Abstract
Background
Alopecia areata (AA), a prevalent form of autoimmune hair loss, has a not well-defined relationship with atopic and allergic disorders, including eczema, hay fever, and asthma.
Objectives
This study aims to elucidate the genetic relationship between atopy, allergies, and alopecia areata (AA) using Mendelian randomization. We hypothesize that atopic and allergic conditions contribute to the genetic predisposition of AA.
Methods
We analyzed extensive genetic data from Genome-wide Association Studies (GWAS) involving over one million individuals. This analysis focused on assessing the genetic correlation between AA and various allergic conditions, including hay fever, eczema, asthma, and allergies to pollen, dust, and cats. The inverse variance weighted method served as our primary analytical tool, complemented by sensitivity analyses to verify the robustness of our results.
Results
Our findings reveal a significant genetic correlation between atopy/allergies and an increased risk of AA. Notably, strong associations were observed for hay fever, eczema, asthma, and specific allergies (pollen, dust, and cats). The sensitivity analyses corroborated these associations, reinforcing the reliability of our primary results.
Conclusions
This study provides compelling genetic evidence of an association between atopic and allergic conditions and the development of AA. These findings suggest that individuals with such conditions may benefit from enhanced surveillance for early signs of AA.
Similar content being viewed by others
Introduction
Alopecia areata (AA), an autoimmune disorder resulting in non-scarring hair loss, impacts about 2% of the global population [1], predominantly affecting individuals under 40 years, with a noticeable prevalence in Asian communities [2]. The condition’s impact on the quality of life is substantial, akin to that of severe dermatological disorders, influencing both physical appearance and psychological health [3]. AA is often accompanied by other autoimmune diseases and a higher predisposition to mental health issues, such as depression, anxiety, and sleep disorders [4]. Recognizing these broad impacts highlights the importance of identifying risk factors and developing targeted treatment approaches to mitigate AA's effects.
Historically, AA has been primarily associated with Th1/IFN-γ overactivation, emphasizing type I over type II inflammatory responses (atopy and allergies) [5]. However, recent studies have increasingly explored the association between AA and allergic conditions, like eczema, hay fever, and asthma [6]. While some studies have found a positive correlation between these conditions and AA, the evidence remains inconclusive, with large-scale studies suggesting a complex interplay between the number of atopic conditions experienced and the risk of developing AA [7]. Additionally, the relationship with common allergens such as dust mites, cats, and pollen warrants further investigation [8]. Currently, there's no definitive evidence establishing a causal link between atopy, allergies, and AA.
Traditional research methods face challenges in fully excluding confounding factors, potentially leading to biased conclusions [9]. The implementation of randomized controlled trials in this context is both logistically challenging and ethically complex. In response, Mendelian randomization (MR) is increasingly being utilized to derive causal inferences between risk factors and diseases [10]. MR uses genetic variants as proxies for environmental exposures, allowing for the evaluation of exposure-disease relationships while minimizing confounding. It provides a framework for assessing the potential causal relationship between atopy, allergies, and AA [11].
This study utilizes MR to investigate the causal impact of atopy and allergies on AA, leveraging data from genome-wide association studies (GWAS) [12]. Our objective is to explore the role of type II inflammatory responses in AA, identify susceptible populations, and inform personalized treatment strategies for AA.
Materials and methods
MR design
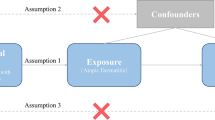
Our study adopted a two-sample MR approach to investigate the causal effects of atopic and allergic conditions—including hay fever, eczema, asthma, and allergies to cats, dust, and pollen—on AA incidence. We used single nucleotide polymorphisms (SNPs) from GWAS summary data as instrumental variables. These SNPs, reflecting the random allocation of genetics, help address confounders like age and sex [13]. Our MR analysis was grounded on three assumptions: the genetic instruments strongly predict the exposure, are free from confounders, and influence the outcome solely through the exposure [14].
Exposure GWAS data
We sourced atopy and allergy GWAS data from the Social Science Genetic Association Consortium (SSGAC) [15], including data from the UK Biobank and 23andMe but excluding FinnGen. We opted for multi-trait GWAS given its superior predictive power over its single-trait counterpart [16]. We focused on six phenotypes: hay fever, eczema, asthma, and allergies to cats, dust mites, and pollen. The effective sample sizes ranged from 254,386 to 1,214,626, with all participants being of European descent. Diagnosis criteria were based on ICD-10 or self-reported surveys [15] (Table 1).
Outcome GWAS data
AA data came from the latest FinnGen database release (DF9—as of May 11, 2023) [17], encompassing 682 AA patients and 361,140 controls of European ancestry (Table 1). Diagnoses adhered to ICD revisions 8 through 10. We noted an average patient age of 42.12 years, with a slight age variance between genders. To mitigate population heterogeneity bias, we only used SNPs from European ancestry GWAS data.
Instrumental variable selection
Our SNP selection involved multiple rigorous steps. Starting with genome-wide significant SNPs (p < 5 × 10–8), we excluded those in linkage disequilibrium (R2 < 0.001, window size = 10,000 kb) [18] or additionally, weak SNPs (F-statistic < 10) [19]. Post-harmonization, we further eliminated palindromic and ambiguous SNPs (those with a minor allele frequency (MAF) > 0.01) [20]. The MR-Pleiotropy Residual Sum and Outlier (MR-PRESSO) analysis [21], helped re-evaluate and exclude SNPs potentially influenced by pleiotropy, ensuring a robust set of instrumental variables for each atopic phenotype.
MR analysis
Addressing pleiotropy concerns, we utilized five MR methods: inverse variance weighted (IVW), MR-Egger regression, weighted median, maximum likelihood ratio, and weighted mode. And the IVW method, our primary analysis technique. Each method offers unique strengths and mitigates specific biases, ensuring a comprehensive examination of the causal effects [22, 23].
Sensitivity analysis
To ensure the validity of our primary results, we performed sensitivity analyses including heterogeneity tests (Cochran’s Q tests and MR-PRESSO global tests) and horizontal pleiotropy tests (MR-Egger intercept tests and leave-one-out analyses). These analyses helped identify sensitivity and the influence of individual SNPs on the causal estimate, enhancing the reliability of our findings [24].
Statistical analysis
Statistical significance was set at p < 0.05. We expressed MR estimates as odds ratios (ORs) with 95% confidence intervals (CIs), indicating the relative risk of AA associated with each atopic phenotype. Analyses were conducted using ‘Two Sample MR’ (version 0.5.7) and ‘MRPRESSO’ (version 1.0) packages in R (version 4.3.1), ensuring rigorous assessment of the MR estimates and identification of pleiotropic outliers [25].
Results
MR estimates
The IVW method, our primary analysis technique, indicated a significant increase in AA risk associated with hay fever (OR = 1.88, 95% CI 1.10–3.20; p = 0.02), eczema (OR = 6.16, 95% CI 2.09–18.13; p < 0.001), asthma (OR = 2.04, 95% CI 1.12–3.73; p = 0.02), pollen allergy (OR = 1.39, 95% CI 1.16–1.68; p < 0.001), dust allergy (OR = 1.25, 95% CI 1.04–1.49; p = 0.02), and cat allergy (OR = 1.24, 95% CI 1.03–1.50; p = 0.02) (Fig. 1). The corresponding scatter plot was shown in Fig. 2, and all analysis methods showed consistent directionality. These associations were corroborated by the maximum likelihood (ML) model, emphasizing the notable link between these conditions and AA. While the weighted median approach found no significant link for hay fever, it did confirm the connection with the other conditions, aligning with the general trend observed in the IVW model. The MR-Egger and weighted mode methods further identified these six conditions as risk factors for AA, although their statistical significance was not as pronounced.
MR estimates for the association between atopy, allergies and AA. “ → ” indicates exceeding the marked range. “Dust allergy” means allergy to dust mites. “*” means p-value is less than 0.05. “**” means p-value is less than 0.01. “***” means p-value is less than 0.001. MR Mendelian randomization, OR odds ratio, CI confidence interval, AA alopecia areata, IVW inverse variance weighted
Sensitivity analyses
The MR-Egger intercept did not indicate the presence of horizontal pleiotropy (p > 0.05 for all exposures), suggesting that our results are free from bias related to pleiotropy. Despite the detection of heterogeneity in eczema exposure (p < 0.05), we applied a random effects IVW model to mitigate this issue (Table 2). Despite the detection of heterogeneity in eczema exposure, the random effects IVW was utilized to perform the MR analysis and balance this heterogeneity. The absence of horizontal pleiotropy bias in this context was reaffirmed by leave-one-out analysis, and forest plots (Additional file 1: Figs. S1–S6) confirmed that no single SNP disproportionately influenced our results, further attesting to the reliability of our findings.
Discussion
This investigation harnessed extensive GWAS data and multiple MR methodologies to explore the potential causality between atopy, allergy, and AA susceptibility. Our data elucidates a significant causal association with conditions like asthma, hay fever, eczema, and allergies to pollen, dust, and cats, each contributing to an elevated AA risk. This emphasizes the importance of integrating a patient’s atopic and allergic history into AA management, highlighting the significant role of Type II inflammatory pathways.
AA's etiology is believed to involve immune events, leading to hair follicle autoantigen exposure [26]. While Th1 and Th2 pathways are implicated in AA's pathogenesis [5], observational studies, limited by their inability to confirm causality, have indicated a correlation between atopy, allergy, and AA. MR offers a more refined approach to determine causality, potentially directing treatment strategies.
Notably, clinical evidence points towards the efficacy of dupilumab, a Type II inflammatory response inhibitor, in treating AA, especially in patients with concurrent allergic conditions [27]. However, instances where dupilumab might have induced AA during atopic dermatitis treatment have been reported, though AA typically resolved with continued treatment [28]. This suggests that atopic and allergic profiles should be considered in AA management, supporting the notion that targeting Th2 pathways could benefit AA treatment, particularly in patients with a history of allergies.
Nevertheless, the study’s scope, limited to participants of European descent and a selection of common diseases and allergens, may restrict its generalizability. Additionally, the nature of MR, while offering insight into causality, does encounter challenges in fully differentiating mediation effects from pleiotropy.
In conclusion, our findings substantiate the causal link between allergies, atopy, and AA, using large-scale genetic data. Further investigations into the specific mechanisms of the Th2 inflammatory pathway in AA and its interaction with the Th1 pathway are warranted. Considering the robust connection between AA and allergies, it's imperative to factor in patients' atopic and allergic histories when devising personalized treatment plans.
Availability of data and materials
Data pertaining to atopy and allergies for the present study was procured from the Social Science Genetic Association Consortium (SSGAC), accessible via their official webpage, https://www.thessgac.com/papers/. Data concerning alopecia areata (AA) can be sourced from the FinnGen Consortium, available at their official website, https://www.finngen.fi/en.
Abbreviations
- AA:
-
Alopecia areata
- GWAS:
-
Genome-wide Association Studies
- MR:
-
Mendelian randomization
- IVW:
-
Inverse variance weighted
- SNPs:
-
Single nucleotide polymorphisms
- SSGAC:
-
Social Science Genetic Association Consortium
- ICD-8, ICD-9, and ICD-10:
-
International Classification of Diseases, encompassing the 8th, 9th, and 10th revisions
- MAF:
-
Minor allele frequency
- MR-PRESSO:
-
MR-Pleiotropy Residual Sum and Outlier
- OR:
-
Odds ratio
- CI:
-
Confidence interval
References
Zhou C, Li X, Wang C, Zhang J. Alopecia areata: an update on etiopathogenesis, diagnosis, and management. Clin Rev Allergy Immunol. 2021;61:403–23.
Sy N, Mastacouris N, Strunk A, Garg A. Overall and racial and ethnic subgroup prevalences of alopecia areata, alopecia totalis, and alopecia universalis. JAMA Dermatol. 2023;159:419–23.
Lee S, Lee H, Lee CH, Lee W-S. Comorbidities in alopecia areata: a systematic review and meta-analysis. J Am Acad Dermatol. 2019;80:466-477.e16.
Okhovat J-P, Marks DH, Manatis-Lornell A, Hagigeorges D, Locascio JJ, Senna MM. Association between alopecia areata, anxiety, and depression: a systematic review and meta-analysis. J Am Acad Dermatol. 2023;88:1040–50.
Suárez-Fariñas M, Ungar B, Noda S, Shroff A, Mansouri Y, Fuentes-Duculan J, et al. Alopecia areata profiling shows TH1, TH2, and IL-23 cytokine activation without parallel TH17/TH22 skewing. J Allergy Clin Immunol. 2015;136:1277–87.
Conic RZ, Miller R, Piliang M, Bergfeld W, Atanaskova MN. Comorbidities in patients with alopecia areata. J Am Acad Dermatol. 2017;76:755–7.
Barahmani N, Schabath MB, Duvic M, National Alopecia Areata Registry. History of atopy or autoimmunity increases risk of alopecia areata. J Am Acad Dermatol. 2009;61:581–91.
Zhang X, McElwee KJ. Allergy promotes alopecia areata in a subset of patients. Exp Dermatol. 2020;29:239–42.
Hernán MA. Methods of public health research—strengthening causal inference from observational data. N Engl J Med. 2021;385:1345–8.
Skrivankova VW, Richmond RC, Woolf BAR, Davies NM, Swanson SA, VanderWeele TJ, et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ. 2021;375: n2233.
Hu X, Zhao J, Lin Z, Wang Y, Peng H, Zhao H, et al. Mendelian randomization for causal inference accounting for pleiotropy and sample structure using genome-wide summary statistics. Proc Natl Acad Sci USA. 2022;119: e2106858119.
Sanjuan MA, Sagar D, Kolbeck R. Role of IgE in autoimmunity. J Allergy Clin Immunol. 2016;137:1651–61.
Burgess S, O’Donnell CJ, Gill D. Expressing results from a Mendelian randomization analysis: separating results from inferences. JAMA Cardiol. 2021;6:7–8.
Davies NM, Holmes MV, Davey SG. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362: k601.
Becker J, Burik CAP, Goldman G, Wang N, Jayashankar H, Bennett M, et al. Resource profile and user guide of the polygenic index repository. Nat Hum Behav. 2021;5:1744–58.
Foley CN, Staley JR, Breen PG, Sun BB, Kirk PDW, Burgess S, et al. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Nat Commun. 2021;12:764.
Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613:508–18.
Sadik A, Dardani C, Pagoni P, Havdahl A, Stergiakouli E, iPSYCH Autism Spectrum Disorder Working Group, et al. Parental inflammatory bowel disease and autism in children. Nat Med. 2022;28:1406–11.
Luo S, Au Yeung SL, Zhao JV, Burgess S, Schooling CM. Association of genetically predicted testosterone with thromboembolism, heart failure, and myocardial infarction: Mendelian randomisation study in UK Biobank. BMJ. 2019;364: l476.
Li P, Wang H, Guo L, Gou X, Chen G, Lin D, et al. Association between gut microbiota and preeclampsia-eclampsia: a two-sample Mendelian randomization study. BMC Med. 2022;20:443.
Chen X, Hong X, Gao W, Luo S, Cai J, Liu G, et al. Causal relationship between physical activity, leisure sedentary behaviors and COVID-19 risk: a Mendelian randomization study. J Transl Med. 2022;20:216.
Kawai VK, Shi M, Feng Q, Chung CP, Liu G, Cox NJ, et al. Pleiotropy in the genetic predisposition to rheumatoid arthritis: a phenome-wide association study and inverse variance-weighted meta-analysis. Arthritis Rheumatol. 2020;72:1483–92.
Disney-Hogg L, Cornish AJ, Sud A, Law PJ, Kinnersley B, Jacobs DI, et al. Impact of atopy on risk of glioma: a Mendelian randomisation study. BMC Med. 2018;16:42.
Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–8.
Long Y, Tang L, Zhou Y, Zhao S, Zhu H. Causal relationship between gut microbiota and cancers: a two-sample Mendelian randomisation study. BMC Med. 2023;21:66.
Xie B, Sun J, Song X. Hair follicle melanocytes initiate autoimmunity in alopecia areata: a trigger point. Clin Rev Allergy Immunol. 2022;63:417–30.
Guttman-Yassky E, Renert-Yuval Y, Bares J, Chima M, Hawkes JE, Gilleaudeau P, et al. Phase 2a randomized clinical trial of dupilumab (anti-IL-4Rα) for alopecia areata patients. Allergy. 2022;77:897–906.
Marks DH, Mesinkovska N, Senna MM. Cause or cure? Review of dupilumab and alopecia areata. J Am Acad Dermatol. 2023;88:651–3.
Acknowledgements
Our team extends its heartfelt acknowledgment to both the FinnGen Consortium and the Social Science Genetic Association Consortium (SSGAC) for their generosity in granting access to the summary data.
Funding
This research endeavor was financially supported by multiple Grants: The Zhejiang Provincial Natural Science Foundation in China (Grant No. LY23H110001); The Major Science and Technology Project of Zhejiang Province in conjunction with the State Administration of Traditional Chinese Medicine (Grant No. GZY-ZJ-KJ-2023035); and the Major Health Science and Technology Project of Hangzhou (Grant No. Z20220040).
Author information
Authors and Affiliations
Contributions
Significant contributions to the analysis and preparation of the manuscript were made by Wen Xu, Hongyan Zhang, Sheng Wan, Bo Xie, and Xiuzu Song. The data evaluations were performed, and the manuscript was composed by Wen Xu, Sheng Wan, and Bo Xie. Valuable discussions enriching the evaluation were offered by Hongyan Zhang and Xiuzu Song. The final version of the manuscript has been reviewed and approved by each author for publication.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The dataset used in this study was acquired from a publicly accessible research database. Prior to inclusion in this database, all data were collected from studies that had obtained informed consent from participants and received ethics approval from appropriate institutional review boards (IRBs). The current study, being a secondary analysis of de-identified data, did not require additional ethical approval.
Consent for publication
Not applicable. This study involves secondary analysis of de-identified data from a publicly accessible research database, where consent for use and publication was obtained in the original studies.
Competing interests
The authors hereby affirm that no conflicts of interest exist in relation to this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
: Figure S1. Leave-one-out analysis (a) and forest plot (b) for hay fever on AA risk. AA alopecia areata, MR Mendelian randomization. Figure S2. Leave-one-out analysis (a) and forest plot (b) for eczema on AA risk. AA alopecia areata, MR Mendelian randomization. Figure S3. Leave-one-out analysis (a) and forest plot (b) for asthma on AA risk. AA alopecia areata, MR Mendelian randomization. Figure S4. Leave-one-out analysis (a) and forest plot (b) for pollen allergy on AA risk. AA alopecia areata, MR Mendelian randomization. Figure S5. Leave-one-out analysis (a) and forest plot (b) for dust mite allergy on AA risk. AA alopecia areata, MR Mendelian randomization. Figure S6. Leave-one-out analysis (a) and forest plot (b) for cat allergy on AA risk. AA alopecia areata, MR Mendelian randomization.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xu, W., Zhang, H., Wan, S. et al. Genetic links between atopy, allergy, and alopecia areata: insights from a Mendelian randomization study. Allergy Asthma Clin Immunol 20, 32 (2024). https://doi.org/10.1186/s13223-024-00892-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13223-024-00892-w