Abstract
Background
The osteogenic differentiation capacity of periodontal mesenchymal stem cells (PDLSCs) can be influenced by different levels of static mechanical strain (SMS) in an inflammatory microenvironment. Long non-coding RNAs (lncRNAs) are involved in various physiological processes. However, the mechanisms by which lncRNAs regulate the osteogenic differentiation of PDLSCs remain unclear.
Methods
We investigated the responses of PDLSCs obtained from periodontitis patients and healthy people to 8% and 12%SMS. Gene microarray and bioinformatics analyses were implemented and identified lncRNA00638 as a target gene for the osteogenesis of PDLSCs from periodontitis patients under SMS. Competing endogenous RNA (ceRNA) network analysis was applied and predicted interactions among lncRNA00638, miRNA-424-5p, and fibroblast growth factor receptor 1 (FGFR1). Gene expression levels were regulated by lentiviral vectors. Cell Counting Kit-8 assays, alkaline phosphatase assays, and Alizarin Red S staining were used to examine the osteogenic potential. RT-qPCR and Western blot were performed to detect the expression levels of related genes and proteins.
Results
We found that 8% and 12% SMS exerted distinct effects on HPDLSCs and PPDLSCs, with 12% SMS having the most significant effect. By microarray analysis, we detected differentially expressed lncRNAs/mRNAs between 12% SMS strained and static PPDLSCs, among which lncRNA00638 was detected as a positive target gene to promote the osteogenic differentiation of PPDLSCs under SMS loading. Mechanistically, lncRNA00638 may act as a ceRNA for miR-424-5p to compete with FGFR1. In this process, lncRNA00638 and miR-424-5p suppress each other and form a network to regulate FGFR1.
Conclusions
Our findings demonstrate that the lncRNA00638/miRNA-424-5p/FGFR1 regulatory network is actively involved in the regulation of PDLSC osteogenic differentiation from periodontitis patients under SMS loading, which may provide evidence for optimizing orthodontic treatments in patients with periodontitis.
Similar content being viewed by others
Background
Periodontitis is an inflammatory infectious disease of tooth-supporting tissues with a high incidence rate that produces aesthetic and functional problems for patients [1]. Orthodontic treatment is the optimal choice for patients with periodontitis to establish the occlusion and restore masticatory function, which are beneficial to periodontal health. In this process, periodontal mesenchymal stem cells (PDLSCs) play an essential role in bone remoulding due to orthodontic tooth movement. However, increasing evidence has shown that the decreased osteogenic ability of PDLSCs in the inflammatory microenvironment may be the cause of intolerance to orthodontic forces in patients with periodontal diseases [2, 3]. Static mechanical strain (SMS) is often used to simulate the stress under which PDLSCs are subjected during OTM in vitro. Our research group found that the osteogenic differentiation ability of PDLSCs can be influenced by different levels of SMS [4]. Exploring the causes and related mechanisms of the different responses of PDLSCs to different levels of SMS can provide a theoretical basis for the rational clinical application of orthodontic forces.
Long non-coding RNAs (lncRNAs) which are more than 200 nt in length and do not encode proteins, are involved in autoimmune, cardiovascular, and neurodegenerative diseases at the posttranscriptional level [5]. In recent years, the biological roles of lncRNAs in osteogenic differentiation and immune inflammation have been confirmed by many researchers [6]. Some lncRNAs, such as POIR, ANCR and XIST, can regulate the osteogenic differentiation of PDLSCs [7,8,9,10], but the underlying molecular mechanisms are still poorly understood. LncRNAs can act as different functional molecules, including molecular signals, decoys, guides and scaffolds, in gene regulation processes [11]. Among these functions, lncRNAs have been proposed to act as competing endogenous RNAs (ceRNAs) in a newly proposed model of gene regulation [12,13,14]. In this novel hypothesis, lncRNAs act as ceRNAs to sponge microRNAs(miRNAs), thereby inhibiting miRNAs from regulating their target genes [15,16,17]. For example, the lncRNA-POIR has been shown to promote the osteogenic differentiation of PDLSCs by competing with the osteogenesis-related target gene FoxO1 for binding miR-182 and blocking the inhibitory effect of miR-182 on FoxO1 [10]. However, whether lncRNAs regulate the osteogenic differentiation process of PDLSCs obtained from the periodontitis patients (PPDLSCs) under SMS is yet to be elucidated.
In this study, we identified a key gene, lncRNA00638, through gene microarray and bioinformatics analysis, and found that lncRNA00638 may positively regulate the osteogenic differentiation of PPDLSCs under mechanically loaded conditions in the periodontitis microenvironment. During this process, lncRNA00638 and miR-424-5p mutually repress one another to regulate fibroblast growth factor receptor 1 (FGFR1), the target gene of miR-424-5p, which affects the osteogenic differentiation of PPDLSCs. This study has identified a new target for improving the osteogenic capability of PPDLSCs under SMS and providing optimized orthodontic treatment in periodontitis patients.
Methods
Sample selection and cell cultures
HPDLSC tooth samples were obtained from 11 volunteers with an average age of 31.9 ± 5.2 years who did not have periodontal diseases. PPDLSC tooth samples were taken from 9 patients with a mean age of 36.1 ± 6.2 years who were diagnosed with moderate chronic periodontitis. The patients with periodontitis all had periodontal probing depth (PD) ≤ 6 mm, an attachment loss (AL) of 3–4 mm, and horizontal resorption of the alveolar bone of up to 1/3–1/2 of the root length. None of the selected volunteers had any systemic diseases, or a history of smoking, radiation therapy or chemotherapy. All procedures were approved by the Ethical Committee of the School of Stomatology, Fourth Military Medical University (IRB-REV-2020005). All participants provided written informed consent.
The periodontal tissues were scraped from the middle 1/3 of the root from the tooth samples. Collagenase type I (Gibco/Invitrogen, Carlsbad, CA, USA) was added to digest the periodontal tissues in a CO2 incubator at 37 °C for 1 h. The digestion was terminated by the addition of an equal volume of α-minimum essential medium (α-MEM, Gibco/Invitrogen, Carlsbad, CA, USA) containing 10% (v/v) foetal bovine serum (FBS), 1% (v/v) penicillin, and 1% (v/v) streptomycin. The obtained cells were inoculated in 6-well plates until they reached 80% confluence and isolated by the limiting dilution technique for subsequent experiments. All subsequent experiments were performed with 3rd-generation cells.
SMS loading
Third-generation cells were inoculated in BioFlex 6-well plates at a density of 2 × 106 cells/well and incubated at 37 °C for 12 h. Cells were synchronized by the addition of serum-free α-MEM medium for 24 h of starvation, and then placed on the FX-4000T Draft Force Loading System (Flexcell, Burlington, USA) for SMS loading. SMS with a force value of 8% or 12% at a frequency of 0.1 Hz were applied to HPDLSCs and PPDLSCs for 12 h. The morphology of the cells before and after SMS loading was observed by inverted microscopy (Olympus, Tokyo, Japan).
Cell viability assays
Cell viability was analysed by Cell Counting Kit-8 (CCK-8, Kanglang Biological Technology Co. Ltd., Shanghai, China) assays. After SMS loading, cells were inoculated into 96-well plates, and 6 replicate wells and 1 blank control well were set for each group. Ten microlitres of CCK-8 solution was added to each well and incubated at 37 °C for 4 h. Then, an enzyme linked immunosorbent assay (ELISA, Biotek, Vermont, USA) was used to detect the absorbance (OD) at 490 nm. The analysis was repeated three times to obtain the statistical data.
Gene microarray and data analysis
Gene microarray was used to analyse differentially expressed lncRNAs and mRNAs before and after 12% SMS loading. PPDLSC samples were divided into the control group and the 12% SMS group. Total RNA was extracted using TRIzol reagent, and the quantity and quality of the RNA were measured with a NanoDrop ND-1000 (Biosystems, USA), followed by RNA purification. The mRNA was extracted from the total RNA, and the mRNA from ach sample was amplified by the random start method and transcribed into fluorescent cRNA along the entire transcriptome length. The labelled cRNA was purified using an RNeasy Mini Kit (Invitrogen, Carlsbad, CA, USA), and the concentration and activity of the labelled cRNA were measured with a Nano Drop ND-1000. After the lncRNA expression microarrays were assembled, they were incubated in an Agilent hybridization oven (Agilent, Santa Clara, CA, USA) at 65 °C for 17 h. The hybridized arrays were washed, fixed and scanned using an Agilent DNA microarray scanner (Axon GenePix 4000B, Axon, USA). The obtained array images were analysed using Agilent Feature Extraction software v11.0.1.1 (Agilent, USA). Raw data were processed using the Gene Spring GX v11.5.1 software package (Agilent, USA), and lncRNAs and mRNAs from at least 2 of the 4 samples with current or marginal “all target values” were selected for further data analysis. Differentially expressed lncRNAs and mRNAs between the two groups were identified by volcano map filtering. Image data analysis was performed using Nimble Scan software (Agilent, USA), and hierarchical clustering was performed using Agilent GeneSpring GX software v11.5.1 (Agilent, USA).
Bioinformatics analysis
We searched the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to determine the biochemical pathways enriched in the differentially expressed mRNAs, Gene Ontology (GO) analysis examines the enrichment of differentially expressed mRNAs in three domains to investigate their potential functions: biological process (BP), cellular component (CC), and molecular function (MF). Through coding-noncoding gene co-expression (CNC) analysis, we found mRNAs whose expression patterns were the same as those of specific lncRNA and predicted the biological functions. Among the studied samples, we screened out several lncRNAs with a fold change > 50.0, P < 0.05 and raw intensity > 100. Then, RT- qPCR analysis of these selected genes was performed to confirm the microarray results and to select the most significantly differentially expressed lncRNA as the target gene. To explore the mechanistic pattern of the target gene, lncRNA00638, we searched miRbase (http://www.mirbase.org/), TargetScan (http://www.targetscan.org/mamm/), and UCSC (http://genome.ucsc.edu/) for information on lncRNA–microRNA interactions. Additionally, ceRNA analysis was carried out to identify potential targeted microRNAs based on TargetScan and MiRanda. By merging the targeted microRNAs, we constructed a ceRNA network. The ceRNA analysis was completed by Kangcheng Biological Engineering (Shanghai, China).
Lentiviral transfection
PPDLSCs were transfected with lentiviral vector to regulate the expression of lncRNA00638. The lentiviral vector was designed and constructed by GeneChem Co. Ltd. (Shanghai, China). Third generation PPDLSCs were seeded on 96-well plates at a density of 1 × 104 cells/ml. Complete medium and ENi.S were added to dilute polybrene respectively and the final concentration was set at 5 µg/ml. When the cell confluence rate approached 1/3, transfection was carried out according to the determined viral titre. The infection efficiency was detected by fluorescence microscopy. mRNA was extracted for RT-qPCR to confirm the expression of lncRNA00638. PPDLSCs were divided into 5 groups after lentiviral transfection and SMS loading. For the transfection of anti-miR424-5p, PPDLSCs were seeded on BioFlex 6-well plates at a density of 1 × 105 cells/ml. Cells were synchronized 24 h before transfection in serum-free medium. Transfection was carried out when the cell confluence rate reached 2/3. RT-qPCR was used to confirm the expression of anti-miR424-5p.
Alizarin red S (ARS) staining and quantification
ARS staining and quantification were used to qualitatively and quantitatively detect osteogenic differentiation ability respectively. After SMS loading, the cell density was adjusted to 1 × 105 cells/well in 6-well plates. When the cells had proliferated to reach 80% confluence, the original culture medium was replaced with osteogenic induction solution (α-MEM with 10% FBS, 10−8 mol/l dexamethasone, 10 mmol/l β-glycerophosphate sodium, 50 µg/ml vitamin C). After 21 days, the cells were rinsed 3 times with PBS, fixed with 4% paraformaldehyde for 30 min, and then rinsed repeatedly with distilled water. ARS solution (Sigma, Shanghai, China) was added to each well and incubated at 37 °C for 20 min. Then the staining solution was discarded, and the cells were rinsed with double-distilled water for 3 times. Images were captured with an inverted microscopy. Additionally, 500 µL/per well cetylpyridinium chloride was added to the plates for quantitative detection and incubated at 37 °C for 30 min, after which 150 µL of the mixture was added to a 96-well plate, and the OD value at 520 nm was measured by ELISA.
Alkaline phosphatase (ALP) assay
Cells were seeded in 96-well plates at a density of 2 × 103 cells/well and repeatedly rinsed with double-distilled water for 3 times, fixed with 4% paraformaldehyde at 37 °C for 45 min, and rinsed again with double distilled water. The cells were stained according to the instructions of an ALP kit (Beyotime, Shanghai, China) and incubated for 45 min in a constant-temperature incubator after addition of the staining solution. Then, the staining solution was removed, and the cells were rinsed with double-distilled water. Images were captured by inverted microscopy, and the OD value at 520 nm was measured by ELISA.
Real-time quantitative polymerase chain reaction (RT-qPCR)
HPDLSCs and PPDLSCs were collected, and total RNA was isolated with TRIzol (Invitrogen, Carlsbad, CA, USA). The total RNA was converted to complementary DNA (cDNA) with a Prime Script™ RT-qPCR reverse transcription kit (Takara, Shiga, Japan). cDNA) was used as a template for the following steps. The SYBR Green method was performed to amplify target fragments. Primer sequences for osteogenesis-related genes and lncRNAs are listed in Table 1.
Western blotting
Radioimmunoprecipitation assay (RIPA) (Bio-Rad, USA) lysis buffer was used to extract the total proteins. The total protein concentration was quantified by the bicinchoninic acid protein assay method (Bio-Rad, USA) according to the manufacturer’s protocol. Protein samples were dissociated by sodium dodecyl sulfate–polyacrylamide (SDS–PAGE) gel electrophoresis and transferred to a PVDF membranes (Millipore, USA). The membranes were washed with blocking solution (TBST) for 45 min and then incubated with antibodies. The target signals were detected by enhanced chemiluminescence (ECL) detection (Peiqing Technology Co., Ltd, Shanghai, China).
Statistical analysis
IBM SPSS statistical package 26.0 (IBM Co., Chicago, USA) was used for statistical analysis. Categorical variables are presented as the mean ± standard deviation (SD). Two independent samples t test was selected for comparisons between two independent groups. One-way ANOVA with Student–Newman–Keuls (SNK) comparison was employed for multiple comparisons among multiple different groups. Pearson’s test was used for correlation analysis. A significance value of 0.05 was chosen.
Results
PPDLSCs and HPDLSCs responded differently to SMS, with 12% SMS having the most significant force value
The proliferative ability of HPDLSCs and PPDLSCs under 12% and 8% SMS were examined by CCK-8 assay. Measurement of the OD value in each group demonstrated that the quantity of HPDLSCs under 12% SMS was significantly higher than that of the 8% SMS group and the control group, whereas compared to that of HPDLSCs, the quantity of PPDSCs was higher under 8% SMS but lower under 12% SMS (Fig. 1A). The osteogenic differentiation ability was detected by RT-qPCR analysis of osteogenic genes and ARS staining. The results showed the following: (1) Under 12% SMS, the relative expression levels of the osteogenic genes Runx2, ALP, and OCN were increased in HPDLSCs compared with the control group, but there was no significant difference in PPDLSCs. After osteogenic induction for 21 days, mineralized nodules in the HPDLSCs + 12%SMS group were more abundant in quantity and more condensed in distribution than those in the control group, but there was no significant difference in the number of mineralized nodules between the PPDLSCs + 12%SMS group and the control group. (2) Under 8% SMS, the expression of osteogenic genes in HPDLSCs was also increased compared with that in the control group but was less than that in the 12% SMS group. Interestingly, in PPDLSCs, the levels of osteogenic genes were significantly higher in the 8% SMS group than in either the control or 12%SMS groups. The ARS staining results were essentially consistent with the RT-qPCR data, that is, mineralized nodules in HPDLSCs + 8% SMS group were more densely distributed than those in the control group but less dense than those in the 12% SMS group, while that the mineralized nodules in the PPDLSCs + 8% SMS group were more densely distributed than those in the control group or the 12% SMS group (Fig. 1B, C). The quantitative ARS assay results were consistent with the changes revealed by RT-qPCR (Fig. 1D). These results suggest that PPDLSCs and HPDLSCs responded differently to SMS. Under 12% SMS, they showed the most significant differences in osteogenic differentiation and proliferation capacities.
Cellular responses to different levels of SMS. A CCK-8 assay was used to analyse the proliferation ability of HPDLSCs and PPDLSCs under 12% and 8% SMS. B RT-PCR was used to detect the expression levels of osteogenesis related genes. C ARS staining was used to detect osteogenic differentiation. Scale bar = 100 µm. D ARS quantitative assay. All experiments were repeated three times. All data are shown as the mean ± SD. a: compared with the HPDLSCs group (P < 0.05); b: compared with the HPDLSCs + 8%SMS group (P < 0.05); c: compared with the HPDLSCs + 12%SMS group (P < 0.05); d: compared with the PPDLSCs group (P < 0.05); e: compared with the PPDLSCs + 8%SMS group (P < 0.05); f: compared with the PDLSCs + 12%SMS group (P < 0.05)
Expression profiles of lncRNAs and mRNAs under SMS
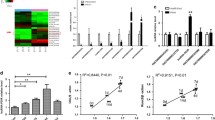
Gene microarray was used to identify the differential lncRNA expression profiles of PPDLSCs after 12% SMS loading. Heatmap (Fig. 2A), scatter plots (Fig. 2B), and volcano plot analyses (Fig. 2C) showed that 4471 lncRNAs and 7077 mRNAs were upregulated; and 6444 lncRNAs and 5816 mRNAs were downregulated after 12% SMS loading. To further explore the putative functions of these lncRNAs and mRNAs, bioinformatic analysis based on KEGG pathway and GO analyses was applied. The results showed that the actin cytoskeletal pathway and MAPK pathway which are mainly involved in cellular biomechanical signalling and osteogenic differentiation regulation, scored high in the KEGG analysis (Fig. 2D). Among the enriched GO terms, “single-multicellular organism process” and “mitotic cell cycle process” among BPs, “cell junction” and “nuclear part” among CCs, and “protein binding” and “RNA binding” among MFs earned the highest enrichment scores. We found that the items identified by the GO analysis were relatively broad and basically belonged to basic cellular BPs, suggesting that 12% SMS inhibited the osteogenic differentiation of PPDLSCs but did not change their mRNA traits (Fig. 2E–J).
Microarray screening and analysis. A Hierarchical clustering and heatmap analysis were used to detect differentially expressed lncRNAs after 12% SMS loading. T1, T2, and T3 are control group samples taken from PPDLSCs without SMS loading, and PP1, PP2, and PP3 are experimental group samples taken from PPDLSCs with SMS loading. There were visible differences between the two groups. B Scatter plot showing variations in the expression of lncRNAs. Black dots are lncRNAs with a differential expression ratio < 2.0, red and green dots are lncRNAs with a differential expression ratio > 2.0. C Volcano plot showing the relationship between the fold changes in the expression of differentially expressed lncRNAs among sample groups and the corresponding statistical significance. Black dots indicate P > 0.05 and red and green dots indicate P < 0.05. D KEGG pathways enriched in mRNAs with increased expression (left) and decreased expression (right). E Top 10 BPs enriched in mRNAs with increased expression. F Top 10 BPs enriched in mRNAs with decreased expression. G Top 10 CCs enriched in mRNAs with increased expression. H Top 10 CCs enriched in mRNAs with decreased expression. I Top 10 MFs enriched in mRNAs with increased expression. J Top 10 MFs enriched in mRNAs with decreased expression
LncRNA00638 was identified as a target lncRNA that is closely associated with the osteogenic differentiation of PPDLSCs under 12% SMS loading
After that, we screened eight alternative lncRNAs with a fold change > 50 and p-value < 0.05, combined with a raw intensity > 100 excluding overlapping sense exon. The RT-qPCR results for seven of the lncRNAs were consistent with the microarray results (Fig. 3A). Then, we assessed whether these lncRNAs could affect the osteogenesis process of PPDLSCs. The RT-qPCR results showed that the expression of lncRNA00638 (ID: ENSG00000258701 in UCSC; ID: NR_024396 in Refseq) changed most significantly after osteogenic induction with 12% SMS (Fig. 3B). In addition, lncRNA00638 expression was continuously elevated with osteogenic induction at 0, 1, 3, 7 and 14 d, peaking at 7 d (Fig. 3C). In addition, the Pearson correlation analysis demonstrated that lncRNA00638 was strongly related to the osteogenic-related genes Runx2 and Col1 during osteogenic induction (Fig. 3D). Therefore, lncRNA00638 was identified as the target lncRNA for subsequent experiments.
LncRNA00638 is associated with osteogenic differentiation. A The microarray results were validated by RT-qPCR. The RT–qPCR and microarray results were consistent for seven of the selected lncRNAs, among which lncRNA00638 showed the most significant difference, while the difference in TCONS_00008000 was not statistically significant. B Among the seven lncRNAs for which relative expression was detected by RT-qPCR after 7 d of osteogenic induction, lncRNA00638 expression changed most significantly. C Expression levels of lncRNA00638 at 0, 1, 3, 7 and 14 d detected by RT-qPCR. D Pearson correlation analysis showed that the expression level of lncRNA00638 was strongly related to Runx2 (R2 = 0.8935) and Col1 (R2 = 0.8935) during osteogenic induction. Osteo: osteogenic induction. Undiff: no osteogenic induction. **: P < 0.01; *: P < 0.05; NS: no significant difference
LncRNA00638 promoted the osteogenic differentiation of PPDLSCs under SMS loading
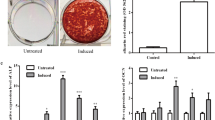
To explore the biological effect of lncRNA00638 on osteogenic differentiation of PPDLSCs under 12% SMS loading, we constructed lentiviruses for lncRNA00638 upregulation and downregulation and carried out lentiviral transfection (Fig. 4A). The upregulation of lncRNA00638 significantly increased the mRNA levels of the osteogenic genes Runx2, ALP and Col1. Conversely, lncRNA00638 RNA interference (RNAi) significantly decreased the expression levels of these genes (Fig. 4B). The results of ALP assay and ARS staining assay also suggested that lncRNA00638 overexpression increased ALP activity and mineralized bone matrix formation, while lncRNA00638 silencing inhibited osteogenic differentiation (Fig. 4C–E). To further investigate the osteogenic functional role of lncRNA00638 in PPDLSCs osteogenesis under 8% SMS, we silenced lncRNA00638 by also transfecting lncRNA00638-RNAi. The results showed that the expression of Runx2, ALP, and Col1 was decreased in the 8% SMS group (Fig. 4F), and the results of the ALP assay and ARS staining corroborated this trend (Fig. 4G–I). These results suggested that the silencing of lncRNA00638 reduced the osteogenic differentiation of PPDLSCs under 8% SMS, although it is a suitable loading for osteogenesis in an inflammatory environment.
LncRNA00638 promotes osteogenic differentiation. A RT-qPCR detection of the infection efficiency of the lncRNA00638-expressing lentivirus. B Expression of Runx2, ALP and Col1 was detected by RT-qPCR after 12% SMS loading and 7 days osteogenic induction. C Images showing ALP and ARS staining. D Quantitative assay of ALP activity. E Quantitative assay of ARS staining. F Expression of Runx2, ALP and Col1 was detected by RT-qPCR after 8% SMS loading and 7 days of osteogenic induction. G Images showing ALP and ARS staining. H Quantitative ALP assay results. I Quantitative ARS staining results. Data are presented as the mean ± SD a: compared with the PPDLSCs group (P < 0.05); b: compared with the PPDLSCs + 12%SMS group (P < 0.05); c: compared with the PPDLSCs + 12% SMS NC group (P < 0.05); d: compared with the PPDLSCs + 12% SMS lncRNA00638 group (P < 0.05); e: compared with the PPDLSCs + 12% SMS lncRNA00638-RNAi (P < 0.05)
LncRNA00638 promoted the osteogenic differentiation of PPDLSCs under SMS loading by sponging miR-424-5p to upregulate FGFR1
CNC analysis revealed several molecules involved in osteogenic regulatory pathways, such as FGFR1, FPR2, and DAPK1, that might be associated with differentially expressed lncRNAs (Fig. 5A). Through the MiRBase, TargetScan, and UCSC databases, we found that lncRNA00638 contains binding sites for nine miRNAs. Additionally, lncRNA00638 can actively act as a ceRNA for USB1, RPL13, NLRX1, and other genes, suggesting that lncRNA00638 is likely to regulate the target genes by adsorbing microRNA (Additional file 1: Figs. S1 and S2). Then through ceRNA analysis, we found that lncRNA00638 can act as a ceRNA for miR-424-5p and FGFR1 (Fig. 5B). To determine whether lncRNA00638 acts as a sponge that competes with FGFR1 for miR-424-5p binding, we transfected miR-424-5p inhibitor (anti-miR-424-5p) and evaluated its efficacy by RT-qPCR. The results showed that anti-miR-424-5p significantly inhibited the expression of miR-424-5p (Fig. 5C). Meanwhile, the expression of lncRNA00638 was obviously increased (Fig. 5D). In contrast, the expression of miR-424-5p was increased after transfection with lncRNA00638-RNAi (Fig. 5E). The results suggest that miR-424-5p and lncRNA00638 may mutually regulate each other. To explore the relationship between miR-424-5p and FGFR1 expression, we transfected anti-miR-424-5p into PPDLSCs and observed the changes in the gene and protein levels of FGFR1. The results showed that anti-miR-424-5p significantly reduced the mRNA and protein levels of FGFR1 in PPDLSCs (Fig. 5F), which indicated that miR-424-5p could promote the expression of FGFR1 in PPDLSCs. To explore the relationship between lncRNA00638 and FGFR1 expression, we monitored FGFR1 expression after upregulating and downregulating lncRNA00638. FGFR1 expression was significantly increased at the mRNA and protein levels after lncRNA00638 overexpression (Fig. 5G). In contrast, lncRNA00638-RNAi transfection negatively regulated the expression level of FGFR1 (Fig. 5H).
Mechanism by which lncRNA00638 regulates osteogenesis. A CNC network mapping for related mRNAs of differentially expressed lncRNAs. B CeRNA lncRNA00638 interaction network diagram. Green circle: lncRNA00638; red circle: miR-424-5p; pink circle: FGFR1. C Transfection efficiency of anti-miR-424-5p. D The expression of miR-424-5p after lncRNA00638-RNAi transfection. E The expression of lncRNA00638 after anti-miR-424-5p transfection. F Anti-miR-424-5p can promoted FGFR1 expression at the gene and protein levels (Full-length blots are presented in Additional file 2: Fig. S1). G FGFR1 gene and protein expression after the overexpression of LncRNA00638 (Full-length blots are presented in Additional file 3: Fig. S2). H FGFR1 gene and protein expression after the silencing of lncRNA00638 (Full-length blots are presented in Additional file 4: Fig. S3)
Discussion
OTM is based on remodelling of the periodontal ligament and alveolar bone under mechanical force. PDLSCs play a vital role in bone resorption on the pressure side and bone regeneration on the tension side during this process [18, 19]. Currently, an increasing number of patients with periodontitis seek orthodontic treatment. Existing evidence shows that combined orthodontic-periodontic treatment can effectively control the levels of inflammatory cytokines and has favourable effects on aesthetic restoration, chewing function and the healing of periodontal tissue [20, 21]. However, the tolerance to the application of excessive orthodontic forces on PDLSCs in the periodontitis microenvironment decreases for several reasons, leading to the destruction of periodontal tissue. The molecular mechanism remains unclear. Accordingly, searching for an effective method to promote the osteogenic capacity of PPDLSCs under SMS is a focus of current research to optimize orthodontic treatment in patients with periodontitis.
In our previous study, different levels of SMS were applied to HPDLSCs and PPDLSCs respectively, and our experiments showed that SMS of 8% was the best for the capability of PPDLSCs, while the best force value for HPDLSCs was 12% SMS [22]. In the present study, we chose these two different levels of SMS and made a lateral comparison between PPDLSCs and HPDLSCs, the results of which are consistent with the previous study indicating that the inflammatory microenvironment attenuates the response of PPDLSCs to mechanical stimuli. Under 12% SMS, the osteogenic capability of HPDLSCs was promoted to a greater extent, while that of PPDLSCs was damaged. However, under conditions of 8% SMS, PPDLSCs showed relatively better osteogenic ability while HPDLSCs did not achieve the best performance. These results fully demonstrate that 12% SMS was the most significant force value.
As a recent research hotspot, lncRNAs play a vital role in many physiological processes and diseases, such as autoimmune, cardiovascular and neurodegenerative diseases. For example, the lncRNA-ANCR was proven to promote osteoblast differentiation by activating the classical Wnt signalling pathway and regulating the expression levels of osteogenic markers such as OCN and Runx2 [8]. Microarray gene chips are one of the important tools to predict and validate the functional roles of lncRNAs. However, existing research represents just the tip of the iceberg. To investigate the mechanisms by which lncRNAs regulate osteogenesis, we performed microarray expression profiling of lncRNAs and screened differentially expressed lncRNAs in PPDLSCs before and after 12% SMS loading. LncRNA00638 was selected as the target lncRNA because it showed the most significant difference in expression by RT-qPCR checking, and the expression levels of lncRNA00638 and the osteogenesis-related genes Runx2/Col1 were highly correlated. To determine the osteogenic capacity of lncRNA00638, we constructed lncRNA00638 overexpression and knockdown lentiviral vectors, and our experiments revealed that lncRNA00638 promoted the osteogenic differentiation of PPDLSCs under 12% SMS. To further investigate the effect of LncRNA00638 on PPDLSCs under appropriate orthodontic force, we silenced the gene after loading 8% SMS loading and found that osteogenesis was also reduced. These results all suggest that lncRNA00638 is a promising target for regulating bone formation in PPDLSCs under mechanical loading conditions.
To further explore the regulatory mechanism of lncRNA00638, bioinformatics analysis and ceRNA network analyses demonstrated that lncRNA00638 may act as a ceRNA with miR-424-5p to regulate FGFR1. There are several molecules involved in osteogenesis regulatory pathways, such as FGFR1, FPR2 and DAPK1. They may be related to differentially expressed lncRNAs, among which FGFR1 plays pivotal roles in bone formation, development, and maturation [23, 24]. Under stimulation from physical signals, the activation of FGFR1/extracellular signal-regulated kinase (FGFR1/ERK) is an important mechanism involved in the regulation of bone formation and remodelling [25,26,27]. In the present study, the microarray expression profile of PPDLSCs with 12% SMS loading revealed a 4.71-fold change in FGFR1 gene expression, indicating a close connection with osteogenesis. It has been reported that miR-424-5p can mediate the FGF2/FGFR1 signalling pathway, thereby affecting the osteogenic differentiation of marrow stromal cells (MSCs) cells under oxidative stress conditions [28]. Yang et al. reported that overexpression of miR-424-5p can downregulated FGFR1 expression in haemangioma endothelial cells, and inhibition of miR-424-5p upregulated FGFR1 expression, suggesting that miR-424-5p can indirectly interfere with ERK1/2 phosphorylation by inhibiting the FGF2/FGFR1 pathway, thereby affecting cell proliferation [29]. In other words, miR-424-5p can actively participate in the functional regulation of FGFR1. Therefore, we hypothesized that both lncRNA00638 and FGFR1 are targets of miR-424-5p and that lncRNA00638 competes with FGFR1 for binding to miR-424-5p. The following experimental results obtained by RT-qPCR and WB corroborated this hypothesis. First, the expression level of miR-424-5p was found to be negatively correlated with that of lncRNA00638. Second, overexpression of lncRNA00638 promoted the gene and protein expression of FGFR1; silencing lncRNA00638 inhibited the gene and protein expression of FGFR1. Third, silencing the expression of miR-424-5p increased the expression of FGFR1 in PPDLSCs. In summary, lncRNA00638 may act as a ceRNA for miR-424-5p to compete with FGFR1 and thus may participate in the regulation of osteogenic differentiation in PPDLSCs under SMS loading.
Although further evidence of direct binding between these genes is needed, our study demonstrates for the first time that the osteogenic differentiation of PPDLSCs is controlled by the lncRNA00638/miRNA-424-5p/FGFR1 regulatory network under SMS, providing new insights for improving osteogenesis ability and orthodontic efficiency in periodontitis patients.
Conclusion
In this study, PPDLSCs and HPDLSCs exhibited significantly different cellular responses to 12% SMS. Then differentially expressed lncRNA/mRNA profiles under 12% SMS loading in PPDLSCs were first identified. We confirmed that lncRNA00638 actively participates in the regulation PPDLSC osteogenic differentiation under SMS loading. Then, mechanistic research indicated that lncRNA00638 may act as a sponge competing with FGFR1 for miR-424-5p binding. These results suggest that lncRNAs can regulate the osteogenic ability of PDLSCs under mechanical strain. Nevertheless, comprehensive analyses are still needed to elucidate the details of the relevant molecular mechanisms.
Availability of data and materials
All data generated or analysed during this study are included in this published article (and its Additional file 1). The datasets generated and analysed during the current study are available in OMIX (https://ngdc.cncb.ac.cn/omix) datasets.
Abbreviations
- PDLSCs:
-
Periodontal mesenchymal stem cells
- PPDLSCs:
-
Periodontal mesenchymal stem cells obtained from the periodontitis patients
- HPDLSCs:
-
Periodontal mesenchymal stem cells obtained from healthy people
- SMS:
-
Static mechanical strain
- lncRNAs:
-
Long non-coding RNAs
- ceRNA:
-
Competing endogenous RNA
- FGFR1:
-
Fibroblast growth factor receptor 1
- α-MEM:
-
α-Minimum essential medium
- FBS:
-
Foetal bovine serum
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- GO:
-
Gene ontology
- BP:
-
Biological process
- CC:
-
Cellular component
- MF:
-
Molecular function
- CNC:
-
Coding-noncoding gene co-expression
- ARS:
-
Alizarin red S
- ALP:
-
Alkaline phosphatase
- RT-qPCR:
-
Real-time quantitative polymerase chain reaction
- cDNA:
-
Complementary DNA
- MSCs:
-
Marrow stromal cells
References
Kinane DF, Stathopoulou PG, Papapanou PN. Periodontal diseases. Nat Rev Dis Primers. 2017;3:17038.
Zhu W, Liang M. Periodontal ligament stem cells: current status, concerns, and future prospects. Stem Cells Int. 2015;2015:972313.
Liu J, Wang L, Liu W, Li Q, Jin Z, Jin Y. Dental follicle cells rescue the regenerative capacity of periodontal ligament stem cells in an inflammatory microenvironment. PLoS ONE. 2014;9(9):e108752.
Zhang L, Liu W, Zhao J, Ma X, Shen L, Zhang Y, Jin F, Jin Y. Mechanical stress regulates osteogenic differentiation and RANKL/OPG ratio in periodontal ligament stem cells by the Wnt/beta-catenin pathway. Biochim Biophys Acta. 2016;1860(10):2211–9.
Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–307.
Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–69.
Feng Y, Wan P, Yin L. Long noncoding RNA X-inactive specific transcript (XIST) promotes osteogenic differentiation of periodontal ligament stem cells by sponging microRNA-214-3p. Med Sci Monit. 2020;26:e918932.
Jia Q, Jiang W, Ni L. Down-regulated non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of periodontal ligament stem cells. Arch Oral Biol. 2015;60(2):234–41.
Tong X, Gu PC, Xu SZ, Lin XJ. Long non-coding RNA-DANCR in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. Biosci Biotechnol Biochem. 2015;79(5):732–7.
Wang L, Wu F, Song Y, Li X, Wu Q, Duan Y, Jin Z. Long noncoding RNA related to periodontitis interacts with miR-182 to upregulate osteogenic differentiation in periodontal mesenchymal stem cells of periodontitis patients. Cell Death Dis. 2016;7(8):e2327.
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–14.
Fan C, Hao Z, Yan J, Li G. Genome-wide identification and functional analysis of lincRNAs acting as miRNA targets or decoys in maize. BMC Genomics. 2015;16:793.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–8.
Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46.
Cao L, Liu W, Zhong Y, Zhang Y, Gao D, He T, Liu Y, Zou Z, Mo Y, Peng S, et al. Linc02349 promotes osteogenesis of human umbilical cord-derived stem cells by acting as a competing endogenous RNA for miR-25-3p and miR-33b-5p. Cell Prolif. 2020;53(5):e12814.
Liang WC, Fu WM, Wang YB, Sun YX, Xu LL, Wong CW, Chan KM, Li G, Waye MM, Zhang JF. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci Rep. 2016;6:20121.
Zhang L, Xie H, Li S. LncRNA LOXL1-AS1 controls osteogenic and adipocytic differentiation of bone marrow mesenchymal stem cells in postmenopausal osteoporosis through regulating the miR-196a-5p/Hmga2 axis. J Bone Miner Metab. 2020;38(6):794–805.
Tanne K, Inoue Y, Sakuda M. Biomechanical behavior of the periodontium before and after orthodontic tooth movement. Angle Orthod. 1995;65(2):123–8.
Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284(23):15676–84.
Zhang J, Zhang AM, Zhang ZM, Jia JL, Sui XX, Yu LR, Liu HT. Efficacy of combined orthodontic-periodontic treatment for patients with periodontitis and its effect on inflammatory cytokines: a comparative study. Am J Orthod Dentofacial Orthop. 2017;152(4):494–500.
Cao L. Orthodontic-periodontic treatment for periodontitis. Am J Orthod Dentofacial Orthop. 2018;153(6):764–5.
Liu J, Li Q, Liu S, Gao J, Qin W, Song Y, Jin Z. Periodontal ligament stem cells in the periodontitis microenvironment are sensitive to static mechanical strain. Stem Cells Int. 2017;2017:1380851.
Jacob AL, Smith C, Partanen J, Ornitz DM. Fibroblast growth factor receptor 1 signaling in the osteo-chondrogenic cell lineage regulates sequential steps of osteoblast maturation. Dev Biol. 2006;296(2):315–28.
Collette JC, Choubey L, Smith KM. Glial and stem cell expression of murine Fibroblast Growth Factor Receptor 1 in the embryonic and perinatal nervous system. PeerJ. 2017;5:e3519.
Zhou W, Zhu Y, Chen S, Xu R, Wang K. Fibroblast growth factor receptor 1 promotes MG63 cell proliferation and is associated with increased expression of cyclin-dependent kinase 1 in osteosarcoma. Mol Med Rep. 2016;13(1):713–9.
Xiao L, Esliger A, Hurley MM. Nuclear fibroblast growth factor 2 (FGF2) isoforms inhibit bone marrow stromal cell mineralization through FGF23/FGFR/MAPK in vitro. J Bone Miner Res. 2013;28(1):35–45.
Weekes D, Kashima TG, Zandueta C, Perurena N, Thomas DP, Sunters A, Vuillier C, Bozec A, El-Emir E, Miletich I, et al. Regulation of osteosarcoma cell lung metastasis by the c-Fos/AP-1 target FGFR1. Oncogene. 2016;35(22):2852–61.
Li L, Qi Q, Luo J, Huang S, Ling Z, Gao M, Zhou Z, Stiehler M, Zou X. FOXO1-suppressed miR-424 regulates the proliferation and osteogenic differentiation of MSCs by targeting FGF2 under oxidative stress. Sci Rep. 2017;7:42331.
Yang L, Dai J, Li F, Cheng H, Yan D, Ruan Q. The expression and function of miR-424 in infantile skin hemangioma and its mechanism. Sci Rep. 2017;7(1):11846.
Acknowledgements
Not applicable.
Funding
The work is supported by a research grant from National Natural Science Foundation of China (No. 82001079).
Author information
Authors and Affiliations
Contributions
XZ contributed to conception and design, data acquisition and analysis, and drafted and critically revised manuscript. QY and XL contributed to data acquisition and analysis, drafted and critically revised manuscript. JG and YX contributed to conception, data analysis, and critically revised manuscript. ZJ contributed to conception and design, data interpretation, and critically revised manuscript. WQ contributed to conception and design, data analysis, and drafted and critically revised manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All procedures were approved by the Ethical Committee of School of Stomatology, Fourth Military Medical University (No. IRB-REV-2020158). The title of the approved project was “LncRNA00638 promotes the osteogenic differentiation of periodontal mesenchymal stem cells from periodontitis patients under static mechanical strain”. The approval date is November 23rd, 2020.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Fig. S1
Structure diagram of microRNA with binding site to lncRNA00638. A mi R-16-5p. B mi R-17-3p. C mi R-21-3p. D mi R-106b-5p. E mi R-195-5p. F mi R-211-3p. G mi R-424-5p. H mi R-503-5P. I mi R-3607-5p. Fig. S2 LncRNA00638 actively participates in the ceRNA process.
Additional file 2
. Full-length blots of Fig. 5F.
Additional file 3
. Full-length blots of Fig. 5G.
Additional file 4
. Full-length blots of Fig. 5H.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, X., Yan, Q., Liu, X. et al. LncRNA00638 promotes the osteogenic differentiation of periodontal mesenchymal stem cells from periodontitis patients under static mechanical strain. Stem Cell Res Ther 14, 177 (2023). https://doi.org/10.1186/s13287-023-03404-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13287-023-03404-6