Abstract
JSRV (Jaagsiekte Sheep Retrovirus) is a retrovirus inducing a transmissible lung adenocarcinoma in sheep and goats with predominantly lepidic and papillary lesions. This naturally occurring lung cancer in large animals shares many features with human pneumonic-type lung adenocarcinomas with predominant lepidic growth. The metastatic spread is rare in both human and animal cancers. This unique feature prompted us to decipher the angiogenesis pathway in these cancers. We focused on the levels of mRNA and proteins of genes implicated in the extension of JSRV-induced lung adenocarcinomas by studying their expression in lung cancers (n = 10) and normal lungs (n = 10) and in primary epithelial alveolar type II cells derived from cancers (n = 10) or normal lungs (n = 6). In parallel, we evaluated the levels of expression of key genes in lung tissues collected from lepidic (n = 13) or papillary (n = 5) human adenocarcinomas and, when available, adjacent normal lungs (n = 11). We measured the expression of the same key genes implicated in angiogenesis, lymphangiogenesis and degradation of the extracellular matrix. In ovine adenocarcinomas, VEGFR2 and VEGFD mRNA were downregulated in cancers; MMP9, TIMP1 and FGFR2 mRNA were overexpressed as compared to normal lungs. Importantly, VEGFA and VEGFR2 proteins were not expressed in JSRV-induced cancers. In human lepidic adenocarcinomas, VEGFA and VEGFR2 mRNA were weakly expressed and no VEGFR2 protein was detectable. Downregulation of key angiogenic players may contribute to the control of extra thoracic invasion of cancer cells in human and ovine pneumonic-type adenocarcinoma with predominant lepidic growth.
Similar content being viewed by others
Introduction
Cancers develop as the result of deregulation of multiple cell processes such as resistance to cell death, independence from growth suppressors and inflammation [1]. Angiogenesis and lymphangiogenesis mechanisms are involved in the spreading of cancer cells. Angiogenesis is the neo-formation of blood vessels from pre-existing ones and is a highly-regulated mechanism [2]. This cell mechanism is crucial for the extension of cancers and is a major target for drug development [3]. When angiogenic pathways are activated, cancer cells communicate with endothelial cells through pro-angiogenic factors that activate the proliferation of endothelial cells. Activation of angiogenesis is controlled by the release of a wide array of intercellular mediators by cancer cells. The main angiogenesis signaling pathway is triggered by VEGF (Vascular Endothelial Growth Factor) and ligation to the VEGF receptors. While VEGFA, B, C and D are secreted by cancer cells, their receptors VEGFR1, R2 and R3 are expressed at the surface of endothelial cells [4]. Ligands bind to their receptors inducing their dimerization and auto-phosphorylation; the activation of signal transduction results in endothelial cell survival and expression of genes involved in cell proliferation [4]. VEGFA was the first-identified member of the VEGF family and is expressed by various cell types including cancer cells [5]. VEGFA is the main inducer of angiogenesis and proliferation and migration of cancer cells; thereby the VEGFA/VEGFR2 complex is a key player for the activation of angiogenesis [6]. VEGFA stimulates the growth of the vascular network that supports cancer growth and metastasis and is often up-regulated in cancers [5]. On the contrary, lymphangiogenesis participates in dissemination of cancer cells by its activation of the growth of lymphatic vessels through the interaction of VEGFC or VEGFD ligands with VEGFR3 receptors expressed at the surface of lymphatic endothelial cells. VEGFC and VEGFD can also activate angiogenesis through their interaction with VEGFR2 [7]. NRP1 and NRP2 (Neuropilin 1 and 2) are VEGFR co-receptors that potentiate VEGF fixation on VEGFRs [8]. Finally, the degradation of the extracellular matrix is essential for cancer growth and progression through proteolytic activities that facilitate the growth or spread of new blood and lymphatic vessels into the primary cancer [2]. The activity of extracellular matrix proteases is under tight control in normal conditions, but cancer cells are prone to activate the proteolytic activities of PLAU (Plasminogen Activator Urokinase), MMP (Matrix Metallopeptidase) 2 and 9, involved in extracellular matrix degradation during cancer invasion process and angiogenesis [9, 10]. SERPINE1 (Serpin family E member 1) and TIMP1 (Tissue Inhibitor of MetalloProteinase 1) are inhibitors of PLAU and MMP9 respectively. Other pathways are implicated in the regulation of angiogenesis such as the FGF pathway controlled by FGF (Fibroblast Growth Factor) ligands and their receptors [11].
Ovine pulmonary adenocarcinoma is a virally induced lung cancer affecting sheep and goats. The JSRV (Jaagsiekte Sheep RetroVirus) β-retrovirus responsible for the disease is transmitted by aerosols, milk, colostrum and in utero [12,13,14]. JSRV infects and transforms cells through its envelope, carrying the oncogenic properties. The retrovirus solely transforms epithelial cells from the distal lung namely AECIIs (Alveolar Epithelial type II Cells) in the alveoli and Club cells in the bronchiole. Ovine pulmonary adenocarcinoma shares striking similarities with human pneumonic-type lung adenocarcinoma (PTLA) with predominant lepidic growth [15,16,17]. This growth pattern refers to the lining of cancer cells along the alveolar septa without disorganization of the alveolar architecture and with no evidence of stromal, vascular or pleural invasion. Lepidic adenocarcinoma affects more non-smokers, women and young subjects than other lung adenocarcinomas [18]. These cancers are slow-growing and as with ovine pulmonary adenocarcinoma, metastatic spread is rare. This unique feature prompted us to decipher the angiogenesis pathway in JSRV-induced pulmonary adenocarcinomas and human lepidic adenocarcinomas especially when considering that most lung cancers in humans are highly invasive [19] and that the activation of angiogenesis is associated with a poor prognosis [20].
Our data showed that the expression of key genes implicated in angiogenesis were dramatically downregulated in JSRV-induced lung cancers. Protein expression of VEGFA and of its receptor VEGFR2 was almost abolished in cancers while VEGFB and VEGFD mRNA expression were significantly reduced. At the same time, expression of TIMP1, inhibiting the metalloproteinases, was increased. Similarly, VEGFA was weakly expressed and VEGFR2 was not expressed in human lepidic adenocarcinomas. Moreover, mRNA expression VEGFD, FGF2 MMP2, NRP1 and NRP2 was decreased in lepidic adenocarcinomas when compared to the normal lungs. Downregulation of key angiogenic players may contribute to the control of extra thoracic invasion of cancer cells in lepidic-predominant adenocarcinomas both in sheep and humans.
Materials and methods
Biological materials
Ovine pulmonary adenocarcinoma specimens were collected post mortem from twenty-five spontaneously infected sheep with clinical evidence of lung cancer obtained from flocks in southern France and from twenty-one sheep without clinical signs of cancer obtained from the Corbas slaughterhouse, nearby Lyon (France) (Table 1). Formal authorization for accessing the facilities was obtained and access was granted under the supervision of a veterinarian. None of the animals used in this study were engaged in an experimental protocol. Clinical status was confirmed by the presence of gross lesions and by the detection of JSRV genome using semi-nested PCR with primers located in env and LTR regions, generated from sequences of field isolates as previously described [21]. All cancer samples were positive for JSRV proviral DNA and were pathologically described as lepidic predominant with papillary subtypes. When possible, primary alveolar epithelial type II cells (AECII) were derived from the tissues. Seventeen primary AECII cultures were obtained from lung cancers and sixteen from normal lungs (Table 1). The lung-derived ovine AECII have been phenotypically characterized for the expression of pro-SP-C and cytokeratin and cultured as previously described [22]. We studied the mRNA and protein expression in lungs and lung-derived AECII from the same animals when possible; unfortunately, due to the limited number of primary cells directly derived from cancer or normal lungs, this was not possible in all cases. When necessary, we used AECII cultures from other animals from our biobank.
Human lung specimens from 18 patients (13 lepidic and five papillary adenocarcinomas) and their associated data were obtained from Cardiobiotec Biobank (CRB-HCL Hospices Civils de Lyon BB-0033-00046), a center for biological resources authorized by the French Ministry of Social Affairs and Health. All samples were collected and used in accordance with the ethical rules of the Biobank and in agreement with French legislation. All patients signed a written informed consent. When available in the Biobank, adjacent normal lung was collected and analyzed (Table 2).
Expression of genes involved in angiogenesis, lymphangiogenesis and extracellular-matrix degradation
Total RNA from 20 ovine and 29 human lung tissues were extracted using the “RNeasy Plus Universal Mini” kit (Qiagen) and total RNA from ovine primary AECII cultures (n = 16) were extracted using the “PureLink RNA mini” kit and treated with the “Turbo DNA free” kit (Ambion) as recommended. One microgram of total RNA was reverse transcribed using the “iScript cDNA synthesis” kit (Biorad). The cDNAs were amplified using the “KAPA SYBR® FAST Universal qPCR kit” (Clinisciences) in triplicate using a MiniOpticon thermocycler (Biorad) with an initial denaturation step at 95 °C for 3 min, followed by 40 cycles of 5 s at 95 °C and 20 s at 60 °C. The melting curves were obtained by a temperature increase from 60 to 95 °C with fluorescence reading every 0.5 °C.
The primers for genes coding for ligands (VEGFA, VEGFB, VEGFC, VEGFD, FGF1, FGF2), receptors (VEGFR1, VEGFR2, VEGFR3, FGFR1, FGFR2, NRP1, NRP2), peptidases and peptidase inhibitors (MMP2, MMP9, PLAU, TIMP1, SERPINE1) and reference genes (PPIA, PSMB2, RPS11, YWHAZ) (Additional file 1) were designed with Primer-BLAST from the available GenBank sequences with a primer spanning an exon–exon junction or with forward and reverse primers into two different exons to avoid interfering amplifications from residual DNA. The efficiency of the primers was evaluated by amplification of 10-fold dilutions of cDNA. All primer sets used in our study have a calculated efficiency of 95 to 110%. Three reference genes were selected from our set of 10 genes (not shown), described as “stably expressed” in humans or animals. Their stability in ovine and human lungs has been evaluated with the geNorm software [23]. This preliminary study (data not shown) allowed us to select PPIA (peptidyl isomerase A), PSMB2 (proteasome subunit beta 2) and YWHAZ (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta) as the most stable reference genes for mRNA quantification in sheep, and PPIA, PSMB2 and RPS11 (ribosomal protein S11) as the most stable ones for humans (Additional file 1). The normalization was performed against the three reference genes selected for their stability in sheep or humans. Normalized relative expression levels were calculated with the 2−ΔΔCq method [24].
Determination of the virus load by quantitative RT-PCR
Virus load was quantified by RT-qPCR from total RNA extracted from lungs or culture AECIIs using primers specific for exogenous JSRV, located at the end of the env gene for the forward primer (5′-GAGTTGAAATGCTGCATATG-3′) and in the LTR for the reverse primer (5′-GGATTCTTACACAATCACC-3′) as we previously described [21]. The virus load was measured in reference to a standard curve generated with a known number of DNA copies of the JSRV target, i.e. the cloned env-LTR amplicon carrying a 50 bp deletion. The standard curve and the total mRNA were amplified using the “KAPA SYBR® FAST Universal qPCR kit” (Clinisciences) in triplicate using a MiniOpticon thermocycler (Biorad) with an initial denaturation step at 95 °C for 3 min, followed by 40 cycles of 5 s at 95 °C and 20 s at 60 °C. The melting curves were obtained by a temperature increase from 60 °C to 95 °C with fluorescence reading every 0.5 °C. Virus load was expressed as the number of viral RNA per ng of total RNA.
Analysis of VEGFA and VEGFR2 protein expression
Total proteins were extracted from ovine and human lung tissues and ovine primary cells. Prior to extraction, lung tissues were dissociated in T-PER lysis buffer (ThermoScientific) using the “FastPrep” device (MP Biomedicals) as recommended. After an incubation of 30 min on ice, lysates were disrupted by four sonication cycles for 20 s at 300 W (Bioblock, Fisher Scientific). Supernatants were collected by centrifugation, and their protein contents quantified using the “Quick Start TM Bradford 1X Dye Reagent” kit (Biorad) as recommended. After heat denaturation, 15–20 µg of total proteins were separated by electrophoretic migration on SDS-PAGE and transferred onto a nitrocellulose membrane. The membranes were pre-incubated with TBST-milk (25 mM Tris pH 7.6; 0.15 M NaCl; 0.05% Tween 20, 5% non-fat dry milk) for 1 h at room temperature. After three washes in TBST, membranes were incubated overnight at 4 °C with specific antibodies. The VEGFA and VEGFR2 proteins were detected with a 1/500 dilution in TBST-milk of rabbit anti-VEGFA antibodies (ab46154, Abcam) and of rabbit anti-VEGFR2 antibodies (ab39256, Abcam). After three washes with TBST, membranes were incubated for 1 h at room temperature with a 1/10 000 dilution of anti-rabbit IgG (whole molecule)-peroxidase antibodies (A0545, Sigma). Detection of β-actin (A3854, Sigma) was performed as a loading control. Immunoreactive bands were revealed with the “SuperSignal West Pico reagent” (Thermofisher). Detection of VEGFA and VEGFR2 was repeated three times and the protein expression was semi-quantified by densitometry using the image processing software UN-SCAN-IT™ (Silk Scientific Corporation).
Statistical analysis
Statistical analyses were performed using Wilcoxon, Student or Welsh tests using R software. All tests were done with a significance threshold of 0.05.
Results
The VEGF pathway is downregulated in JSRV-induced adenocarcinomas
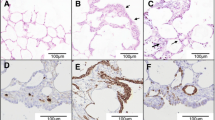
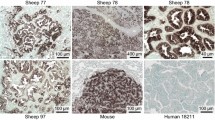
We analyzed the levels of mRNA expression of VEGFA using RT-qPCR and its protein expression by Western blot in normal lungs, cancers, and lung-derived AECII cultures. The level of VEGFA mRNAs was not significantly altered in JSRV-induced cancers or the derived-AECII cultures when compared to normal samples (Figure 1). Interestingly, the expression of VEGFA protein was abolished in most cancers (90%), while the protein was expressed in 80% normal lungs (Figures 2A and C). AECII cultures derived from normal lungs or cancers both expressed VEGFA at a similar level (Figures 2B and C), suggesting that the in vitro conditions might have modified the expression of VEGFA proteins.
VEGFA and VEGFR2 mRNA expression in JSRV-induced adenocarcinoma. The mRNA expression of ovine VEGFA and VEGFR2 was analyzed by RT-qPCR in lungs (10 normal lungs and 10 cancers) and lung-derived AECII cultures (6 from normal lungs and 10 from cancers). Statistical analysis was performed using the Wilcoxon test with **p < 0.005. The mRNA expression of the cellular genes was normalized with expression of the PPIA, PSMB2 and YWHAZ reference genes.
Downregulation of VEGFA and VEGFR2 protein expression in JSRV-induced adenocarcinoma. Levels of protein expression of VEGFA and VEGFR2 were analyzed by Western blot in lungs (A) and cultured AECIIs (B). C Protein expression of each sample was semi-quantified by densitometry and a ratio [protein of interest/β-actin] was calculated. Statistical analysis was performed using Student t-test or the Wilcoxon test with ***p < 0.0005.
The mRNA expression of VEGFR2, the main receptor of VEGFA, was significantly reduced in cancers as compared to normal lungs and undetectable in AECII cultures derived from cancers or normal lungs (Figure 1). Correlating with the decrease of mRNA expression, VEGFR2 proteins were undetectable in cancers but present in 80% of normal lungs (Figures 2A and C). VEGFR2 protein expression was null in primary AECIIs derived from normal lungs or cancers (Figure 2B). These results highlight the loss of expression of VEGFA and of its main receptor VEGFR2 in JSRV-induced pulmonary adenocarcinomas.
We measured mRNA expression of VEGFB, VEGFC, VEGFD ligands, of their receptors VEGFR1, VEGFR3 and of their coreceptors NRP1 and NRP2. Except for the expression of VEGFD, which was significantly reduced in JSRV-induced adenocarcinomas, none of the other genes were differentially expressed in cancers (Figure 3). Expression of VEGFB and VEGFC was low to undetectable in most tested tissues (Figure 3A). Expression of the VEGFR1 and VEGFR3 receptors (Figure 3B) and NRP1 and NRP2 (data not shown) was not modified in JSRV-induced cancers when compared to normal lungs. Expression of VEGFB mRNA was significantly reduced by 2.5-fold in primary AECII derived from cancers as compared to cells derived from normal lungs and VEGFC expression was downregulated in cultured AECII (p value at 0.056). These results indicate that expression of genes involved in lymphangiogenesis was altered in JSRV-induced lung cancers with VEGFC and VEGFD under-expressed in cancers as compared to normal lungs.
Alteration of VEGF/VEGFR mRNA expression in JSRV- induced adenocarcinomas. The mRNA expression of ligands VEGFB, VEGFC and VEGFD (A), and their receptors VEGFR1 and VEGFR3 (B) were analyzed by RT-qPCR in lungs and lung-derived AECII cultures. Statistical analysis was performed using Student t-test or the Wilcoxon test with *p < 0.05.
Expression of genes from the FGF pathway is unchanged in ovine pulmonary adenocarcinoma except for the overexpression of FGFR2 mRNA
We analyzed the expression of genes involved in the FGF pathway by quantification of mRNA expression of the FGF1 and FGF2 ligands and of their receptors (FGFR1 and FGFR2) in lungs and primary AECII cultures. No difference of mRNA expression was measured for FGF1, FGF2 and FGFR1 between normal lungs and cancers (Figure 4). FGFR2 mRNA were over-expressed in cancers when compared to normal lungs (Figure 4B). These results highlight that the FGF pathway was unchanged in ovine pulmonary adenocarcinoma except for the over-expression of FGFR2.
The metalloprotease/antiprotease pathway is impaired in JSRV-induced cancers
Degradation of the extracellular matrix is a major step for cancer extension. Among key genes, mRNA expression of MMP2, MMP9 and PLAU peptidases and TIMP1 and SERPINE1 peptidase inhibitors has been measured in lungs and cultured AECIIs (Figure 5). No difference in mRNA levels was evidenced in cancers when compared to normal lungs for MMP2 metalloproteinase, PLAU or its inhibitor SERPINE1 (Figures 5A and B). No expression of MMP9 was observed in normal samples and only four cancers expressed this gene (Figure 5A). Interestingly, TIMP1, one of the metallopeptidase inhibitors, was significantly over-expressed in cancers as compared to normal lungs (Figure 5B). Expression of MMP2 and MMP9 metalloproteinases was retained in primary normal AECII (Figure 5A). These results suggest that in JSRV-induced lung adenocarcinomas, mRNA expression of MMP2 and MMP9 was reduced in cancer-derived AECII, while not statistically significant, compared to AECII derived from normal lungs. We show that metalloproteinases, that can enhance cell migration by degrading the extracellular matrix, were not overexpressed in cancer except for MMP9 and that the expression of TIMP1, one of their inhibitors, was increased suggesting a blockade of this pathway leading to the degradation of the extracellular matrix.
Expression of genes involved in extracellular matrix degradation in JSRV-induced adenocarcinomas. Expression of PLAU, MMP2 and MMP9 (A) and SERPINE1 and TIMP1 (B) mRNA was quantified by RT-qPCR in lungs and lung-derived AECII. Statistical analysis was performed using the Wilcoxon test with *p < 0.05.
Virus load was quantified by quantitative RT-PCR and expressed as the number of copies per ng of total RNA (Figure 6). There was no significant correlation between the expression level of genes implicated in the angiogenic pathway and the number of JSRV RNA copies in lungs (data not shown). This suggests that the observed deregulation of the cellular genes was part of the tumoral process and not directly linked to the level of JSRV replication.
Quantification of JSRV viral genome in cancers and cancer-derived AECIIs. JSRV viral RNA have been quantified by RT-qPCR in cancers (n = 10) (filled diamond) and cancer-derived AECII cultures (n = 10) (filled square). The level of JSRV RNA was expressed as the number of copies per ng of total cell RNA.
Dysregulation of the expression of genes implicated in angiogenesis in human lepidic adenocarcinomas
We evaluated the levels of expression of key genes in lung tissues collected from lepidic or papillary adenocarcinomas and when available adjacent, non-tumoral lungs from the same patients. Unlike JSRV-induced adenocarcinomas, mRNA (Figure 7A) and proteins (Figure 7B) of VEGFA were expressed in human lepidic adenocarcinomas but at very low levels in cancers and in their normal counterparts. VEGFA protein expression was detected in 6/13 (~46%) lepidic adenocarcinomas, 3/5 (60%) papillary adenocarcinoma and 9/11 (~81%) normal lungs. We did not detect differences in the levels of VEGFA mRNA or proteins between lepidic and papillary adenocarcinomas (Figure 7).
Lack of VEGFA and VEGFR2 mRNA expression in human lepidic and papillary adenocarcinomas. A mRNA expression of VEGFA and VEGFR2 analyzed by RT-qPCR in normal lungs and lepidic or papillary adenocarcinomas. The mRNA expression of the cellular genes has been normalized with expression of the PPIA, PSMB2 and RPS11 reference genes. B VEGFA and VEGFR2 protein expression by Western blot.
Interestingly, we detected a weak (not significant) expression of VEGFR2, the main VEGFA receptor, in lepidic and papillary tumors (Figure 7). No VEGFR2 protein was detected in lepidic, papillary and normal lung samples (Figure 7). The low mRNA expression of VEGFA and the absence of expression of VEGFR2 in lepidic adenocarcinomas suggest the lack of activation of the angiogenic pathway.
The expression of VEGFD and FGF2 mRNA was significantly downregulated in lepidic and papillary adenocarcinomas when compared to adjacent normal lung tissues (Figure 8). Interestingly, mRNA expression of the MMP2, NRP1 and NRP2 genes was lower (but not significant) in lepidic-type adenocarcinomas as compared to normal tissues from the same patients (Figure 8). Similarly, MMP2 mRNA expression was significantly lower in papillary-type cancers as compared to normal lungs (Figure 8). Every other gene analyzed in JSRV induced adenocarcinoma was analyzed in human adenocarcinomas; their expression was not different in cancers and normal lungs (data not shown).
Expression of other genes involved in angiogenesis in human lepidic and papillary adenocarcinomas. Expression of human VEGFD, FGF2, MMP2, NRP1 and NRP2 mRNA was analyzed by RT-qPCR in normal lungs and lepidic or papillary lung adenocarcinomas. Statistical analysis was performed using Wilcoxon test with *p < 0.05 and **p < 0.005.
Taking all together, this study shows the absence of significant over-expression of genes implicated in metastasis and/or degradation of the extracellular matrix. On the contrary, we observed decreased mRNA expression of VEGFD, FGF2 and MMP2 and the absence of VEGFR2 proteins in human lepidic adenocarcinomas.
Discussion
Lung cancer is not a unique disease and can be divided according to different pathological types and subtypes [25]; they may evolve differently in patients and this does influence prognosis and treatment. Most animal models of lung cancers are developed in small rodents. Our work is focused on ovine pulmonary adenocarcinoma, a virally-induced lung cancer in large animals, as a model of human lepidic adenocarcinomas with pneumonic presentation. Both cancers share clinical, radiological and pathological similarities [15]. Predominantly lepidic pulmonary adenocarcinomas are characterized by their development into the lung parenchyma without evidence of stromal, vascular or pleural invasion [15]. Recently, a transgenic mouse model expressing JSRV-Env was developed and exhibited spontaneous lung adenocarcinomas with a metastatic phenotype [26]. These lung cancers were associated with epithelial-mesenchymal transition gene expression upregulation. Our work focused on lepidic-predominant lung adenocarcinomas in sheep and humans, two cancers for which metastatic spread is rare [15], suggesting that the mouse model may not perfectly reproduce the naturally-induced disease.
Cancers need nutrients, oxygen and a way to evacuate metabolic waste to survive just like normal tissues. These functions rely on blood and lymphatic vessels. In the peculiar context of a limited extra-thoracic extension of the cancer cells in predominantly lepidic-type adenocarcinomas, we analyzed the expression of genes involved in angiogenesis, lymphangiogenesis and degradation of the extracellular matrix to decipher this specific feature and we conducted a comparative study in human and ovine lung cancers. The analysis of the VEGFA/VEGFR2 pathway of the main angiogenesis pathway gave some leads to understanding these features. We compared the mRNA expression of the genes of interest in lepidic and in papillary subtypes of human lung cancers, and when available in adjacent normal lungs from lepidic-type adenocarcinomas. For the ovine virally-induced adenocarcinomas, we compared tissues from tumors with positive detection of JSRV genome and JSRV-negative tissues without lesions. When available, we measured the mRNA expression in primary alveolar epithelial type II cells, derived from tumoral or normal tissues as previously described [22]. We identified modulations of the mRNA and protein levels of key genes that might participate in the limitation of extra thoracic metastatic extension in lepidic adenocarcinoma.
We report the absence of VEGFA proteins in JSRV-induced adenocarcinomas in sheep, while the mRNA level of VEGFA was maintained and not different in normal lungs and cancers. VEGFA is a key gene during the development of vessels during organogenesis or cancer. Its expression is highly regulated at the translational level [27]. In humans, it has been reported that miRNA regulate VEGFA expression. miRNA, such as miRNA-297, miRNA-299, miRNA-567 and miRNA-609 are able to control its expression by preventing protein production without inhibition of the transcription [28]. The existence and activity of this VEGFA-specific miRNA must be explored in sheep, but our data might suggest a similar control in small ruminants. Interestingly, VEGFA protein expression was restored in primary cultures of AECIIs derived from both normal lungs and cancers, proving that these cells retained their potential to produce VEGFA proteins. Our observation may be simply due to the in vitro conditions used to culture and maintain the cells, conditions that do not strictly mimic the in vivo cancer environment. The miRNA activity may also be only efficient in the cancer context since the microenvironment has been reported as an important factor for miRNA inhibition of VEGFA in pancreatic or human colon cancers [29].
We analyzed the mRNA expression of VEGFR2, a key receptor for angiogenesis. Interestingly, we observed a significant reduction of VEGFR2 mRNA and protein expression in ovine lung cancers as compared to normal lungs suggesting a negative control of the receptor in JSRV-induced cancers. The VEGFR2 receptor is mainly present at the surface of the endothelial cells and logically, primary AECII did not express the proteins while the level of mRNA was low to undetectable. Taken together, without VEGFA and VEGFR2, angiogenesis is not activated [4]. Comparatively, we did not see differences in VEGFA expression in human lepidic adenocarcinomas. The VEGFR2 protein was absent in lepidic as well as in papillary adenocarcinomas, while the levels of mRNA stayed very low. To summarize, we show that the main angiogenesis pathway was blocked in ovine JSRV-induced tumors with the absence of VEGFA and VEGFR2 proteins. In human cancers, VEGFA expression was not altered but VEGFR2 was not expressed. Blocking VEGFA and VEGFR2 might contribute to the limitation of extra thoracic dissemination of lepidic-predominant adenocarcinomas in humans and animals. These two proteins, essential for angiogenesis, are targeted by anti-metastasis drugs in various cancers including lung cancers [3].
Alternative pathways play a role in angiogenesis. We then analyzed the expression of other genes implicated in angiogenesis, lymphangiogenesis and degradation of the extracellular matrix. Overall, we measured a reduced expression of VEGFB mRNA in cancer-derived AECIIs when compared to cells derived from normal lungs. The expression in tissues was inconstant and low in all cases. Only half of tissues had detectable VEGFB mRNA expression. VEGFB is a peculiar angiogenesis ligand. It interacts with VEGFR1 but this interaction does not necessarily activate angiogenesis; the effects of VEGFB vary under specific conditions from induction of endothelial cell permeability to anti-growth and anti-angiogenic effects [30]. Interestingly, the level of VEGFD mRNA was lower in cancers than in normal lungs both in human and ovine adenocarcinomas. A significant mRNA down-expression of VEGFC and VEGFD has been reported in human lung adenocarcinomas [31]. In both tumors, the lower expression of VEGFD and the stable expression of VEGFR3 in tumoral tissues when compared to normal tissues might contribute to the control of metastatic extension.
Aside from the neo-vascularization, tumoral extension necessitates the degradation of the extracellular matrix by cellular peptidases. We measured mRNA expression of peptidases such as MMP2 and MMP9, PLAU and of their inhibitors, SERPINE1 and TIMP1. Less than 50% of the JSRV-induced tumors expressed MMP9 mRNA, otherwise absent in normal tissues, suggesting a blocking of this protease. But interestingly TIMP1, an MMP inhibitor, was strongly expressed in tumors. Besides its inhibitory properties, TIMP1 can block the response of endothelial cells to angiogenic factors and participate in the inhibition of endothelial cell migration [32]. We hypothesize that even if some tumors express MMP9, the expression of TIMP1 should block their protease function.
The FGF pathway participates in angiogenesis through its action on cancer cell proliferation [33]. Our results showed perturbation of the mRNA expression of FGF2 and FGFR2. The FGFR2 expression was significantly higher in tumoral lungs than in normal tissues in sheep and the FGF2 ligand is down-regulated in human lepidic tumors. Aside from its role in angiogenesis, FGFR2 has a role in cell proliferation [33]. Our results show that key genes implicated in angiogenesis were impaired during sheep lung tumors and that comparable results were obtained when analyzing this pathway in human lepidic adenocarcinoma.
The ovine pulmonary adenocarcinoma is induced by a retrovirus while the etiology of human lepidic adenocarcinoma is still unknown although a viral etiology has been questioned for decades [16]. Some oncogenic viruses have the ability to directly activate angiogenesis [34] such as EBV (Epstein Barr Virus), HPV (Human Papilloma Virus) and HBV (Hepatitis B Virus) that carry viral proteins with proangiogenic activities [35,36,37]. To explore a potential direct effect of JSRV proteins on the mRNA levels of the studied genes, we quantified JSRV RNA copies in lungs and AECII cultures. We found no correlation between the level of JSRV expression and the observed deregulation of genes involved in the angiogenesis pathway.
We performed similar analyses in lepidic adenocarcinoma and papillary adenocarcinomas. The results were comparable: VEGFA and VEGFR2 patterns of expression were similar; VEGFD, FGF2 were significantly downregulated as in lepidic adenocarcinomas and MMP2 was also significantly downregulated. This may be explained by the non-invasive pattern of the cancers analyzed in our study, as shown by their TNM staging.
Taken together, our results indicate that angiogenesis does not represent a hallmark of human and ovine pulmonary adenocarcinomas with predominant lepidic growth. In humans, lepidic adenocarcinoma is a rare lung cancer with no extra thoracic extension. We show that the absence of VEGFA/VEGR2 activation could presumably contribute to the control of extra-thoracic spreading of cancer cells in the virally induced cancer in sheep and in lepidic adenocarcinomas in humans.
The reported benefit of anti-angiogenic agents for the treatment of lung adenocarcinomas at large, may not be observed for patients with lepidic adenocarcinomas or at least for those with no detectable expression of VEGFA and VEGFR2; patients may then avoid potential toxicities that may occur with those agents, that would not be delivered if limited benefit is expected. Ultimately, as we show, JSRV-induced lung adenocarcinoma which naturally occurs in sheep provides a unique model to help to decipher the specific features of lepidic- predominant human lung adenocarcinomas.
References
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674
Otrock ZK, Mahfouz RA, Makarem JA, Shamseddine AI (2007) Understanding the biology of angiogenesis: review of the most important molecular mechanisms. Blood Cells Mol Dis 39:212–220
Hall RD, Le TM, Haggstrom DE, Gentzler RD (2015) Angiogenesis inhibition as a therapeutic strategy in non-small cell lung cancer (NSCLC). Transl Lung Cancer Res 4:515–523
Simons M, Gordon E, Claesson-Welsh L (2016) Mechanisms and regulation of endothelial VEGF receptor signalling. Nat Rev Mol Cell Biol 17:611–625
Alevizakos M, Kaltsas S, Syrigos KN (2013) The VEGF pathway in lung cancer. Cancer Chemother Pharmacol 72:1169–1181
Lee SH, Jeong D, Han YS, Baek MJ (2015) Pivotal role of vascular endothelial growth factor pathway in tumor angiogenesis. Ann Surg Treat Res 89:1–8
Yoshimatsu Y, Miyazaki H, Watabe T (2016) Roles of signaling and transcriptional networks in pathological lymphangiogenesis. Adv Drug Deliv Rev 99:161–171
Raimondi C, Ruhrberg C (2013) Neuropilin signalling in vessels, neurons and tumours. Semin Cell Dev Biol 24:172–178
Deryugina EI, Quigley JP (2015) Tumor angiogenesis: MMP-mediated induction of intravasation- and metastasis-sustaining neovasculature. Matrix Biol 44–46:94–112
Montuori N, Ragno P (2013) Role of uPA/uPAR in the modulation of angiogenesis. Chem Immunol Allergy 99:105–122
Brewer JR, Mazot P, Soriano P (2016) Genetic insights into the mechanisms of Fgf signaling. Genes Dev 30:751–771
Cousens C, Thonur L, Imlach S, Crawford J, Sales J, Griffiths DJ (2009) Jaagsiekte sheep retrovirus is present at high concentration in lung fluid produced by ovine pulmonary adenocarcinoma-affected sheep and can survive for several weeks at ambient temperatures. Res Vet Sci 87:154–156
Grego E, De Meneghi D, Alvarez V, Benito AA, Minguijon E, Ortin A, Mattoni M, Moreno B, Perez de Villarreal M, Alberti A, Capucchio MT, Caporale M, Juste R, Rosati S, De las Heras M (2008) Colostrum and milk can transmit jaagsiekte retrovirus to lambs. Vet Microbiol 130:247–257
Caporale M, Centorame P, Giovannini A, Sacchini F, Di Ventura M, De las Heras M, Palmarini M (2005) Infection of lung epithelial cells and induction of pulmonary adenocarcinoma is not the most common outcome of naturally occurring JSRV infection during the commercial lifespan of sheep. Virology 338:144–153
Mornex JF, Thivolet F, De las Heras M, Leroux C (2003) Pathology of human bronchioloalveolar carcinoma and its relationship to the ovine disease. Curr Top Microbiol Immunol 275:225–248
Monot M, Archer F, Gomes M, Mornex JF, Leroux C (2015) Advances in the study of transmissible respiratory tumours in small ruminants. Vet Microbiol 181:170–177
Detterbeck FC, Marom EM, Arenberg DA, Franklin WA, Nicholson AG, Travis WD, Girard N, Mazzone PJ, Donington JS, Tanoue LT, Rusch VW, Asamura H, Rami-Porta R, Staging I, Prognostic Factors C, Advisory B, Multiple Pulmonary Sites W (2016) The IASLC lung cancer staging project: background data and proposals for the application of TNM staging rules to lung cancer presenting as multiple nodules with ground glass or lepidic features or a pneumonic type of involvement in the forthcoming eighth edition of the TNM classification. J Thorac Oncol 11:666–680
Breathnach OS, Kwiatkowski DJ, Finkelstein DM, Godleski J, Sugarbaker DJ, Johnson BE, Mentzer S (2001) Bronchioloalveolar carcinoma of the lung: recurrences and survival in patients with stage I disease. J Thorac Cardiovasc Surg 121:42–47
D’Antonio C, Passaro A, Gori B, Del Signore E, Migliorino MR, Ricciardi S, Fulvi A, de Marinis F (2014) Bone and brain metastasis in lung cancer: recent advances in therapeutic strategies. Ther Adv Med Oncol 6:101–114
Sanmartin E, Sirera R, Uso M, Blasco A, Gallach S, Figueroa S, Martinez N, Hernando C, Honguero A, Martorell M, Guijarro R, Rosell R, Jantus-Lewintre E, Camps C (2014) A gene signature combining the tissue expression of three angiogenic factors is a prognostic marker in early-stage non-small cell lung cancer. Ann Surg Oncol 21:612–620
Monot M, Erny A, Gineys B, Desloire S, Dolmazon C, Aublin-Gex A, Lotteau V, Archer F, Leroux C (2015) Early steps of Jaagsiekte sheep retrovirus-mediated cell transformation involve the interaction between Env and the RALBP1 cellular protein. J Virol 89:8462–8473
Archer F, Jacquier E, Lyon M, Chastang J, Cottin V, Mornex JF, Leroux C (2007) Alveolar type II cells isolated from pulmonary adenocarcinoma: a model for JSRV expression in vitro. Am J Respir Cell Mol Biol 36:534–540
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JH, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, Geisinger K, Hirsch FR, Ishikawa Y, Kerr KM, Noguchi M, Pelosi G, Powell CA, Tsao MS, Wistuba I, WHO Panel (2015) The 2015 World Health Organization Classification Of Lung Tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 10:1243–1260
Chang HW, Lin ZM, Wu MJ, Wang LY, Chow YH, Jiang SS, Ch’ang HJ, Chang VH (2017) Characterization of a transgenic mouse model exhibiting spontaneous lung adenocarcinomas with a metastatic phenotype. PLoS One 12:e0175586
Arcondeguy T, Lacazette E, Millevoi S, Prats H, Touriol C (2013) VEGF-A mRNA processing, stability and translation: a paradigm for intricate regulation of gene expression at the post-transcriptional level. Nucleic Acids Res 41:7997–8010
Jafarifar F, Yao P, Eswarappa SM, Fox PL (2011) Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J 30:1324–1334
Zhang Y, Yang P, Wang XF (2016) Microenvironmental regulation of cancer metastasis by miRNAs. Trends Cell Biol 24:153–160
Li X, Kumar A, Zhang F, Lee C, Tang Z (2012) Complicated life, complicated VEGF-B. Trends Mol Med 18:119–127
Niki T, Iba S, Tokunou M, Yamada T, Matsuno Y, Hirohashi S (2000) Expression of vascular endothelial growth factors A, B, C, and D and their relationships to lymph node status in lung adenocarcinoma. Clin Cancer Res 6:2431–2439
Jackson HW, Defamie V, Waterhouse P, Khokha R (2016) TIMPs: versatile extracellular regulators in cancer. Nat Rev Cancer 17:38–53
Turner N, Grose R (2010) Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 10:116–129
Vrancken K, Vervaeke P, Balzarini J, Liekens S (2011) Viruses as key regulators of angiogenesis. Rev Med Virol 21:181–200
Lee SW, Lee YM, Bae SK, Murakami S, Yun Y, Kim KW (2000) Human hepatitis B virus X protein is a possible mediator of hypoxia-induced angiogenesis in hepatocarcinogenesis. Biochem Biophys Res Commun 268:456–461
Lopez-Ocejo O, Viloria-Petit A, Bequet-Romero M, Mukhopadhyay D, Rak J, Kerbel RS (2000) Oncogenes and tumor angiogenesis: the HPV-16 E6 oncoprotein activates the vascular endothelial growth factor (VEGF) gene promoter in a p53 independent manner. Oncogene 19:4611–4620
Murono S, Inoue H, Tanabe T, Joab I, Yoshizaki T, Furukawa M, Pagano JS (2001) Induction of cyclooxygenase-2 by Epstein–Barr virus latent membrane protein 1 is involved in vascular endothelial growth factor production in nasopharyngeal carcinoma cells. Proc Natl Acad Sci USA 98:6905–6910
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MG, FA, CL made substantial contributions to conception design, acquisition, analysis and interpretation of data. BG, CD, and AE made major contributions to data acquisition. MG and CL drafted the manuscript. NG and JFM have been involved in drafting the manuscript and revising it critically. All the authors read and approved the final manuscript.
Acknowledgements
We thank François Guiguen, DVM, Jean-Christophe Natorp, DVM, and Rémy Falguière for the collection of tissues. We thank Prof. Jacques Cadranel, Dr Madeleine Duc Dodon and Dr Catherine Legras-Lachuer for helpful discussions.
Funding
The work was funded by grants from the “Plan Cancer 2009–2013” and “Ligue contre le Cancer, Ardèche”. MG is the recipient of a Ph.D. fellowship from the “Plan Cancer 2009–2013”.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional file
13567_2017_480_MOESM1_ESM.docx
Additional file 1. Primers used for detecting mRNAs by quantitative RT-PCR in human and sheep lung adenocarcinomas. PCR primers for mRNA quantification of ligands, ligand receptor and reference genes have been designed for human and ovine. X: species specificity. Reference sequences are designated by their GenBank #.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Gomes, M., Archer, F., Girard, N. et al. Blocked expression of key genes of the angiogenic pathway in JSRV-induced pulmonary adenocarcinomas. Vet Res 48, 76 (2017). https://doi.org/10.1186/s13567-017-0480-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13567-017-0480-z