Abstract
Background
The relationship between metformin use and prostate cancer (PCa) risk has yet to be clear despite more than a decade of debate on this topic. Hence, we aimed to investigate the causal role of metformin in reducing PCa risk through an up-to-date comprehensive genome-wide analysis.
Methods
We employed validated instrument variables of metformin use derived from a prior high-quality study, including five potential targets (AMPK, GCG, GDF15, MCI and MG3). Mendelian randomization (MR) analysis was performed to harmonize genetically predicted metformin use and PCa phenotypes. PCa phenotypes were from two large genome-wide association studies (GWAS), the Prostate Cancer Association Group to Investigate Cancer-Associated Alterations in the Genome (PRACTICAL) and the FinnGen cohort. Seven methods were applied to generate MR results: the inverse variance weighted (IVW), IVW with multiplicative random effects, MR-Egger, MR-Egger (bootstrap), weighted median, simple mode and weighted mode. Strict sensitivity analysis was conducted to satisfy core assumptions of MR design.
Results
We enrolled 32 significant single nucleotide polymorphisms (SNPs) that involved with metformin use. Nearly all targets yielded insignificant primary results (IVW with multiplicative random effects), except that AMPK target posed a positive effect on PCa risk from FinnGen cohort [odds ratio (OR): 6.09, 95% confidence interval (CI): 1.10-33.53, P value: 0.038]. The general effect of metformin use, comprising all 5 targets, also yielded negative results (random-effect meta-analysis with OR: 1.09, 95% CI: 0.76–1.58, P value: 0.637 for PRACTICAL; OR: 2.55, 95% CI: 0.58–11.16, P value: 0.215 for FinnGen). None of the sensitivity analyses provided support for a causal association between metformin use and PCa risk.
Conclusion
This up-to-date study did not support the protective role of metformin in reducing PCa risk, considering each target, overall effect, and sensitivity analysis. It is imperative to reflect on the presumed “almighty medicine” and ongoing phase III trials are anticipated to assess the anti-neoplasm effect of metformin.
Similar content being viewed by others
Introduction
Metformin is a widely used pharmacological agent for the management of type 2 diabetes mellitus, which has attracted growing attention due to its potential anti-tumorigenic characters [1]. Although the contentious conclusions have left this matter unresolved, the efficacy of metformin in diverse cancer types has been elucidated in an escalating number of clinical investigations [2,3,4,5,6,7,8,9,10,11,12,13,14].
The association between metformin use and prostate cancer (PCa) risk is of great interest to urologists. Previous studies indicated the protective effect of metformin in at least a certain part of population [8, 15,16,17,18,19,20,21,22,23], while some others contradicted the results [24,25,26,27,28,29,30,31,32,33]. As observational studies are rife with numerous disadvantages [34], a genetic tool is appropriate to investigate the causal association between metformin and PCa. Mendelian randomization (MR) analysis is an effective method to solve such issues [35,36,37]. But the frustrating reality is that only one study focused on this topic [38]. The study utilized the target of adenosine 5’-monophosphate-activated protein kinase (AMPK) to proxy metformin effect on HbA1c reduction, which was inaccurate and biased due to its complicated effect [38].
Recently, a comprehensive research summarized the distinct drug target impacts of metformin through genome-wide analysis [39]. As a result, we are able to exploit such instruments to explore the effect of metformin on PCa, which is the aim of this study.
Methods
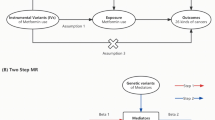
The objective of this study is to test the causal effect of metformin use on PCa risk through a comprehensive MR analysis. Figure 1 showed the study flowchart.
The study flowchart. AMPK: adenosine 5’-monophosphate-activated protein kinase; GCG: glucagon; GDF15: growth differentiation factor 15; MCI: mitochondrial complex I; MG3: mitochondrial glycerol 3; PRACTICAL: Prostate Cancer Association Group to Investigate Cancer-Associated Alterations in the Genome
Metformin proxied instrument variables
We utilized the certified variants of metformin use from a previous high-quality study [39]. Briefly, the authors determined 5 targets (AMPK, GCG, GDF15, MCI, MG3) with 32 variants through a series of validation. They conducted a thorough literature review to identify the drug targets of metformin (AMPK, GCG, GDF15, MCI, MG3). The five metformin-related targets were then mapped to the related genes through the ChEMBL database [40, 41]. Furthermore, the related genes were mapped to the related genetic variants based on recent comprehensive data [42,43,44,45,46,47,48]. The related genetic variants were then associated with the glycemic trait HbA1c from 344,182 UK Biobank participants (served as the exposure variable, Fig. 1) and we provided the summary-level data in Table S1.
Prostate cancer outcomes
We selected PCa outcomes from the Prostate Cancer Association Group to Investigate Cancer-Associated Alterations in the Genome (PRACTICAL) and the FinnGen cohort. PRACTICAL is a consortium to investigate the genetic susceptibility of PCa, consisting of 79,148 cases and 61,106 controls [49]. As for FinnGen cohort, we extracted release 5 version data, consisting of 6,311 cases and 88,902 controls [50]. Only European ancestry was included and overlap was avoided between the exposure and outcome variables. Details were provided in Table S2.
Statistical analysis
All the analysis was completed in R (version 4.2.0). TwoSampleMR and ieugwasr were the main R packages. Two-sample MR analysis was the primary results in our study. F-statistic, calculated as beta2/se2, was implemented to test the power of instrument variables, with F-statistic > 10 thought as strong variants [51]. When conducting MR analysis, proxy with r2 > 0.8 was considered if a single nucleotide polymorphism (SNP) was not matched between the exposure and outcome variables. We enrolled seven methods to generate MR results: the inverse variance weighted (IVW), IVW with multiplicative random effects, MR-Egger, MR-Egger (bootstrap), weighted median, simple mode and weighted mode. The IVW with multiplicative random effects method was considered as our primary result. If there was only one SNP in the exposure and outcome variables, Wald ratio was calculated as the primary result. We would report MR results based on each target mentioned above and then give the whole results of the five targets. Additionally, heterogeneity and pleiotropy tests were conducted. All results were reported as odds ratio (OR) or beta value with 95% confidence interval (95% CI). On the other hand, Steiger tests were conducted to certify whether the assumption that exposure caused outcome was valid. A reverse MR analysis was also performed to examine if the reverse causality existed.
Three assumptions should be met during MR analysis (Fig. 1). First, all SNPs were associated with the exposure variable. Second, any SNP associated with any potential confounder should be excluded. Third, SNPs should not be associated with the outcome variable directly. To satisfy all these assumptions, we intended to perform the following sensitivity analysis. Sensitivity analysis 1 removed SNPs with F-statistic < 10. Sensitivity analysis 2 removed SNPs associated with hypertension additionally. Sensitivity analysis 3 removed SNPs associated with hypertension and dyslipidemia additionally. Sensitivity analysis 4 removed SNPs associated with hypertension, dyslipidemia and body mass index (BMI) additionally. Sensitivity analysis 5 removed SNPs associated with hypertension, dyslipidemia, BMI and any cancer outcome additionally.
Results
We enrolled 32 significant SNPs from the previous research [39], including five targets (AMPK, GCG, GDF15, MCI, MG3) that involved with metformin use. Those 32 SNPs were associated with the genome-wide association study (GWAS) of a glycemic marker, HbA1c, from UK Biobank (18,242 diabetic cases/325,940 controls). To prevent overlap bias between the exposure and outcome variables that could induce false positive rate, we selected prostate cancer GWAS from another two UK Biobank-unrelated cohorts, PRACTICAL (79,148 cases/61,106 controls) and FinnGen (6,331 cases/88,902 controls), to perform MR analysis (Fig. 1).
The MR effect of metformin targets on PCa risk
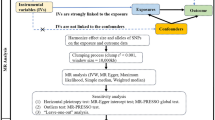
We conducted the MR analysis of metformin use effect on PCa cancer risk based on each target (Fig. 2).Nearly all targets yielded insignificant primary results (IVW with multiplicative random effects), except that AMPK target posed a positive effect on PCa risk from FinnGen cohort (OR: 6.09, 95% CI: 1.10-33.53, P value: 0.038, Table S3B). The effect of AMPK target on PCa risk from PRACTICAL was of no significance (OR: 0.87, 95% CI: 0.23–3.33, P value: 0.835, Table S3A). The other four targets, including GCG (OR: 1.33, 95% CI: 0.05–34.81, P value: 0.865 for PRACTICAL; OR: 4.67, 95% CI: 0.02-1176.74, P value: 0.585 for FinnGen), GDF15 (OR: 1.13, 95% CI: 0.16–7.78, P value: 0.901 for PRACTICAL; OR: 3.17, 95% CI: 0.01-756.47, P value: 0.680 for FinnGen), MCI (OR: 1.16, 95% CI: 0.77–1.74, P value: 0.477 for PRACTICAL; OR: 0.89, 95% CI: 0.50–1.60, P value: 0.699 for FinnGen) and MG3 (OR: 0.45, 95% CI: 0.07–2.95, P value: 0.402 for PRACTICAL; OR: 29.76, 95% CI: 0.22-4085.54, P value: 0.177 for FinnGen), all showed negative results (Fig. 2, Table S3A and Table S3B).
The MR effect of metformin use on PCa risk from PRACTICAL and FinnGen, based on each target and general effect. MR: Mendelian randomization; PCa: prostate cancer risk; PRACTICAL: Prostate Cancer Association Group to Investigate Cancer-Associated Alterations in the Genome; SNP: single nucleotide polymorphism; OR: odds ratio; CI: confidence interval; SD: standard deviation
The general effect of metformin use, comprising all 5 targets, also yielded negative results when we utilized methods of IVW with multiplicative random effects (OR: 1.12, 95% CI: 0.78–1.60, P value: 0.542 for PRACTICAL; OR: 1.09, 95% CI: 0.62–1.91, P value: 0.760 for FinnGen), fixed-effect meta-analysis (OR: 1.09, 95% CI: 0.76–1.58, P value: 0.637 for PRACTICAL; OR: 1.16, 95% CI: 0.68–2.01, P value: 0.584 for FinnGen) and random-effect meta-analysis (OR: 1.09, 95% CI: 0.76–1.58, P value: 0.637 for PRACTICAL; OR: 2.55, 95% CI: 0.58–11.16, P value: 0.215 for FinnGen; Fig. 2; Table 1, Table S3A and Table S3B). Steiger analysis showed correct causal direction from metformin use to PCa risk (Table S4A and Table S4B). In addition, the reverse MR analysis to examine if the reverse causality existed indicated no causal association between PCa and metformin use (P value of IVW with multiplicative random effects: 0.383 for PRACTICAL and 0.779 for FinnGen, Table S5A and Table S5B).
Sensitivity analysis
To meet the three core assumptions of MR analysis, we performed five sensitivity analysis mentioned in the method section. There were 23, 22, 18, 16 and 16 SNPs enrolled in sensitivity analysis 1–5 respectively. The detailed SNPs information was provided in Table S6A-E.
All the sensitivity analysis did not support a causal association between metformin use and PCa risk. The effect sizes based on the IVW with multiplicative random effects were: sensitivity analysis 1 (OR: 1.08, 95% CI: 0.73–1.60, P value: 0.698 for PRACTICAL; OR: 1.03, 95% CI: 0.63–1.70, P value: 0.901 for FinnGen), sensitivity analysis 2 (OR: 1.11, 95% CI: 0.74–1.67, P value: 0.614 for PRACTICAL; OR: 1.00, 95% CI: 0.59–1.67, P value: 0.991 for FinnGen), sensitivity analysis 3 (OR: 1.47, 95% CI: 0.84–2.56, P value: 0.174 for PRACTICAL; OR: 1.24, 95% CI: 0.60–2.58, P value: 0.567 for FinnGen), sensitivity analysis 4 (OR: 1.10, 95% CI: 0.73–1.67, P value: 0.649 for PRACTICAL; OR: 0.96, 95% CI: 0.49–1.87, P value: 0.905 for FinnGen) and sensitivity analysis 5 OR: 1.10, 95% CI: 0.73–1.67, P value: 0.649 for PRACTICAL; OR: 0.96, 95% CI: 0.49–1.87, P value: 0.905 for FinnGen; Fig. 3, Table S7A and Table S7B).
The MR effect of metformin use on PCa risk from PRACTICAL and FinnGen, based on sensitivity analysis. MR: Mendelian randomization; PCa: prostate cancer risk; PRACTICAL: Prostate Cancer Association Group to Investigate Cancer-Associated Alterations in the Genome; SNP: single nucleotide polymorphism; OR: odds ratio; CI: confidence interval; SD: standard deviation
Discussion
In this study, we confirmed that no protective effect of metformin use on PCa risk. The association between metformin use and PCa risk reduction has been debated for over fifteen years [52, 53]. Most results were derived from in vitro or observational studies, as randomized controlled trials were impractical and the follow-up time was too long to gain enough events. Contradictory results were expected due to potential known or unknown confounders. Therefore, we conducted such a genetic epidemiological study to examine the hypothesis that whether metformin use causally reduced PCa risk, which had the advantage of test causality if all assumptions were satisfied. Unfortunately, we were unable to validate the preventive effect of metformin on PCa risk based on MR analysis of each target, all five targets and all the sensitivity analysis. Maybe we should re-examine the relationship between metformin use and PCa risk.
An early study has explored the impact of genetic-predicted metformin use on PCa risk [38]. However, the research just utilized the AMPK-proxied HbA1c reduction as a substitute of metformin, which was obsolete and far from comprehensive, as metformin exerted its effect not only through AMPK pathway activation. Also, the way it included SNPs in the MR analysis was with inferior priority to the recent study [39]. Au Yeung and colleagues found no causal association between metformin use and PCa risk in their conclusion [38], which was similar with our results. From this point of view, the utilization of metformin for PCa prevention should be cautious, at least metformin might not reduce PCa risk in a blood sugar dependent way.
Interestingly, one randomized controlled trial regarding the protective effect of metformin on anthropometric and metabolic complications in patients receiving radical radiotherapy and androgen deprivation therapy was completed and reported its preliminary results [20]. This phase II trial discovered that metformin did not attenuate the complication rate, which was frustrating. Nevertheless, metformin is currently under investigation in the further phase 3 trial to evaluate its potential anti-tumor effects. As far as we know, this is the first randomized controlled trial to investigate only the effect of metformin in prostate disease, although the aim is to evaluate its preventive impact in decreasing complication rate. But it did provide some information. Maybe metformin actually does not have the potency as we expected in antagonizing PCa. We ought to be vigilant when considering the effect of such an “almighty medicine”. We are also looking forward to the further results of the phase 3 trial [20].
The study tried to solve the long discussed issue. We utilized the design of MR to avoid confounder bias and and intended to establish a causal association. We incorporated an up-to-dated comprehensive genetic proxy of metformin into our study to explore its role in PCa risk. Apart from the above advantages, we divided the metformin effect into several targets and calculated the specific effect of each target. Nearly all targets yielded no significant results, which indeed confirmed no causal relationship between metformin use and PCa risk. Moreover, all the three key assumptions of MR analysis were met and we conducted several sensitivity analysis to validate our results. We believe our research could offer information to those urologists who are interested in medical treatment of PCa.
Some limitations should be admitted. First, we just enrolled European ancestry in this study resulting from a lack of summary statistics from other ancestries. Additionally, there might be some other targets through which metformin functioned, but we have not discovered till now. Notwithstanding, we summarized the current evidence of metformin effect on PCa. The results might alter as further targets of metformin are found.
To conclude, the study did not find a reliable causality between metformin use and PCa risk, based on each target, general effect or sensitivity analysis. We should reflect on the “almighty medicine” and doubt its protective effect of PCa risk. Perhaps metformin influences PCa through other rather than glycemic pathway. The ongoing phase III trial is anticipated as it would assess the anti-neoplasm effect.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Mallik R, Chowdhury TA. Metformin in cancer. Diabetes Res Clin Pract. 2018;143:409–19.
Lai SW, Liao KF, Chen PC, Tsai PY, Hsieh DP, Chen CC. Antidiabetes drugs correlate with decreased risk of lung cancer: a population-based observation in Taiwan. Clin Lung Cancer. 2012;13(2):143–8.
Bragagnoli AC, Araujo RLC, Ferraz MW, Dos Santos LV, Abdalla KC, Comar F, Santos FA, Oliveira MA, Carvalheira JBC. Cárcano FM, Da Silveira Nogueira Lima JP. Metformin plus Lrinotecan in patients with refractory colorectal cancer: a phase 2 clinical trial. Br J Cancer. 2021;124(6):1072–8.
Barakat HE, Hussein RRS, Elberry AA, Zaki MA, Ramadan ME. The impact of metformin use on the outcomes of locally advanced breast cancer patients receiving neoadjuvant chemotherapy: an open-labelled randomized controlled trial. Sci Rep. 2022;12(1):7656.
Krebs M, Kotlyar MJ, Fahl J, Janaki Raman S, Röhrig F, Marquardt A, Kübler H, Kneitz B, Schulze A, Kalogirou C. Metformin regulates the miR-205/VEGFA Axis in Renal Cell Carcinoma cells - exploring a clinical synergism with tyrosine kinase inhibitors. Urol Int. 2023 Nov 30.
Kuusiniemi E, Karihtala P, Puistola U, Ahtikoski A, Urpilainen E. Oxidative stress-regulating enzymes and endometrial Cancer survival in relation to Metformin Intake in Diabetic patients. Anticancer Res. 2023;43(12):5545–54.
Khajeh E, Aminizadeh E, Moghadam AD, Ramouz A, Klotz R, Golriz M, Merle U, Springfeld C, Chang DH, Longerich T, Büchler MW, Mehrabi A. Association of perioperative use of statins, metformin, and aspirin with recurrence after curative liver resection in patients with hepatocellular carcinoma: a propensity score matching analysis. Cancer Med. 2023;12(19):19548–59.
Papachristodoulou A, Heidegger I, Virk RK, Di Bernardo M, Kim JY, Laplaca C, Picech F, Schäfer G, De Castro GJ, Hibshoosh H, Loda M, Klocker H, Rubin MA, Zheng T, Benson MC, McKiernan JM, Dutta A, Abate-Shen C. Metformin overcomes the consequences of NKX3.1 loss to suppress prostate Cancer progression. Eur Urol. 2023 Aug 31:S0302-2838(23)03016-6.
Löfling LL, Støer NC, Andreassen BK, Ursin G, Botteri E. Low-dose aspirin, statins, and metformin and survival in patients with breast cancers: a Norwegian population-based cohort study. Breast Cancer Res. 2023;25(1):101.
Joo JH, Zhang HS, Chun J, Park EC, Park S. Association of Metformin Treatment with Risk for Death in Diabetic patients with concomitant gastric Cancer. Cancers (Basel). 2023;15(16):4134.
Kemnade JO, Florez M, Sabichi A, Zhang J, Jhaveri P, Chen G, Chen A, Miller-Chism C, Shaun B, Hilsenbeck SG, Hernandez DJ, Skinner HD, Sandulache VC. Phase I / II trial of metformin as a chemo-radiosensitizer in a head and neck cancer patient population. Oral Oncol. 2023;145:106536.
Goodwin PJ, Chen BE, Gelmon KA, Whelan TJ, Ennis M, Lemieux J, Ligibel JA, Hershman DL, Mayer IA, Hobday TJ, Bliss JM, Rastogi P, Rabaglio-Poretti M, Mukherjee SD, Mackey JR, Abramson VG, Oja C, Wesolowski R, Thompson AM, Rea DW, Stos PM, Shepherd LE, Stambolic V, Parulekar WR. Effect of Metformin vs Placebo on Invasive Disease-Free Survival in patients with breast Cancer: the MA.32 Randomized Clinical Trial. JAMA. 2022;327(20):1963–73.
Huang J, Tong Y, Hong J, Huang O, Wu J, He J, Chen W, Li Y, Chen X, Shen K. Neoadjuvant docetaxel, epirubicin, and cyclophosphamide with or without metformin in breast cancer patients with metabolic abnormality: results from the randomized phase II NeoMET trial. Breast Cancer Res Treat. 2023;197(3):525–33.
Essa NM, Salem HF, Elgendy MO, Gabr A, Omran MM, Hassan NA, Tashkandi HM, Harakeh S, Boshra MS. Efficacy of Metformin as Adjuvant Therapy in metastatic breast Cancer Treatment. J Clin Med. 2022;11(19):5505.
Al Shareef Z, Al-Shahrabi R, Saheb Sharif-Askari F, AlDhmanie A, Alshamsi Y, Zarooni AA, Mahmoud RA, Soliman SSM, Halwani R, Bendardaf R. Incidence and risk factors of prostate cancer among the Northern and Eastern parts of the United Arab Emirates population. Prostate. 2024;84(2):185–92.
Khan S, Chang SH, Seyerle AA, Wang M, Hicks V, Drake BF. Post-diagnostic metformin and statin use and risk of biochemical recurrence in veterans diagnosed with prostate cancer. Prostate. 2023;83(12):1150–7.
Jo JK, Song HK, Heo Y, Kim MJ, Kim YJ. Risk analysis of metformin use in prostate cancer: a national population-based study. Aging Male. 2023;26(1):2156497.
Lee YHA, Hui JMH, Chung CT, Liu K, Dee EC, Ng K, Tse G, Chan JSK, Ng CF. Metformin use and hospital attendance-related resources utilization among diabetic patients with prostate cancer on androgen deprivation therapy: a population-based cohort study. Cancer Med. 2023;12(8):9128–32.
Lee YHA, Hui JMH, Chan JSK, Liu K, Dee EC, Ng K, Tang P, Tse G, Ng CF. Metformin use and mortality in Asian, diabetic patients with prostate cancer on androgen deprivation therapy: a population-based study. Prostate. 2023;83(1):119–27.
Usmani N, Ghosh S, Sanghera KP, Ong AD, Koul R, Dubey A, Ahmed S, Quon H, Yee D, Parliament M, Sivananthan G, Hunter W, Danielson B, Rowe L, McDonald M, Kim JO. Metformin for Prevention of Anthropometric and metabolic complications of androgen deprivation therapy in prostate Cancer patients receiving Radical Radiotherapy: a phase II randomized controlled trial. Int J Radiat Oncol Biol Phys. 2023;115(2):317–26.
Lopez DS, Malagaris I, Polychronopoulou E, Tsilidis KK, Milani SA, Kristen Peek M, Villasante-Tezanos A, Alzweri L, Baillargeon J, Kuo YF, Canfield S. Metformin and testosterone replacement therapy inversely associated with hormone-associated cancers (prostate, colorectal and male breast cancers) among older White and black men. Clin Endocrinol (Oxf). 2022;97(6):792–803.
Lee YHA, Zhou J, Hui JMH, Liu X, Lee TTL, Hui K, Chan JSK, Wai AKC, Wong WT, Liu T, Ng K, Lee S, Dee EC, Zhang Q, Tse G. Risk of new-onset prostate Cancer for Metformin Versus Sulfonylurea Use in type 2 diabetes Mellitus: a propensity score-matched study. J Natl Compr Canc Netw. 2022;20(6):674–e68215.
Nair-Shalliker V, Bang A, Egger S, Yu XQ, Chiam K, Steinberg J, Patel MI, Banks E, O’Connell DL, Armstrong BK, Smith DP. Family history, obesity, urological factors and diabetic medications and their associations with risk of prostate cancer diagnosis in a large prospective study. Br J Cancer. 2022;127(4):735–46.
Najafi F, Rajati F, Sarokhani D, Bavandpour M, Moradinazar M. The relationship between Metformin Consumption and Cancer Risk: an updated umbrella review of systematic reviews and Meta-analyses. Int J Prev Med. 2023;14:90.
Dickerman BA, García-Albéniz X, Logan RW, Denaxas S, Hernán MA. Evaluating metformin strategies for Cancer Prevention: a Target Trial Emulation using Electronic Health records. Epidemiology. 2023;34(5):690–9.
Freedman LS, Agay N, Farmer R, Murad H, Olmer L, Dankner R. Metformin Treatment among men with Diabetes and the risk of prostate Cancer: a Population-based historical cohort study. Am J Epidemiol. 2022;191(4):626–35.
Chen CB, Eskin M, Eurich DT, Majumdar SR, Johnson JA, Metformin. Asian ethnicity and risk of prostate cancer in type 2 diabetes: a systematic review and meta-analysis. BMC Cancer. 2018;18(1):65.
Wang Y, Liu X, Yan P, Tang J, Chen T, Sun Y, Zhou W, Bi Y, Zhang ZJ. Effect of metformin on the risk of prostate cancer in patients with type 2 diabetes by considering different confounding factors: a meta-analysis of observational studies. Eur J Cancer Prev. 2020;29(1):42–52.
He K, Hu H, Ye S, Wang H, Cui R, Yi L. The effect of metformin therapy on incidence and prognosis in prostate cancer: a systematic review and meta-analysis. Sci Rep. 2019;9(1):2218.
Ruiter R, Visser LE, van Herk-Sukel MP, Coebergh JW, Haak HR, Geelhoed-Duijvestijn PH, Straus SM, Herings RM, Stricker BH. Lower risk of cancer in patients on metformin in comparison with those on sulfonylurea derivatives: results from a large population-based follow-up study. Diabetes Care. 2012;35(1):119–24.
Margel D, Urbach DR, Lipscombe LL, Bell CM, Kulkarni G, Austin PC, Fleshner N. Metformin use and all-cause and prostate cancer-specific mortality among men with diabetes. J Clin Oncol. 2013;31(25):3069–75.
van Staa TP, Patel D, Gallagher AM, de Bruin ML. Glucose-lowering agents and the patterns of risk for cancer: a study with the General Practice Research Database and secondary care data. Diabetologia. 2012;55(3):654–65.
Kuo YJ, Sung FC, Hsieh PF, Chang HP, Wu KL, Wu HC. Metformin reduces prostate cancer risk among men with benign prostatic hyperplasia: a nationwide population-based cohort study. Cancer Med. 2019;8(5):2514–23.
Suissa S, Azoulay L. Metformin and the risk of cancer: time-related biases in observational studies. Diabetes Care. 2012;35(12):2665–73.
Lin L, Tang Y, Ning K, Li X, Hu X. Investigating the causal associations between metabolic biomarkers and the risk of kidney cancer. Commun Biol. 2024;7(1):398. https://doi.org/10.1038/s42003-024-06114-8.
Lin L, Ning K, Xiang L, Peng L, Li X. SGLT2 inhibition and three urological cancers: Up-to-date results. Diabetes/Met Res Rev. 2024;40(3):e3797. https://doi.org/10.1002/dmrr.v40.3.
Lin L, Ma Y, Li Z, Liu L, Hu Q, Zhou L. Genetic susceptibility of urolithiasis: comprehensive results from genome-wide analysis. World J Urol. 2024;42(1):230. https://doi.org/10.1007/s00345-024-04937-y.
Au Yeung SL, Schooling CM. Impact of glycemic traits, type 2 diabetes and metformin use on breast and prostate cancer risk: a mendelian randomization study. BMJ Open Diabetes Res Care. 2019;7(1):e000872.
Zheng J, Xu M, Walker V, Yuan J, Korologou-Linden R, Robinson J, Huang P, Burgess S, Au Yeung SL, Luo S, Holmes MV, Davey Smith G, Ning G, Wang W, Gaunt TR, Bi Y. Evaluating the efficacy and mechanism of metformin targets on reducing Alzheimer’s disease risk in the general population: a mendelian randomisation study. Diabetologia. 2022;65(10):1664–75.
Mendez D, Gaulton A, Bento AP, Chambers J, De Veij M, Félix E, Magariños MP, Mosquera JF, Mutowo P, Nowotka M, Gordillo-Marañón M, Hunter F, Junco L, Mugumbate G, Rodriguez-Lopez M, Atkinson F, Bosc N, Radoux CJ, Segura-Cabrera A, Hersey A, Leach AR. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019;47(D1):D930–40.
Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(Database issue):D668–72.
Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran P, Oliver-Williams C, Kamat MA, Prins BP, Wilcox SK, Zimmerman ES, Chi A, Bansal N, Spain SL, Wood AM, Morrell NW, Bradley JR, Janjic N, Roberts DJ, Ouwehand WH, Todd JA, Soranzo N, Suhre K, Paul DS, Fox CS, Plenge RM, Danesh J, Runz H, Butterworth AS. Genomic atlas of the human plasma proteome. Nature. 2018;558(7708):73–9.
Folkersen L, Fauman E, Sabater-Lleal M, Strawbridge RJ, Frånberg M, Sennblad B, Baldassarre D, Veglia F, Humphries SE, Rauramaa R, de Faire U, Smit AJ, Giral P, Kurl S, Mannarino E, Enroth S, Johansson Å, Enroth SB, Gustafsson S, Lind L, Lindgren C, Morris AP, Giedraitis V, Silveira A, Franco-Cereceda A, Tremoli E, Gyllensten U, Ingelsson E, Brunak S, Eriksson P, Ziemek D, Hamsten A, Mälarstig A. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet. 2017;13(4):e1006706.
Suhre K, Arnold M, Bhagwat AM, Cotton RJ, Engelke R, Raffler J, Sarwath H, Thareja G, Wahl A, DeLisle RK, Gold L, Pezer M, Lauc G, El-Din Selim MA, Mook-Kanamori DO, Al-Dous EK, Mohamoud YA, Malek J, Strauch K, Grallert H, Peters A, Kastenmüller G, Gieger C, Graumann J. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357.
Yao C, Chen G, Song C, Keefe J, Mendelson M, Huan T, Sun BB, Laser A, Maranville JC, Wu H, Ho JE, Courchesne P, Lyass A, Larson MG, Gieger C, Graumann J, Johnson AD, Danesh J, Runz H, Hwang SJ, Liu C, Butterworth AS, Suhre K, Levy D. Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat Commun. 2018;9(1):3268.
Emilsson V, Ilkov M, Lamb JR, Finkel N, Gudmundsson EF, Pitts R, Hoover H, Gudmundsdottir V, Horman SR, Aspelund T, Shu L, Trifonov V, Sigurdsson S, Manolescu A, Zhu J, Olafsson Ö, Jakobsdottir J, Lesley SA, To J, Zhang J, Harris TB, Launer LJ, Zhang B, Eiriksdottir G, Yang X, Orth AP, Jennings LL, Gudnason V. Co-regulatory networks of human serum proteins link genetics to disease. Science. 2018;361(6404):769–73.
Võsa U, Claringbould A, Westra HJ, Bonder MJ, Deelen P, Zeng B, Kirsten H, Saha A, Kreuzhuber R, Yazar S, Brugge H, Oelen R, de Vries DH, van der Wijst MGP, Kasela S, Pervjakova N, Alves I, Favé MJ, Agbessi M, Christiansen MW, Jansen R, Seppälä I, Tong L, Teumer A, Schramm K, Hemani G, Verlouw J, Yaghootkar H, Sönmez Flitman R, Brown A, Kukushkina V, Kalnapenkis A, Rüeger S, Porcu E, Kronberg J, Kettunen J, Lee B, Zhang F, Qi T, Hernandez JA, Arindrarto W, Beutner F, BIOS Consortium; i2QTL Consortium, Dmitrieva J, Elansary M, Fairfax BP, Georges M, Heijmans BT, Hewitt AW, Kähönen M, Kim Y, Knight JC, Kovacs P, Krohn K, Li S, Loeffler M, Marigorta UM, Mei H, Momozawa Y, Müller-Nurasyid M, Nauck M, Nivard MG, Penninx BWJH, Pritchard JK, Raitakari OT, Rotzschke O, Slagboom EP, Stehouwer CDA, Stumvoll M, Sullivan P, ‘t Hoen PAC, Thiery J, Tönjes A, van Dongen J, van Iterson M, Veldink JH, Völker U, Warmerdam R, Wijmenga C, Swertz M, Andiappan A, Montgomery GW, Ripatti S, Perola M, Kutalik Z, Dermitzakis E, Bergmann S, Frayling T, van Meurs J, Prokisch H, Ahsan H, Pierce BL, Lehtimäki T, Boomsma DI, Psaty BM, Gharib SA, Awadalla P, Milani L, Ouwehand WH, Downes K, Stegle O, Battle A, Visscher PM, Yang J, Scholz M, Powell J, Gibson G, Esko T, Franke L. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021;53(9):1300–1310.
GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369(6509):1318–30.
Schumacher FR, Al Olama AA, Berndt SI, Benlloch S, Ahmed M, Saunders EJ, Dadaev T, Leongamornlert D, Anokian E, Cieza-Borrella C, Goh C, Brook MN, Sheng X, Fachal L, Dennis J, Tyrer J, Muir K, Lophatananon A, Stevens VL, Gapstur SM, Carter BD, Tangen CM, Goodman PJ, Thompson IM Jr, Batra J, Chambers S, Moya L, Clements J, Horvath L, Tilley W, Risbridger GP, Gronberg H, Aly M, Nordström T, Pharoah P, Pashayan N, Schleutker J, Tammela TLJ, Sipeky C, Auvinen A, Albanes D, Weinstein S, Wolk A, Håkansson N, West CML, Dunning AM, Burnet N, Mucci LA, Giovannucci E, Andriole GL, Cussenot O, Cancel-Tassin G, Koutros S, Beane Freeman LE, Sorensen KD, Orntoft TF, Borre M, Maehle L, Grindedal EM, Neal DE, Donovan JL, Hamdy FC, Martin RM, Travis RC, Key TJ, Hamilton RJ, Fleshner NE, Finelli A, Ingles SA, Stern MC, Rosenstein BS, Kerns SL, Ostrer H, Lu YJ, Zhang HW, Feng N, Mao X, Guo X, Wang G, Sun Z, Giles GG, Southey MC, MacInnis RJ, FitzGerald LM, Kibel AS, Drake BF, Vega A, Gómez-Caamaño A, Szulkin R, Eklund M, Kogevinas M, Llorca J, Castaño-Vinyals G, Penney KL, Stampfer M, Park JY, Sellers TA, Lin HY, Stanford JL, Cybulski C, Wokolorczyk D, Lubinski J, Ostrander EA, Geybels MS, Nordestgaard BG, Nielsen SF, Weischer M, Bisbjerg R, Røder MA, Iversen P, Brenner H, Cuk K, Holleczek B, Maier C, Luedeke M, Schnoeller T, Kim J, Logothetis CJ, John EM, Teixeira MR, Paulo P, Cardoso M, Neuhausen SL, Steele L, Ding YC, De Ruyck K, De Meerleer G, Ost P, Razack A, Lim J, Teo SH, Lin DW, Newcomb LF, Lessel D, Gamulin M, Kulis T, Kaneva R, Usmani N, Singhal S, Slavov C, Mitev V, Parliament M, Claessens F, Joniau S, Van den Broeck T, Larkin S, Townsend PA, Aukim-Hastie C, Gago-Dominguez M, Castelao JE, Martinez ME, Roobol MJ, Jenster G, van Schaik RHN, Menegaux F, Truong T, Koudou YA, Xu J, Khaw KT, Cannon-Albright L, Pandha H, Michael A, Thibodeau SN, McDonnell SK, Schaid DJ, Lindstrom S, Turman C, Ma J, Hunter DJ, Riboli E, Siddiq A, Canzian F, Kolonel LN, Le Marchand L, Hoover RN, Machiela MJ, Cui Z, Kraft P, Amos CI, Conti DV, Easton DF, Wiklund F, Chanock SJ, Henderson BE, Kote-Jarai Z, Haiman CA, Eeles RA. Profile Study; Australian Prostate Cancer BioResource (APCB); IMPACT Study; Canary PASS Investigators; Breast and Prostate Cancer Cohort Consortium (BPC3); PRACTICAL (Prostate Cancer Association Group to Investigate Cancer-Associated Alterations in the Genome) Consortium; Cancer of the Prostate in Sweden (CAPS); Prostate Cancer Genome-wide Association Study of Uncommon Susceptibility Loci (PEGASUS); Genetic Associations and Mechanisms in Oncology (GAME-ON)/Elucidating Loci Involved in Prostate Cancer Susceptibility (ELLIPSE) Consortium. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018;50(7):928–936.
Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, Reeve MP, Laivuori H, Aavikko M, Kaunisto MA, Loukola A, Lahtela E, Mattsson H, Laiho P, Della Briotta Parolo P, Lehisto AA, Kanai M, Mars N, Rämö J, Kiiskinen T, Heyne HO, Veerapen K, Rüeger S, Lemmelä S, Zhou W, Ruotsalainen S, Pärn K, Hiekkalinna T, Koskelainen S, Paajanen T, Llorens V, Gracia-Tabuenca J, Siirtola H, Reis K, Elnahas AG, Sun B, Foley CN, Aalto-Setälä K, Alasoo K, Arvas M, Auro K, Biswas S, Bizaki-Vallaskangas A, Carpen O, Chen CY, Dada OA, Ding Z, Ehm MG, Eklund K, Färkkilä M, Finucane H, Ganna A, Ghazal A, Graham RR, Green EM, Hakanen A, Hautalahti M, Hedman ÅK, Hiltunen M, Hinttala R, Hovatta I, Hu X, Huertas-Vazquez A, Huilaja L, Hunkapiller J, Jacob H, Jensen JN, Joensuu H, John S, Julkunen V, Jung M, Junttila J, Kaarniranta K, Kähönen M, Kajanne R, Kallio L, Kälviäinen R, Kaprio J, FinnGen; Kerimov N, Kettunen J, Kilpeläinen E, Kilpi T, Klinger K, Kosma VM, Kuopio T, Kurra V, Laisk T, Laukkanen J, Lawless N, Liu A, Longerich S, Mägi R, Mäkelä J, Mäkitie A, Malarstig A, Mannermaa A, Maranville J, Matakidou A, Meretoja T, Mozaffari SV, Niemi MEK, Niemi M, Niiranen T, O Donnell CJ, Obeidat ME, Okafo G, Ollila HM, Palomäki A, Palotie T, Partanen J, Paul DS, Pelkonen M, Pendergrass RK, Petrovski S, Pitkäranta A, Platt A, Pulford D, Punkka E, Pussinen P, Raghavan N, Rahimov F, Rajpal D, Renaud NA, Riley-Gillis B, Rodosthenous R, Saarentaus E, Salminen A, Salminen E, Salomaa V, Schleutker J, Serpi R, Shen HY, Siegel R, Silander K, Siltanen S, Soini S, Soininen H, Sul JH, Tachmazidou I, Tasanen K, Tienari P, Toppila-Salmi S, Tukiainen T, Tuomi T, Turunen JA, Ulirsch JC, Vaura F, Virolainen P, Waring J, Waterworth D, Yang R, Nelis M, Reigo A, Metspalu A, Milani L, Esko T, Fox C, Havulinna AS, Perola M, Ripatti S, Jalanko A, Laitinen T, Mäkelä TP, Plenge R, McCarthy M, Runz H, Daly MJ, Palotie A. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613(7944):508–518.
Lin L, Wang W, Xiao K, Guo X, Zhou L. Genetically elevated bioavailable testosterone level was associated with the occurrence of benign prostatic hyperplasia. J Endocrinol Invest. 2023;46(10):2095–102.
Zakikhani M, Dowling RJ, Sonenberg N, Pollak MN. The effects of adiponectin and metformin on prostate and colon neoplasia involve activation of AMP-activated protein kinase. Cancer Prev Res (Phila). 2008;1(5):369–75.
Ben Sahra I, Laurent K, Loubat A, Giorgetti-Peraldi S, Colosetti P, Auberger P, Tanti JF, Le Marchand-Brustel Y, Bost F. The antidiabetic drug metformin exerts an antitumoral effect in vitro and in vivo through a decrease of cyclin D1 level. Oncogene. 2008;27(25):3576–86.
Acknowledgements
We express our thanks to those people who have contributed to the IEU GWAS database project and the MRC Integrative Epidemiology Unit (IEU) at the University of Bristol. Sincere thanks also go to the many GWAS consortia who have made the GWAS data that they generated publicly available, and many members of the IEU who have contributed to curating these data.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Conception and design of study: X Z and Z L.Acquisition of data: X Z.Data analysis and/or interpretation: X Z and Z L.Drafting of manuscript and/or critical revision: X Z.Approval of final version of manuscript: X Z and Z L.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, X., Li, Z. Does metformin really reduce prostate cancer risk: an up-to-date comprehensive genome-wide analysis. Diabetol Metab Syndr 16, 159 (2024). https://doi.org/10.1186/s13098-024-01397-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13098-024-01397-7