Abstract
Objectives
Previous studies have analyzed the potential effect of KCNQ1 rs2237892 polymorphism on the predisposition to type 2 diabetes mellitus, but the findings are inconclusive and the subject of debate. The purpose of our study was to provide further insight into the potential association between KCNQ1 rs2237892 polymorphism and the risk of type 2 diabetes mellitus.
Methods
In total, 50 articles (60 studies) with 77,276 cases and 76,054 controls were utilized in our analysis. The pooled odds ratio (OR), 95% confidence interval (95% CI), and p value were used to evaluate the significance of our findings. Funnel plots and Beggar’s regression tests were utilized to determine the presence of publication bias.
Results
Our meta-analysis results indicated that KCNQ1 rs2237892 polymorphism could be correlated with the risk of type 2 diabetes mellitus under the C allelic, recessive, and dominant genetic models (OR = 1.25, 95% 1.19–1.32, p < 0.001; OR = 1.50, 95% CI 1.34–1.68, p < 0.001; OR = 1.26, 95% CI 1.14–1.40, p < 0.001, respectively). Additionally, ethnicity analysis revealed that the source of control, case size, and Hardy–Weinberg Equilibrium status were correlated to the polymorphism in the three genetic models.
Conclusions
Our meta-analysis demonstrated significant evidence to support the association between KCNQ1 rs2237892 polymorphism and predisposition to type 2 diabetes mellitus.
Similar content being viewed by others
Background
The worldwide prevalence of type 2 diabetes mellitus (T2DM) is increasing, along with associated comorbidities such as cardiovascular disease [1]. The International Diabetes Federation (IDF) reports that there were 9.3% (463 million) adults with diabetes in 2019, and 700 million people will have diabetes by 2045 [2]. Researchers consider T2DM to be a polygenic metabolic disorder with genetic heterogeneity that is affected by nongenetic (environmental), genetic, and lifestyle factors. However, the pathogenesis of T2DM still remains unclear [3].
Previous studies have reported that the potassium voltage-gated channel KQT-like sub-family, member 1 gene (KCNQ1) is associated with T2DM in Japanese, Korean, Chinese, Indian, and European populations [4,5,6,7]. Case–control studies investigating the role of KCNQ1 polymorphisms in T2DM, have indicated that rs2237892, a single nucleotide polymorphism (SNP) located on intron 15, has a strong association with T2DM. Therefore, rs2237892 has been widely investigated in subsequent studies. However, there are disagreements between the different studies, and their validity has been limited by insufficient sample size and lack of ethnic diversity in the study populations [8,9,10,11]. Although a previous meta-analysis in 2012 investigated the association between KCNQ1 rs2237892 polymorphism and T2DM risk, the authors only utilized 25 articles [12]. Therefore, our objective in the present meta-analysis, was to further examine and elucidate the connection between KCNQ1 rs2237892 polymorphism and an increased risk of T2DM.
Methods
Publication search
We systematically searched for relevant publications published through March 11, 2021 using Cochrane Library, PubMed, EMBASE, Web of Science, and China National Knowledge Infrastructure. We used the following search terms: (“KCNQ1”, OR “potassium voltage-gated channel”, OR “KQT-like subfamily, member 1”, OR “rs2237892”) AND (“variant”, OR “polymorphism”, OR “mutation”) AND (“T2DM”, OR “type 2 diabetes mellitus”, OR “type 2 diabetes”, OR “T2D”). Two investigators independently checked the references of retrieved articles to select the publications they would include in the analysis.
Selection criteria
Studies were selected according to the following inclusion criteria: full text could be found; the case–control studies focused on the relevance of KCNQ1 rs2237892 polymorphism and T2DM risk; the KCNQ1 rs2237892 genotype was obtained, and association between the KCNQ1 rs2237892 SNP and T2DM prevalence was assessed. Studies were excluded if they met the following exclusion criteria: they were repetitions of other articles; they were not case–control studies; they were unpublished studies, conference articles, meta-analyses, systematic evaluations, and they were pedigree studies. We consulted the Preferred Reporting Project (PRISMA) Guide for Systematic Evaluation and Meta-Analysis to comply with standards for conducting and presenting results from meta-analyses [13].
Data extraction
Referring to the inclusion/exclusion criteria, two investigators independently extracted data that included: first author, country, publication year, amount of cases and controls, Hardy–Weinberg equilibrium (HWE), control group source, and the availability of KCNQ1 rs2237892 genotype. Only articles with maximum sample size were selected when similar data appeared in multiple publications. A third investigator reviewed the final results to ensure data accuracy, and discussions were held to resolve any conflicts.
Study quality assessment
Two investigators performed independent quality assessments for each eligible article according to the 9-point Newcastle–Ottawa Scale [14]. The third investigator resolved any conflicting results produced by the two investigators. The assessment score included these criteria: case and control selection (4 points); confounding factor quality corrected in cases and controls (2 points), exposure ascertainment (3 points). The total scores ranged from 0 to 9, and scores > 6 were indicative of high-quality articles.
Statistical analysis
We estimated the significance of the data describing KCNQ1 rs2237892 SNP and T2DM risk using the OR and 95% CI. The Chi-Square-Based Q-test and I-Squared test were utilized to analyze the heterogeneity with p < 0.1 suggesting heterogeneity [15, 16]. We estimated the pooled OR by fixed effect model (Mantel–Haenszel) when no heterogeneity existed, or by the DerSimonian and Laird random effects model [17, 18]. We performed the Chi-squared test in controls, to examine HWE. To estimate the influence of the pooled ORs caused by an individual data set, we performed sensitivity analysis for each of the comparison models. The publication bias was tested by Funnel plot and Begg linear regression (19, 20), and Stata 12.0 was used to perform all analyses.
Results
Study characteristics
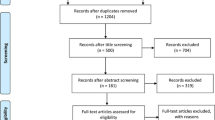
Figure 1 shows flowcharts of the selection of publications for the present study. There were 535 publications located in several electronic databases. After examining the research title, content, and abstract of the publications, the two investigators excluded 169 duplicate documents, 298 irrelevant papers, and examined the remaining 68 articles in full. Finally, our meta-analysis included 50 (60 case–control) publications. Among the 60 case–control studies, 51 included Asian populations, 4 included Caucasian, and 5 involved other populations. Of the studies in our meta-analysis, 24 were based on population (PB), 19 were based on hospital (HB), and 17 studies were based on no report (NR). The sample group of 21 studies was less than 500 patients, 10 studies included between 500 and 1000 patients, 4 studies included between 1001 and 2000 patients, and the remaining 6 studies had a sample group of greater than 2000. HWE balance (p < 0.05) was not met in 5 of the control groups. Due to lack of control group descriptions, 19 studies did not meet HWE assessment. Table 1 shows the main features of the study and the genotype distribution results of the HWE test.
Meta-analysis results
The meta-analysis included 153,330 participants (77,276 cases and 76,054 controls). KCNQ1 rs2237892 polymorphism was significantly associated with T2DM risk under the C allelic, recessive, and dominant genetic models (OR:1.25, 1.50 and 1.26; 95% CI 1.19–1.32, 1.34–1.68, and 1.14–1.40; p < 0.001, respectively). In ethnic subgroup analysis shown in Table 2, KCNQ1 rs2237892 polymorphism was correlated with increased risk of T2DM in the dominant genetic model of East Asians, in the C allelic genetic model of East Asians, and in the C allelic genetic model of West Asian populations (OR = 1.39, 1.32 and 1.25; 95% CI 1.31–1.49, 1.27–1.37 and 1.19–1.32; p < 0.001, respectively). In the stratified analysis by source of control, marked correlation was found in the C allelic genetic model (HB, PB, and NR: OR = 1.24, 1.25 and 1.16; 95% CI 1.14–1.37, 1.19–1.32 and 1.02–1.32; p < 0.001, respectively) and the dominant genetic model (HB and PB: OR = 1.25 and 1.48; 95% CI 1.08–1.46 and 1.38–1.59, p < 0.05, respectively). In the case size stratification, the C allelic genetic model (OR = 1.23, 1.14, 1.25 and 1.33; 95% CI 1.09–1.38, 0.88–1.48, 1.19–1.32 and 1.27–1.39; p < 0.001, respectively), the dominant genetic model (OR = 1.24, 1.13, 1.41 and 1.43; 95% CI 1.05–1.46, 0.81–1.58 and 1.33–1.53; p < 0.001, respectively) and the recessive genetic model (500–1000: OR = 1.32, 95% CI 0.91–1.91, p < 0.001) found notable association between KCNQ1 rs2237892 polymorphism and increased T2DM risk. Finally, we stratified by sample size—significant correlation was found in the C allelic genetic model (< 500, 1001–2000 and > 2000: OR = 1.23, 1.25 and 1.33; 95% CI 1.09–1.38, 1.19–1.32 and 1.27–1.39; p < 0.001, respectively) and the dominant genetic model (< 500, 1001–2000 and > 2000: OR = 1.24, 1.41 and 1.43; 95% CI 1.05–1.46, 1.14–1.75 and 1.33–1.53; p < 0.001, respectively).
Discussion
The association of KCNQ1 rs2237892 polymorphism with T2DM has been reported in many previous studies [21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62]. In 2008, two independently conducted genome-wide association studies (GWAS) in Japanese populations identified KCNQ1 as a novel T2DM susceptibility gene [5, 6, 8]. Subsequently, the SNP locus rs2237892 of this gene was found to be correlated with the incidence of T2DM in Korean population [15]. In our present meta-analysis, there were 60 studies, 77,276 cases and 76,057 controls, that we evaluated for the possible association between KCNQ1 rs2237892 polymorphism and T2DM risk. Our results showed that KCNQ1 rs2237892 polymorphism could be associated with T2DM in the dominant (CC vs CT + TT), recessive (CC + CT vs TT) and allele models (C vs T). In a stratified analysis based on ethnicity, source of control, and case size, we found that KCNQ1 rs2237892 polymorphism was significantly associated with T2DM in the dominant model, the allele model of East Asians, and in the allele model of West Asian populations. In Southeast Asian, South Asian, Caucasian, and other populations, KCNQ1 rs2237892 polymorphism was not significantly related to T2DM. In the stratified analysis according to the source of control, we found that KCNQ1 rs2237892 polymorphism was significantly correlated with T2DM in the dominant model and the allele model of HB and PB group, and in the allele model of NR group. But the correlation between KCNQ1 rs2237892 polymorphism and T2DM in children lacked corresponding evidence. The stratified analysis of the sample size showed that the correlation between populations occurred when the number of samples in the case group was less than 500, within 1001–2000, and > 2000. The above analysis shows that the ethnicity, the source of the control group, and the sample size of the case group may be the factors in the association occurred (Fig. 2).
Previously, a meta-analysis was performed in 2012 to investigate the association between KCNQ1 rs2237892 polymorphism and T2DM risk; however, only 25 articles were included in the analysis. Recently, a meta-analysis was performed to investigate the relationship between several KCNQ1 SNPs and T2DM risk, and a significant relationship between KCNQ1 polymorphism rs2237892 and T2DM risk was found [63]. However, the analysis was limited to 38 articles and incomplete sample size as well as selective bias are potential limitations of that study [63] (Fig. 3).
Compared to previous studies, our results demonstrate robust evidence to support a correlation between KCNQ1 rs2237892 polymorphism and T2DM risk. Scientists do not currently understand the biological mechanisms that cause an association between KCNQ1 and T2DM. There is biological evidence supporting the hypothesis that KCNQ1 might play a role in the predisposition to T2DM. KCNQ1, encoding the alpha subunit of the IKsK + channel, is expressed in the tissues or cells of the heart [64], as well as in pancreas islets, which play an important role in the regulation of insulin secretion [23] (Fig. 4).
This meta-analysis has several limitations. Firstly, most of the articles included in the meta-analysis involved the Asian population, while there were few articles involving Caucasian and other populations. Therefore, we could not perform the analysis grouped by different populations, and the ability to apply our results to a more general population is subsequently limited. Secondly, T2DM is caused by complex interactions between genetic, lifestyle, and environmental factors. Our study focused exclusively on the impact of genetic factors on T2DM risk. In the future, further studies should be conducted to determine interconnection between KCNQ1 rs2237892, lifestyle factors, and environmental factors on T2DM.
Conclusion
Our meta-analysis demonstrated an association between KCNQ1 rs2237892 polymorphism and the predisposition to T2DM. There was notable correlation between KCNQ1 rs2237892 and T2DM in East Asian populations and West Asian populations. However, for the Southeast Asian, South Asian, Caucasian, and other populations, the relevance of the KCNQ1 rs2237892 SNP was not confirmed because of the relatively limited sample size and the sparse amount of research into this subject. In addition, the source of the control group and the sample size of the case would also have an impact on the study results in the stratified analysis of this study. Therefore, in future research, we suggest exploring the relationship between KCNQ1 rs2237892 polymorphism and T2DM in a wide variety of populations. Although two meta-analyses were performed previously, the number of articles included in these was less than that in our study. Therefore, we believe that our study is superior than the two previous meta- studies.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- T2DM:
-
Type 2 diabetes mellitus
- IDF:
-
International Diabetes Federation
- PRISMA:
-
Preferred Reporting Project
- HWE:
-
Hardy–Weinberg equilibrium
- SNP:
-
Single nucleotide polymorphism
References
Standards of medical care in diabetes--2013. Diabetes care. 2013;36 Suppl 1(Suppl 1):S11–66.
Saeedi P, Petersohn I, Salpea P, Malanda B, Karuranga S, Unwin N, et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9(th) edition. Diabetes research and clinical practice. 2019;157:107843.
Li YY, Wang XM, Lu XZ. KCNQ1 rs2237892 C→T gene polymorphism and type 2 diabetes mellitus in the Asian population: a meta-analysis of 15,736 patients. J Cell Mol Med. 2014;18(2):274–82.
Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature. 1996;384(6604):78–80.
Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, Andersen G, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40(9):1098–102.
Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, Furuta H, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40(9):1092–7.
Been LF, Ralhan S, Wander GS, Mehra NK, Singh J, Mulvihill JJ, et al. Variants in KCNQ1 increase type II diabetes susceptibility in South Asians: a study of 3,310 subjects from India and the US. BMC Med Genet. 2011;12:18.
Lee YH, Kang ES, Kim SH, Han SJ, Kim CH, Kim HJ, et al. Association between polymorphisms in SLC30A8, HHEX, CDKN2A/B, IGF2BP2, FTO, WFS1, CDKAL1, KCNQ1 and type 2 diabetes in the Korean population. J Hum Genet. 2008;53(11–12):991–8.
Liu Y, Zhou DZ, Zhang D, Chen Z, Zhao T, Zhang Z, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes in the population of mainland China. Diabetologia. 2009;52(7):1315–21.
Chen Z, Yin Q, Ma G, Qian Q. KCNQ1 gene polymorphisms are associated with lipid parameters in a Chinese Han population. Cardiovasc Diabetol. 2010;9:35.
Holmkvist J, Banasik K, Andersen G, Unoki H, Jensen TS, Pisinger C, et al. The type 2 diabetes associated minor allele of rs2237895 KCNQ1 associates with reduced insulin release following an oral glucose load. PloS one. 2009;4(6):e5872.
Sun Q, Song K, Shen X, Cai Y. The association between KCNQ1 gene polymorphism and type 2 diabetes risk: a meta-analysis. PLoS One. 2012; 7(11): e48578.
Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–9.
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–5.
Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Statistics in Medicine.21(11):1539–58.
Zintzaras E, Lau J. Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J Clin Epidemiol. 2008;61(7):634–45.
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ (Clinical research ed). 2003;327(7414):557–60.
McKenzie JE, Beller EM, Forbes AB. Introduction to systematic reviews and meta-analysis. Respirology (Carlton, Vic). 2016;21(4):626–37.
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clinical research ed). 1997;315(7109):629–34.
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101.
Dai XP, Huang Q, Yin JY, Guo Y, Gong ZC, Lei MX, et al. KCNQ1 gene polymorphisms are associated with the therapeutic efficacy of repaglinide in Chinese type 2 diabetic patients. Clin Exp Pharmacol Physiol. 2012;39(5):462–8.
Takeuchi F, Serizawa M, Yamamoto K, Fujisawa T, Nakashima E, Ohnaka K, et al. Confirmation of multiple risk Loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes. 2009;58(7):1690–9.
Qi Q, Li H, Loos RJ, Liu C, Wu Y, Hu FB, et al. Common variants in KCNQ1 are associated with type 2 diabetes and impaired fasting glucose in a Chinese Han population. Hum Mol Genet. 2009;18(18):3508–15.
Yamauchi T, Hara K, Maeda S, Yasuda K, Takahashi A, Horikoshi M, et al. A genome-wide association study in the Japanese population identifies susceptibility loci for type 2 diabetes at UBE2E2 and C2CD4A-C2CD4B. Nat Genet. 2010;42(10):864–8.
Han X, Luo Y, Ren Q, Zhang X, Wang F, Sun X, et al. Implication of genetic variants near SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, FTO, TCF2, KCNQ1, and WFS1 in type 2 diabetes in a Chinese population. BMC Med Genet. 2010;11:81.
Tan JT, Ng DP, Nurbaya S, Ye S, Lim XL, Leong H, et al. Polymorphisms identified through genome-wide association studies and their associations with type 2 diabetes in Chinese, Malays, and Asian-Indians in Singapore. J Clin Endocrinol Metab. 2010;95(1):390–7.
Xu M, Bi Y, Xu Y, Yu B, Huang Y, Gu L, et al. Combined effects of 19 common variations on type 2 diabetes in Chinese: results from two community-based studies. PloS one. 2010;5(11):e14022.
Zhou JB, Yang JK, Zhao L, Xin Z. Variants in KCNQ1, AP3S1, MAN2A1, and ALDH7A1 and the risk of type 2 diabetes in the Chinese Northern Han population: a case-control study and meta-analysis. Medical science monitor : international medical journal of experimental and clinical research. 2010;16(6):Br179–83.
Saif-Ali R, Muniandy S, Al-Hamodi Z, Lee CS, Ahmed KA, Al-Mekhlafi AM, et al. KCNQ1 variants associate with type 2 diabetes in Malaysian Malay subjects. Ann Acad Med Singapore. 2011;40(11):488–92.
Tabara Y, Osawa H, Kawamoto R, Onuma H, Shimizu I, Makino H, et al. Genotype risk score of common susceptible variants for prediction of type 2 diabetes mellitus in Japanese: the Shimanami Health Promoting Program (J-SHIPP study). Development of type 2 diabetes mellitus and genotype risk score. Metabolism: clinical and experimental. 2011;60(11):1634–40.
Yu W, Ma RC, Hu C, So WY, Zhang R, Wang C, et al. Association between KCNQ1 genetic variants and obesity in Chinese patients with type 2 diabetes. Diabetologia. 2012;55(10):2655–9.
Plengvidhya N, Chanprasert C, Chongjaroen N, Yenchitsomanus PT, Homsanit M, Tangjittipokin W. Impact of KCNQ1, CDKN2A/2B, CDKAL1, HHEX, MTNR1B, SLC30A8, TCF7L2, and UBE2E2 on risk of developing type 2 diabetes in Thai population. BMC Med Genet. 2018;19(1):93.
Gamboa-Meléndez MA, Huerta-Chagoya A, Moreno-Macías H, Vázquez-Cárdenas P, Ordóñez-Sánchez ML, Rodríguez-Guillén R, et al. Contribution of common genetic variation to the risk of type 2 diabetes in the Mexican Mestizo population. Diabetes. 2012;61(12):3314–21.
Cui LJ, Chang XY, Zhu LY, Feng G, Zhou T, Zhang CX, et al. Relationship between the polymorphisms in KCNQ1 and type 2 diabetes in Chinese Kazakh population. Genetics and molecular research : GMR. 2016;15(2).
Bazzi MD, Nasr FA, Alanazi MS, Alamri A, Turjoman AA, Moustafa AS, et al. Association between FTO, MC4R, SLC30A8, and KCNQ1 gene variants and type 2 diabetes in Saudi population. Genetics and molecular research : GMR. 2014;13(4):10194–203.
Zhang W, Wang H, Guan X, Niu Q, Li W. Variant rs2237892 of KCNQ1 Is Potentially Associated with Hypertension and Macrovascular Complications in Type 2 Diabetes Mellitus in A Chinese Han Population. Genomics Proteomics Bioinformatics. 2015;13(6):364–70.
Turki A, Mtiraoui N, Al-Busaidi AS, Khirallah M, Mahjoub T, Almawi WY. Lack of association between genetic polymorphisms within KCNQ1 locus and type 2 diabetes in Tunisian Arabs. Diabetes Res Clin Pract. 2012;98(3):452–8.
Zhou X, Zhu J, Bao Z, Shang Z, Wang T, Song J, et al. A variation in KCNQ1 gene is associated with repaglinide efficacy on insulin resistance in Chinese Type 2 Diabetes Mellitus Patients. Sci Rep. 2016;6:37293.
Riobello C, Gómez J, Gil-Peña H, Tranche S, Reguero JR, de la Hera JM, et al. KCNQ1 gene variants in the risk for type 2 diabetes and impaired renal function in the Spanish Renastur cohort. Mol Cell Endocrinol. 2016;427:86–91.
Huerta-Chagoya A, Vázquez-Cárdenas P, Moreno-Macías H, Tapia-Maruri L, Rodríguez-Guillén R, López-Vite E, et al. Genetic determinants for gestational diabetes mellitus and related metabolic traits in Mexican women. PloS one. 2015;10(5):e0126408.
Williams AL, Jacobs SB, Moreno-Macías H, Huerta-Chagoya A, Churchhouse C, Márquez-Luna C, et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506(7486):97–101.
Qian Y, Lu F, Dong M, Lin Y, Li H, Dai J, et al. Cumulative effect and predictive value of genetic variants associated with type 2 diabetes in Han Chinese: a case-control study. PloS one. 2015;10(1):e0116537.
Tam CH, Ho JS, Wang Y, Lam VK, Lee HM, Jiang G, et al. Use of net reclassification improvement (NRI) method confirms the utility of combined genetic risk score to predict type 2 diabetes. PloS one. 2013;8(12):e83093.
Almawi WY, Nemr R, Keleshian SH, Echtay A, Saldanha FL, AlDoseri FA, et al. A replication study of 19 GWAS-validated type 2 diabetes at-risk variants in the Lebanese population. Diabetes Res Clin Pract. 2013;102(2):117–22.
Long J, Edwards T, Signorello LB, Cai Q, Zheng W, Shu XO, et al. Evaluation of genome-wide association study-identified type 2 diabetes loci in African Americans. Am J Epidemiol. 2012;176(11):995–1001.
Iwata M, Maeda S, Kamura Y, Takano A, Kato H, Murakami S, et al. Genetic risk score constructed using 14 susceptibility alleles for type 2 diabetes is associated with the early onset of diabetes and may predict the future requirement of insulin injections among Japanese individuals. Diabetes Care. 2012;35(8):1763–70.
van Vliet-Ostaptchouk JV, van Haeften TW, Landman GW, Reiling E, Kleefstra N, Bilo HJ, et al. Common variants in the type 2 diabetes KCNQ1 gene are associated with impairments in insulin secretion during hyperglycaemic glucose clamp. PloS one. 2012;7(3):e32148.
Odgerel Z, Lee HS, Erdenebileg N, Gandbold S, Luvsanjamba M, Sambuughin N, et al. Genetic variants in potassium channels are associated with type 2 diabetes in a Mongolian population. J Diabetes. 2012;4(3):238–42.
Saif-Ali R, Ismail IS, Al-Hamodi Z, Al-Mekhlafi HM, Siang LC, Alabsi AM, et al. KCNQ1 haplotypes associate with type 2 diabetes in Malaysian Chinese Subjects. Int J Mol Sci. 2011;12(9):5705–18.
Lin YD, Qian Y, Dong MH, Lu F, Shen C, Jin GF, et al. Association of polymorphisms of potassium voltage-gated channel, KQT-like subfamily, member 1 and type 2 diabetes in Jiangsu province. China Chinese journal of preventive medicine. 2013;47(6):538–41.
Yamakawa-Kobayashi K, Natsume M, Aoki S, Nakano S, Inamori T, Kasezawa N, et al. The combined effect of the T2DM susceptibility genes is an important risk factor for T2DM in non-obese Japanese: a population based case-control study. BMC Med Genet. 2012;13:11.
Al-Shammari MS, Al-Ali R, Al-Balawi N, Al-Enazi MS, Al-Muraikhi AA, Busaleh FN, et al. Type 2 diabetes associated variants of KCNQ1 strongly confer the risk of cardiovascular disease among the Saudi Arabian population. Genet Mol Biol. 2017;40(3):586–90.
Totomoch-Serra A, Muñoz ML, Burgueño J, Revilla-Monsalve MC, Perez-Muñoz A, Diaz-Badillo Á. The ADRA2A rs553668 variant is associated with type 2 diabetes and five variants were associated at nominal significance levels in a population-based case-control study from Mexico City. Gene. 2018;669:28–34.
Chen J, Pan T. Association between transcription KCNQ1 rs2237892 polymorphism and development of type 2 diabetes in Hefei. Acta Universitatis Medicinalis Anhui. 2018;53(1):110–3.
Da H. Genetic epidemiology of type 2 diabetes mellitus and metabolic syndrome in Hubei Han population: Central China Normal University; 2011.
Gao X, Lv DM, Wang Y, Wang T, Li W, Zhang Y. Association Studies of KCNQ1 Gene Polymorphism and Type 2 Diabetes Mellitus in Huaihai Region. Chinese general practice. 2012;15(27):3139–42.
Li Y, Zhang T, Liu H. Association of KCNQ1 and SRR gene polymorphisms with type 2 diabetes mellitus in Uygur and Han nationality. Chin J Diabetes. 2018;26(07):534–42.
Xu T, Pan B, Ding P. Correlation study between gene polymorphism of KCNQ1 and SLC30A8 and type 2 diabetes mellitus. China Modern Doctor. 2018;56(09):1–4.
Yang M. Early screening of type 2 diabetes mellitus in Chinese Han population: Ningbo University; 2013.
Yang K. Association between KCNQ1 gene polymorphism and susceptibility to type 2 diabetes mellitus in Han population: Zhengzhou University; 2018.
Zhang S. Association of IL6R, APM1 and KCNQ1 genes with type 2 diabetes mellitus in Hubei Han population. Central China Normal University, 2009.
Zhu A, Yang X, Wu Y, Zhang Z, Li M. Association study of single nucleotide polymorphisms in KCNQ1 and susceptibility of type 2 diabetes. J Trop Med. 2014;14(01):41–5.
Yu XX, Liao MQ, Zeng YF, Gao XP, Liu YH, Sun W, Zhu S, Zeng FF, Ye YB. Associations of KCNQ1 Polymorphisms with the Risk of Type 2 Diabetes Mellitus: An Updated Meta-Analysis with Trial Sequential Analysis. J Diabetes Res. 2020;2020:7145139.
Lee MP, Ravenel JD, Hu RJ, Lustig LR, Tomaselli G, Berger RD, Brandenburg SA, Litzi TJ, Bunton TE, Limb C, Francis H, Gorelikow M, Gu H, Washington K, Argani P, Goldenring JR, Coffey RJ, Feinberg AP. Targeted disruption of the Kvlqt1 gene causes deafness and gastric hyperplasia in mice. J Clin Invest. 2000;106(12):1447–55.
Acknowledgements
We appreciate the cooperation of the partners and staffs cooperated in this study.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Manuscript writing, editing and review were conducted by HLJ; YJD and XL participated in the articles search; HLJ and HD performed data analysis and evaluation the quality of the selected studies. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Jiang, HL., Du, H., Deng, YJ. et al. Effect of KCNQ1 rs2237892 polymorphism on the predisposition to type 2 diabetes mellitus: An updated meta-analysis. Diabetol Metab Syndr 13, 75 (2021). https://doi.org/10.1186/s13098-021-00683-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13098-021-00683-y