included two groups of critically ill patients who showed no statistically significant mortality difference. We thus cannot exclude any populational difference in disease severity, with associated risks of hospitalization, ICU admission and death, related to the infecting Omicron sublineage. Combined with the observational nature of our study design, we also did not define a priori the sample size of the study, making our findings exploratory. This is because SEVARVIR aims at capturing the dynamics of emerging SARS-CoV-2 sublineages and analyzing their phenotype and relationship with mortality in real time. However, our study also has major strengths, in particular the constitution of a unique national prospective multicenter cohort of well-phenotyped critically ill patients and the availability of full-length SARS-CoV-2 genome sequences, allowing for prospective exploration of the clinical consequences of emerging and spreading SARS-CoV-2 sublineages.
In conclusion, our exploratory analysis of critically-ill patients with Omicron JN.1 infection suggested a different clinical phenotype than patients infected with the earlier XBB sublineage, including more frequent obesity and less immunosuppression. Compared with XBB, JN.1 infection was not associated with higher day-28 mortality.
Availability of data and materials
Raw data are available on reasonable request to the corresponding author.
Abbreviations
- ARDS:
-
Acute respiratory distress syndrome
- COVID-19:
-
Coronavirus disease 2019
- ICU:
-
Intensive care unit
- RT-PCR:
-
Reverse transcriptase-polymerase chain reaction
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronavirus 2
- SAPS II:
-
Simplified acute physiology score II score
- SOFA:
-
Sequential organ failure assessment
- WHO:
-
World Health Organization
References
Wang Q, Guo Y, Liu L, Schwanz LT, Li Z, Nair MS, et al. Antigenicity and receptor affinity of SARS-CoV-2 BA.2.86 spike. Nature. 2023;624:639–44.
Kaku Y, Okumura K, Padilla-Blanco M, Kosugi Y, Uriu K, Hinay AA, et al. Virological characteristics of the SARS-CoV-2 JN1 variant. Lancet Infect Dis. 2024;24:e82.
Rubin R. As COVID-19 cases surge, here’s what to know about JN1, the latest SARS-CoV-2 “variant of interest.” JAMA 2024;331:382–83.
de Prost N, Audureau E, Heming N, Gault E, Pham T, Chaghouri A, et al. Clinical phenotypes and outcomes associated with SARS-CoV-2 variant omicron in critically ill French patients with COVID-19. Nat Commun. 2022;13:6025.
de Prost N, Audureau E, Préau S, Favory R, Guigon A, Bay P, et al. Clinical phenotypes and outcomes associated with SARS-CoV-2 Omicron variants BA.2, BA5 and BQ.1.1 in critically ill patients with COVID-19: a prospective, multicenter cohort study. Intensive Care Med Exp. 2023;11:48.
Rockwood K, Song X, MacKnight C, Bergman H, Hogan DB, McDowell I, et al. A global clinical measure of fitness and frailty in elderly people. CMAJ. 2005;173:489–95.
WHO Working Group on the Clinical Characterisation and Management of COVID-19 infection. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect Dis. 2020;20:e192–7.
Vincent JL, Moreno R, Takala J, Willatts S, De Mendonça A, Bruining H, et al. The SOFA (Sepsis-related organ failure assessment) score to describe organ dysfunction/failure. On behalf of the working group on sepsis-related problems of the European society of intensive care medicine. Intensive Care Med. 1996;22:707–10.
Le Gall JR, Lemeshow S, Saulnier F. A new simplified acute physiology score (SAPS II) based on a European/North American multicenter study. JAMA. 1993;270:2957–63.
ARDS Definition Task Force, Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, et al. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;307:2526–33.
CDC. COVID Data Tracker. Centers for disease control and prevention. 2020. https://covid.cdc.gov/covid-data-tracker. Accessed 7 Feb 2024.
SPF. Infections respiratoires aiguës (grippe, bronchiolite, COVID-19). Bulletin du 31 janvier 2024. https://www.santepubliquefrance.fr/maladies-et-traumatismes/maladies-et-infections-respiratoires/grippe/documents/bulletin-national/infections-respiratoires-aigues-grippe-bronchiolite-covid-19-.-bulletin-du-31-janvier-2024. Accessed 7 Feb 2024.
Vieillard-Baron A, Flicoteaux R, Salmona M, Chariot A, De Maupeou DB, Darmon M, et al. Omicron variant in the critical care units of paris metropolitan area the reality research group. Am J Respir Crit Care Med. 2022;206(3):349–63.
Tamura T, Mizuma K, Nasser H, Deguchi S, Padilla-Blanco M, Oda Y, et al. Virological characteristics of the SARS-CoV-2 BA.2.86 variant. Cell Host & Microbe. 2024. https://www.sciencedirect.com/science/article/pii/S1931312824000052. Accessed 6 Feb 2024.
Williamson EJ, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–6.
Yang S, Yu Y, Xu Y, Jian F, Song W, Yisimayi A, et al. Fast evolution of SARS-CoV-2 BA.2.86 to JN.1 under heavy immune pressure. Lancet Infect Dis. 2024;24:e70–2.
Link-Gelles R. Early Estimates of Updated 2023–2024 (Monovalent XBB.1.5) COVID-19 Vaccine Effectiveness Against Symptomatic SARS-CoV-2 Infection Attributable to Co-Circulating Omicron Variants Among Immunocompetent Adults —Increasing Community Access to Testing Program, United States, September 2023–January 2024. MMWR Morb Mortal Wkly Rep. 2024. https://www.cdc.gov/mmwr/volumes/73/wr/mm7304a2.htm. Accessed 7 Feb 2024.
COVID-ICU Group on behalf of the REVA Network and the COVID-ICU Investigators. Clinical characteristics and day-90 outcomes of 4244 critically ill adults with COVID-19: a prospective cohort study. Intensive Care Med. 2021;47:60–73.
Fourati S, Audureau E, Arrestier R, Marot S, Dubois C, Voiriot G, et al. SARS-CoV-2 genomic characteristics and clinical impact of SARS-CoV-2 viral diversity in critically Ill COVID-19 patients: a prospective multicenter cohort study. Viruses. 2022;14:1529.
den Uil CA, Termorshuizen F, Rietdijk WJR, Sablerolles RSG, van der Kuy HPM, Haas LEM, et al. Age moderates the effect of obesity on mortality risk in critically Ill patients With COVID-19: a nationwide observational cohort study. Crit Care Med. 2023;51:484–91.
Acknowledgements
The authors would like to thank all staff involved in the study, Dr Pierre-André Natella, Ms. Nolwenn Bombenger for taking care of regulatory aspects, Mr. Léo Graca for taking care of data management, Mr. Mohamed Ader for clinical data abstraction, the nurses and physicians who took care of the patients, the laboratory staff who took care of virological samples and the patients and their family for agreeing to participate in the study. Assistance Publique—Hôpitaux de Paris is the sponsor of the study.
Keyvan Razazi, Armand Mekontso Dessap, Raphaël Bellaïche, Lucile Picard, Alexandre Soulier, Mélissa N’Debi, Sarah Seng, Christophe Rodriguez, Frédéric Pene, Anne-Sophie L’Honneur, Adrien Joseph, Elie Azoulay, Maud Salmona, Marie-Laure Chaix, Charles-Edouard Luyt, David Levy, Julien Mayaux, Stéphane Marot, Juliette Bernier; Maxime Gasperment, Tomas Urbina, Hafid Ait-Oufella, Eric Maury, Laurence Morand-Joubert, Djeneba Bocar Fofana, Jean-François Timsit, Diane Descamps, Guillaume Voiriot, Nina de Montmollin, Mathieu Turpin, Stéphane Gaudry, Ségolène Brichler, Tài Olivier Pham, Elyanne Gault, Sébastien Jochmans, Aurélia Pitsch, Guillaume Chevrel, Céline Clergue, Kubab Sabah, Laurence Courdavault Vagh Weinmann, Claudio Garcia-Sanchez, Ferhat Meziani, Louis-Marie Jandeaux, Samira Fafi-Kremer, Elodie Laugel, Sébastien Preau, Aurélie Guignon, Antoine Kimmoun, Evelyne Schvoerer, Cédric Hartard, Charles Damoisel, Nicolas Brechot, Helene Péré, François Beloncle, Francoise Lunel Fabiani, Rémi Coudroy, Arnaud W. Thille, François Arrive, Sylvain le Pape, Laura Marchasson, Luc Deroche, Nicolas Leveque, Vincent Thibaut, Béatrice la Combe, Séverine Haouisee, Alexandre Boyer, Sonia Burrel, Gaetan Beduneau, Christophe Girault, Maximillien Grall, Dorothée Carpentier, Jean-Christophe Plantier, Emmanuel Canet, Audrey Rodallec, Berthe Marie Imbert, Sami Hraeich, Pierre-Edouard Fournier, Philippe Colson, Anaïs Dartevel, Sylvie Larrat, Guillaume Thiery, Sylvie Pillet, Kada Klouche, Edouard Tuaillon, Cécile Aubron, Adissa Tran, Sophie Vallet, Pierre-Emmanuel Charles, Alexis le Rougemont, Bertrand Souweine, Cecile Henquell, Audrey Mirand, Bruno Mourvillier, Laurent Andreoletti, Clément Lier, Damien du Cheyron, Nefert Candace Dossou, Astrid Vabret, Gaël Piton, Quentin Lepiller, Sylvie Roger
Funding
This work was supported by the EMERGEN consortium—ANRS Maladies Infectieuses Emergentes (ANRS0153). This study has been labelled as a National Research Priority by the National Orientation Committee for Therapeutic Trials and other researches on Covid-19 (CAPNET). The investigators would like to acknowledge ANRS | Emerging infectious diseases for their scientific support, the French Ministry of Health and Prevention and the French Ministry of Higher Education, Research and Innovation for their funding and support.
Author information
Authors and Affiliations
Consortia
Contributions
N.D.P., E.A., J.M.P., and S.F., designed the study and obtained funding; E.A. performed statistical analyses; N.D.P., A.G., S.P., F.U., F.D., F.T., C.D., D.C., T.D., C.S., T.P., P.B., included the patient and were responsible for clinical data collection; L.H., A.G., Q.L.H., V.T., A.M., J.T., A.H., S.H., V.G., A.C., and S.F. were responsible of the management of virological samples; J.-M.P., and S.F. were responsible of virological analyses; N.D.P., E.A., and S.F. wrote the first draft of the article; All authors revised and approved the article. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted. N.D.P. and S.F. are the guarantors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Comité de Protection des Personnes Sud-Méditerranée I (N° EudraCT/ID-RCB: 2021-A02914-37). Informed consent was obtained from all patients or their relatives.
Consent for publication
Consent to publish has been obtained from patients or their relatives.
Competing interests
S.F. has served as a speaker for GlaxoSmithKline, AstraZeneca, MSD, Pfeizer, Cepheid and Moderna; J.-M.P. has served as an advisor or speaker for Abbvie, Arbutus, Assembly Biosciences, Gilead and Merck; E.A. has received fees for lectures from Alexion, Sanofi, Gilead and Pfizer. His hospital has received research grant from Pfizer, MSD and Alexion. D.D. served as an advisor for Gilead-Sciences, ViiV Health care, and Merck. N.D.P has served as an advisor or speaker for Moderna and AstraZeneca. Other authors and investigators have no competing interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
de Prost, N., Audureau, E., Guillon, A. et al. Clinical phenotypes and outcomes associated with SARS-CoV-2 Omicron sublineage JN.1 in critically ill COVID-19 patients: a prospective, multicenter cohort study in France, November 2022 to January 2024. Ann. Intensive Care 14, 101 (2024). https://doi.org/10.1186/s13613-024-01319-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13613-024-01319-w