Abstract
Background
Several studies have reported the roles of Trichinella spiralis extracellular vesicles in immune regulation and pathogen diagnosis. Currently, the T. spiralis muscle larvae excretory/secretory product (Ts-ML-ES) is the antigen recommended by the International Commission on Trichinellosis (ICT) for serological diagnosis of trichinellosis. However, it can only be used to detect middle and late stages of infections, and cross-reactions with other parasite detections occur. Therefore, there is a need to identify antigens for specific detection of early stage trichinellosis.
Methods
Extracellular vesicles of T. spiralis muscle larvae (Ts-ML-EVs) were isolated by ultracentrifugation and characterized by transmission electron microscopy, nanoparticle tracking analysis, flow cytometry and western blot. Ts-ML-EVs protein profiles were analyzed by LC-MS/MS proteomics for identification of potential antigens (Ts-TTPA). Ts-TTPA were cloned into pMAL-c5X vector and expressed as recombinant proteins for evaluation of potential as detected antigens by western blot and ELISA.
Results
Isolated Ts-ML-EVs were round or elliptic (with diameters between 110.1 and 307.6 nm), showing a bilayer membrane structure. The specific surface markers on the Ts-ML-EVs were CD81, CD63, enolase and the 14-3-3 protein. A total of 53 proteins were identified by LC-MS/MS, including a variety of molecules that have been reported as potential detection and vaccine candidates. The cDNA of Ts-TTPA selected in this study has a total length of 1152 bp, encoding 384 amino acids with a molecular weight of 44.19 kDa. It contains a trypsin domain and can be recognized by anti-His antibody. It reacted with swine sera infected with 10,000 T. spiralis at 15, 25, 35 and 60 days post-infection (dpi). At 10 μg/ml, this antigen could detect T. spiralis antibodies from the swine sera at 13 dpi. There were no cross-reactions with the swine sera infected with other parasites including Clonorchis sinensis, Toxoplasma gondii, Taenia suis, Ascaris suis and Trichuris suis.
Conclusions
This study identifies potential early stage detection antigens and more thoroughly characterizes a serine protease domain-containing protein. Extracellular vesicle proteins may be explored as effective antigens for the early stage detection of trichinellosis.
Graphical Abstract

Similar content being viewed by others
Backgrounds
Trichinella spiralis is an important foodborne parasite that causes trichinellosis in a wide variety of hosts, causing significant public health and economic impacts. Hosts are usually infected by ingestion of raw or undercooked meats containing the cysts of muscle larvae [1]. After digestion in the stomach, the cysts develop into intestinal infective larvae and undergo four molts to grow into adults in the intestine. The adult worms travel through the intestinal epithelium where they mate and produce offspring. The newborn larvae pass through the host circulatory system to reach the muscle cells for permanent parasitism [2, 3].
The most commonly used Ts-ML-ES antigen is recommended by the International Commission on Trichinellosis (ICT) for serological detection for trichinellosis. However, previous studies have shown that 100% detection of anti-T. spiralis IgG antibodies by ES-ELISA are unlikely for at least 1–3 months after the primary T. spiralis infection [4]. There is a clear window of 3–4 weeks between infection and specific antibody positivity [5]. Therefore, there is a need to identify antigens for detecting early stage T. spiralis infection. Some studies have reported screening of early stage antigens from adult worm crude extracts and excretory-secretory products [6]. A number of early antigens have been identified from cDNA subtractive libraries and proteomics, but they generally lack specificity and sensitivity [7, 8].
Extracellular vesicles (EVs) are vesicles secreted by a variety of cells and can be classified into micro-vesicles, exosomes, apoptotic bodies and oncosomes [9]. EVs may participate in parasite-host interactions and host immune regulation. EV proteins are also explored for detection and diagnosis, including those from the parasites [10,11,12,13]. For example, Kifle et al. identified 15,120-kDa EV proteins from Schistosoma mansoni and showed their potential to serve as vaccine candidates for schistosomiasis [14]. Our previous study also showed that T. spiralis muscle larval EVs (Ts-ML-EVs) contained fewer proteins than the muscle larval excretory-secretory products (Ts-ML-ES) but produced stronger host immunological responses [15].
In this study, we used LC-MS/MS technology combined with bioinformatics analysis to analyze EV proteins from T. spiralis muscle larvae, aiming to identify antigens that could detect antibodies from the swine sera at the early stage of T. spiralis infection. We found that Ts-TTPA protein with trypsin domain identified swine sera at 13 dpi most quickly and is a candidate antigen for early stage detection of trichinellosis.
Methods
Animals and T. spiralis-infected swine sera
Female Wistar rats (~ 200 g) were purchased from the Experimental Animal Center of Jilin University.
The swine sera used in the experiment included 7, 9, 11, 13, 15, 17, 19, 21, 25, 30, 35, 45, 60, 90 and 120 dpi of swine serum infected with 10,000 T. spiralis. The serum of pigs not infected with T. spiralis was used as negative control. The above swine serum is from our team's previous research [5]. Similarly, the swine serum used in the cross-reactions was derived from previous studies in the laboratory [16].
Parasite maintenance and collection
The T. spiralis (ISS 534 strain) used in this study was maintained in female Wistar rats in our laboratory (3500 ML/rat, maintained for about 6 weeks after oral infection), and T. spiralis muscle larvae were recovered from the muscles of infected rats with a standard pepsin/hydrochloric acid digestion method [17]. After washing with normal saline containing 2 × penicillin-streptomycin (Biological Industries, Kibbutz Beit Haemek, Israel), 5000 muscle larvae/ml were cultured in RIPM1640 medium (Gibco, California, USA) containing 2 × penicillin-streptomycin at a 37 °C for 18 h [5].
Isolation of extracellular vesicles of T. spiralis muscle larvae (Ts-ML-EVs)
The above cultures were collected in 50-ml tubes and centrifuged at 800 g for 10 min followed by 5000 g for 20 min to remove excess worm fragments and other large impurities. The supernatant was then passed through a 0.22-μm filter (Millipore, Darmstadt, Germany) to remove larger vesicles and bacteria. Centrifugation was performed using a 10-kDa ultrafiltration tube (Millipore, Darmstadt, Germany) at 5000 g to concentrate the liquid to the appropriate volume. The supernatant was then ultracentrifuged at 4 °C, 120,000 g for 2 h with an ultracentrifuge (Hitachi, Tokyo, Japan). After the supernatant was discarded carefully, sterile PBS (phosphate buffer saline, PH = 7.4) was used to suspend the bottom vesicles, and the protein concentration of the collected Ts-ML-EVs was measured with a bicinchoninic acid (BCA) protein assay kit (Beyotime, Beijing, China). Samples were individually packaged in tubes and stored at − 80 °C for the next steps.
Characterization of Ts-ML-EVs by transmission electron microscopy, nanosight, western blot and flow cytometry
Negative staining transmission electron microscopy (TEM) was used to analyze the morphology, structure and size of the Ts-ML-EVs. Briefly, 10 μl of the Ts-ML-EVs suspension was dropped on the copper mesh, adsorbed for 1 min and then soaked in 2% glutaraldehyde at room temperature. The grids were then negatively stained with 2% phosphotungstic acid for 1 min and air-dried at room temperature. The Ts-ML-EVs were examined at 80 kV using a Hitachi H-7650 transmission electron microscope (Hitachi, Tokyo, Japan). The Ts-ML-EVs particles were characterized for size and concentration using a NanoSight NS300 (Malvern Panalytical, UK). Then, western blotting (WB) and flow cytometry were used for surface marker identification. For WB, 15 μg Ts-ML-EVs and 15 μg Ts-ML-ES) were electrophoresed in 12% SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) and transferred to PVDF membranes (Immobilon, Millipore, USA). The membranes were blocked with 5% skim milk and incubated with primary antibodies including polyclonal rabbit anti-CD63 (1:1000, Abcam, Cambridge, UK) and polyclonal goat anti-enolase (1:200, Abcam, Cambridge, UK). Subsequently, two different horseradish peroxidase (HRP)-conjugated secondary antibodies were used: goat anti-rabbit IgG (1:2000, Cell Signaling Technology, USA) and donkey anti-goat IgG (1:50,000, Jackson ImmunoResearch, USA). The membrane was visualized using an ECL luminescence chromogenic solution (Thermofisher Scientific, Waltham, MA, USA) and a chemiluminescence instrument (Analytik Jena, Thuringia, Germany). For flow cytometry, the Ts-ML-EVs were diluted to 100 μg/ml in 100 μl PBS, and PE-Mouse Anti-Human CD63 (Clone: H5C6, BD Biosciences, New Jersey, USA) and APC-Mouse Anti-Human CD81 (Clone: JS-81, BD Biosciences, NJ, USA) antibodies were added. The samples were incubated at room temperature for 30 min in the dark and then ultracentrifuged for 15 min at 4 °C, 120,000 g. The supernatant was discarded, and PBS was added to resuspend the bottom vesicles. Finally, a flow cytometer was used (BD FACS Aria, BD Biosciences, NJ, USA) for vesicle detection.
SDS-PAGE and western blot analysis
The SDS-PAGE analyses were performed on the Ts-ML-ES (15 μg) and Ts-ML-EVs (15 μg) on three gels, respectively. One of these was transferred to a membrane for blocking, and the bands were displayed by hybridization using swine serum (1:200) of uninfected or 25 dpi with 10,000 T. spiralis muscle larvae. One of the gels was stained with Coomassie Bright Blue Fast Staining solution (P1300, Solarbio, Beijing, China) and destained with destaining solution. Then, a band at the same position as the hybridized band was cut out and sequenced. Another gel was used for silver staining. Specific protocols refer to the manual provided by the manufacturer (P0017S, Beyotime, Beijing, China).
The recombinant protein (rTs-TTPA) was performed on 12% SDS-PAGE. After protein transfer and blocking, swine sera at 15, 25, 35, 60 and 120 dpi with 10,000 T. spiralis and negative serum (1:200) or mouse His-tag antibody (1:1000) were used as primary antibodies, while HRP-goat anti-pig IgG (Abcam, Massachusetts, UK) and HRP-goat anti-mouse IgG were used as secondary antibodies (1:2000). Chemiluminescence substrates were used to detect protein bands through UVP ChemStudio (Analytik Jena, Thuringia, Germany).
In-gel digestion
For in-gel tryptic digestion, gel pieces were destained in 50 mM NH4HCO3 in 50% acetonitrile (v/v) until clear. Gel pieces were dehydrated with 100 μl of 100% acetonitrile for 5 min, the liquid removed and the gel pieces rehydrated in 10 mM dithiothreitol before being incubated at 56 °C for 60 min. Gel pieces were again dehydrated in 100% acetonitrile. The liquid was removed and gel pieces rehydrated with 55 mM iodoacetamide. Samples were then incubated at room temperature, in the dark, for 45 min. Gel pieces were washed with 50 mM NH4HCO3 and dehydrated with 100% acetonitrile. Gel pieces were rehydrated with 10 ng/μl trypsin, and resuspended in 50 mM NH4HCO3 on ice for 1 h. Excess liquid was removed, and the gel pieces were digested with trypsin at 37 °C overnight. Peptides were extracted with 50% acetonitrile/5% formic acid, followed by 100% acetonitrile. Peptides were dried to completion and resuspended in 2% acetonitrile/0.1% formic acid for further analysis.
LC-MS/MS analysis
The tryptic peptides were dissolved in 0.1% formic acid (Fluka, Seelze, Germany) (solvent A) and directly loaded onto a homemade reversed-phase analytical column (15 cm long, 75 μm i.d.). The gradient was comprised of an increase from 6 to 23% solvent B [0.1% formic acid in 98% acetonitrile (Fisher Chemical, Waltham, USA)] and ran over 16 min, 23% to 35% over 8 min, finally climbing to 80% over 3 min and then holding at 80% for the last 3 min. This was performed at a constant flow rate of 400 nl/min on an EASY-nLC 1000 UPLC system (Thermo Fisher Scientific, Waltham, MA, USA).
The peptides were subjected to an NSI source followed by tandem mass spectrometry (MS/MS) in a Q ExactiveTM Plus (Thermo Fisher Scientific, Waltham, MA, USA) coupled online to the UPLC. The electrospray voltage applied was 2.0 kV. The m/z scan range was 350–1800 for a full scan, and intact peptides were detected in the Orbitrap (Thermo Fisher Scientific, Waltham, USA) at a resolution of 70,000. Peptides were then selected for MS/MS using an NCE setting of 28, and the fragments were detected in the Orbitrap at a resolution of 17,500. This data-dependent procedure alternated between one MS scan followed by 20 MS/MS scans with 15.0 s dynamic exclusion. Automatic gain control (AGC) was set at 5E4.
Data processing
The resulting MS/MS data were processed using Proteome Discoverer 2.4. Tandem mass spectra were searched blasted against the T. spiralis database (18,572) sequences. Trypsin/P was specified as a cleavage enzyme allowing up to two missing cleavages. A mass error was set to 10 ppm for precursor ions and 0.02 Da for fragment ions. Carbamidomethyl on Cys was specified as a fixed modification, and oxidation was specified as a variable modification on Met. Peptide confidence was set at high, and peptide ion score was set at > 20.
ELISA
Ten μg/ml rTs-TTPA in 100 μl CBS (carbonate buffer solution) was coated on an EIA-ELISA plate at 4 °C overnight. After the encapsulation solution was discarded, it was blocked with 200 μl 5% skim milk at 37 °C for 1 h, and then PBST (phosphate buffer saline with 2% Tween-20) was used for three 1-min washes. After patting the liquid dry, serum collected from pigs on different days post-infection with 10,000 T. spiralis or serum infected with Clonorchis sinensis, Toxoplasma gondii, Taenia suis, Ascaris suis and Trichuris suis, diluted with a blocking solution (1:100), was added for the binding reaction. After the same washing and patting, HRP-goat-anti-pig secondary antibodies (Abcam, Cambridge, UK) in a blocking solution (1:5000) were incubated at 37 °C for 45 min. After washing and termination, TMB substrate (Tiangen, Beijing, China) was added to detect absorbance at 450 nm (Biotek, Vermont, USA).
Protein expression and purification
The protein (named tissue-type plasminogen activator, Ts-TTPA) was identified after LC-MS/MS analysis. The expression vector pMAL-c5X was cloned with BamHI-EcoRI, and these were transformed into BL21 (DE3) for protein expression. By optimizing the conditions, the soluble rTs-TTPA was obtained. The induction conditions were 1 M IPTG at 16 °C for 20 h. The purified protein was obtained by repeated freeze-thawing, ultrasonic crushing and Ni-affinity chromatography (Qiagen, Dusseldorf, Germany) according to the manufacturer’s instructions.
Bioinformatics analysis
Secondary structure prediction of proteins was performed with Protein Structure Prediction Server [18]. Domain analysis was performed with Pfam [19]. A Swiss model was used to build the 3D protein structure [20]. Isoelectric point prediction was completed in ProtParam [21]. The GO analysis was completed in Gene Ontology Resource [22]. Molecular Evolutionary Genetics Analysis software (MEGA10) [23] was used to draw the maximum likelihood tree.
Statistical analysis
GraphPad Prism 9 (9.0.0) was used for statistical analysis and visualization with one-way ANOVA followed by Tukey’s multiple comparison post-test.
Results
Characterization of Ts-ML-EVs
The Ts-ML-EVs were characterized as typical circular or oval bilayer membrane-like vesicles with different sizes (Fig. 1a), ranging from 110.1–307.6 nm, and peak sizes were 181 nm (Fig. 1b). We confirmed by western blot, the Ts-ML-EVs contained the EV marker proteins Enolase and 14-3-3 (Fig. 1c). In addition, flow cytometry showed that CD63 and CD81 were the specific markers on the surface of the Ts-ML-EVs, and the positive rates were 48.8% and 77.7%, respectively (Fig. 1d). These results suggest that the extracellular vesicles collected in this study conformed to the expected characteristics of extracellular vesicles and can be used in further experiments.
Characterization and identification of Ts-ML-EVs. A The morphology and size of T. spiralis extracellular vesicles visualized by scanning electron microscopy. White arrow indicates the extracellular vesicles (scale bar 100 μM). B NTA shows the size distribution of T. spiralis extracellular vesicle by intensity. The illustration is a combination of histograms and peak charts. C Identification of marker proteins 14-3-3, enolase in the excretory-secretory product (40 μg) and extracellular vesicles (40 μg) of T. spiralis. D CD81 and CD63 were identified on the extracellular vesicles of T. spiralis by flow cytometry
Ts-ML-EVs can be recognized by swine serum at 25 days post-infection
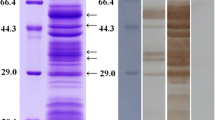
Both Coomassie Blue and silver stainings showed that Ts-ML-EVs had many bands, with molecular weights ranging from 10 to 180 kDa (Fig. 2a). Western blot analysis showed that the bands of Ts-ML-EVs that could be reacted with 25 dpi swine serum were concentrated between 40 and 180 kDa. However, negative swine serum did not recognize these protein bands (Fig. 2b).
Protein identification by LC-MS/MS analysis
The protein bands identified by 25 dpi with 10,000 T. spiralis early infection sera were separately analyzed by LC-MS/MS, and a total of 200 unique T. spiralis extracellular vesicle proteins with a pep count ≥ 2 were identified by searching the T. spiralis database in UniProt. Except for the redundant sequences, they were clustered into 53 unique proteins. The molecular weight (MW) of 53 proteins ranged from 10 to 484 kDa, of which 35 (66.03%) proteins were distributed in the range of 30–60 kDa (Table 1, Fig. 3a). The enrichment analysis of these proteins shows that these proteins have cellular components, molecular functions and biological processes (Fig. 3b). There are 25 proteins involved in the organic substance metabolic process, metabolic process and primary metabolic process. Also, 32 proteins (60.37%) were involved in the catalytic activity process, which was the function that contained the most proteins of all the GO (Gene Ontology) annotations.
Bioinformatic analysis of Ts-TTPA sequences
Bioinformatics analysis revealed that the full-length cDNA sequence of the Ts-TTPA gene was 1152 bp and contained 384 amino acids. The predicted MW of Ts-TTPA was 44.19 kDa and the isoelectric point was 8.81. Amino acids 1–24 (ATGAATAATAGAAATAAAATAAAA) were predicted to be its signal peptide, and the Pfam database showed the presence of trypsin domains at 118–371 aa. In addition, the Swiss model was used to predict 3D structures, showing secondary structures including α-helix, β-folding and curling (Fig. 4a). In addition, we performed an evolutionary analysis of this protein and found that it has the highest homology with Trichinella nelsoni (Fig. 4b).
Bioinformatic analysis of Ts-TTPA. A Three-dimensional structural model of Ts-TTPA protein with secondary structure display. The alpha helix is covered in purple, the beta fold is covered in green, and the ring area is covered in gray. B Cladogram of analysis of Ts-TTPA. The maximum parsimony tree of tissue-type plasminogen activator protein in 12 species of Trichinella spiralis and Trichuris suis was drawn in MEGA. Protein sequence thumbnails of Ts-TTPA for each species are shown on the right. Different-colored squares represent different amino acids
Expression and purification of rTs-TTPA
The expression vectors, PMAL-c5X-tissue-type plasminogen activator (UniProt: A0A0V1AVL6), were constructed successfully with His and MBP labels (Fig. 5a). The purified recombinant proteins were obtained by inducible expression and named rTs-TTPA (Fig. 5b). The purified protein was identified by mouse-His tag antibody (Fig. 5c).
Expression and purification of rTs-TTPA. A The constructed vector was identified by double restriction enzyme digestion. From right to left are DNA markers, pMAL-c5X-Ts-TTPA plasmid digested with BamHI/EcoRI and pMAL-c5X-Ts-TTPA plasmid. The red arrow shows the Ts-TTPA gene. B The purified Ts-TTPA was stained with Coomassie Bright Blue. The red arrow shows the Ts-TTPA protein. C The purified rTs-TTPA was identified by monoclonal His-tag antibody
Recognition of rTs-TTPA by infected serum
Subsequently, hybridization with swine serum post-infection on different days showed that there was no reaction with negative serum and serum at 120 dpi with 10,000 T. spiralis, while swine serum at 15, 25, 35 and 60 dpi with 10,000 T. spiralis could recognize this protein, with the earliest detection time being 15 dpi (Fig. 6).
Dynamics of serum anti-Trichinella IgG in experimentally infected pigs
Serum anti-Trichinella IgG levels of pigs at different time intervals were detected by ELISA using rTs-TTPA. By testing 86 negative samples, we determined that the cut-off value was 0.48278 (Fig. 7a). The rTs-TTPA-ELISA can detect the specific anti-Trichinella IgG at 13 dpi, with a positive rate of 100% in the detected samples (Fig. 7b). In addition, the results from the cross-reaction experiments showed that the detection value was lower than the cut-off line, which indicated that the rTs-TTPA-ELISA had no cross-reactions with other common swine parasites and had good specificity (Fig. 7c).
Establishment of rTs-TTPA-ELISA for early stage detection. A Eighty-six negative pig serum samples (1:100) were used to calculate the cut-off, with line representing mean + 3 SD (blue). B Kinetics of serum anti-Trichinella IgG in pigs experimentally infected with 10,000 muscle larvae (n = 3). C Cross-reactions assay of serum infected with Clonorchis sinensis, Toxoplasma gondii, Taenia suis, Ascaris suis and Trichuris suis (n = 3)
Discussion
Trichinella spiralis is an important zoonotic disease, and its detection has always been a hot topic in research. Although molecular detections have been widely reported, serological detection is the most widely used detection method for trichinellosis and is recommended by the World Health Organization (WHO) and the International Commission on Trichinellosis (ICT) [24]. Previous studies have reported the detection of trichinellosis using circulating antigens such as crude extract protein and excretory-secretory products as detected molecules, but there is an obvious detection window period limited to within 2 or 3 weeks of infection [25]. The ML-ES antigen-ELISA has been used consistently in our laboratory to detect IgG in serum from pigs infected with T. spiralis, and IgG detection is not complete until 19 dpi [5]. Therefore, improving detected antigens has clinical value, and increasingly studies are focusing on screening antigens for early stage detection [4].
In recent years, research on parasite-derived extracellular vesicles has attracted much attention [26]. Parasite-derived extracellular vesicles can be used as signaling molecules to participate in parasite-host interactions, maintain the parasitism of parasites and cause host diseases. Extracellular vesicles also participate in host defense responses through antigen presentation [27,28,29]. Proteomic and transcriptomic analysis revealed that parasite-derived extracellular vesicles contain a large number of proteins and non-coding RNAs, which are involved in parasite reproduction, survival and host immune regulation, and also elucidated a novel parasite-host communication mode mediated by parasite-derived extracellular vesicles [30, 31]. In addition, compared with other sources, the extracellular vesicles collected in this study are enriched and may detect antigenic proteins that have not been previously reported, providing new candidate molecules for the diagnosis of T. spiralis infection.
We screened 53 proteins from Ts-ML-EVs by LC-MS/MS, and besides being involved in some basic metabolic pathways, they are also widely involved in the catalytic and hydrolase activity of T. spiralis, which have been reported to play important roles in the growth, development and invasion of T. spiralis [32, 33]. In addition, some proteins in Ts-ML-EVs have also been reported to be useful for early stage detection and candidate vaccines. Furthermore, we conducted extensive screening of unreported proteins to obtain new detected antigens or vaccine candidates.
The candidate molecule Ts-TTPA identified here has the trypsin domain, which belongs to the peptidase family S1, in which serine serves as the nucleophilic amino acid at the active site. It is the largest protease family, including catalytic active and non-catalytic active enzymes, known as "serine protease" and "serine protease homologs," respectively [34]. Some proteins with serine protease activity isolated from T. spiralis have been reported to be involved in parasite-host interactions, including immune regulation and the invasion of intestinal epithelium [35,36,37]. In addition, these antigenic proteins have been reported as candidate molecules for the diagnosis of antigens. Sun et al. used the recombinant expression of Ts-SP as an antigen to detect the serum of mice infected with 300 muscle larvae and found that they could detect the infected serum at 12 dpi at the earliest [6]. The Ts-TTPA we reported also had the same effect, and it was able to detect infected serum as early as 7 dpi. Three sera infected with 10,000 T. spiralis were fully detected at 13 dpi. The identification of these detected antigens partially supplemented the window period for the detection of IgG and provided a new reference for the early stage detection of T. spiralis. However, the sample size of this experiment is not large, which carries certain limitations, and relevant research and detection need to be further carried out. In addition, this Ts-TTPA can be expressed in large quantities and is easier to obtain than excretory-secretory product antigens. Due to the trypsin activity of this protein, future functional experiments and host interaction studies should be done.
Previous studies have also reported the protective efficacy of Ts-ML-EVs as a vaccine, but which molecules were not specified [15], although 53-kDa excretory/secretory antigens and a deoxyribonuclease-2-alpha antigen have been reported to have protective potential [38, 39]. In addition, whether some other proteins, including our candidate proteins, also play a role in the protective immunity related to Ts-ML-EVs needs to be further investigated.
Conclusions
The extracellular vesicles of T. spiralis muscle larvae successfully isolated in this study have specific markers of extracellular vesicles. The protein binds in extracellular vesicles of T. spiralis identified by 25 dpi serum; especially the Ts-TTPA protein with trypsin domain can be the fastest to identify swine serum at 13 dpi and is a candidate antigen for early stage detection of trichinellosis.
Availability of data and materials
All data in this study are available in the data presented and additional files.
References
Bai X, Hu X, Liu X, Tang B, Liu M. Current research of trichinellosis in China. Front Microbiol. 2017;8:1472. https://doi.org/10.3389/fmicb.2017.01472.
Wang N, Bai X, Tang B, Yang Y, Wang X, Zhu H, et al. Primary characterization of the immune response in pigs infected with Trichinella spiralis. Vet Res. 2020;51:17. https://doi.org/10.1186/s13567-020-0741-0.
Brodaczewska K, Wolaniuk N, Lewandowska K, Donskow-Lysoniewska K, Doligalska M. Biodegradable chitosan decreases the immune response to Trichinella spiralis in mice. Molecules. 2017;22:11. https://doi.org/10.3390/molecules22112008.
Sun GG, Wang ZQ, Liu CY, Jiang P, Liu RD, Wen H, et al. Early serodiagnosis of trichinellosis by ELISA using excretory-secretory antigens of Trichinella spiralis adult worms. Parasit Vectors. 2015;8:484. https://doi.org/10.1186/s13071-015-1094-9.
Wang N, Bai X, Ding J, Lin J, Zhu H, Luo X, et al. Trichinella infectivity and antibody response in experimentally infected pigs. Vet Parasitol. 2021;297:109111. https://doi.org/10.1016/j.vetpar.2020.109111.
Sun GG, Song YY, Jiang P, Ren HN, Yan SW, Han Y, et al. Characterization of a Trichinella spiralis putative serine protease. Study of its potential as sero-diagnostic tool. PLoS Neglect Trop Dis. 2018;12:e0006485. https://doi.org/10.1371/journal.pntd.0006485.
Wang ZQ, Liu RD, Sun GG, Song YY, Jiang P, Zhang X, et al. Proteomic analysis of Trichinella spiralis adult worm excretory-secretory proteins recognized by sera of patients with early trichinellosis. Front Microbiol. 2017;8:986. https://doi.org/10.3389/fmicb.2017.00986.
Gao H, Tang B, Bai X, Wang L, Wu X, Shi H, et al. Characterization of an antigenic serine protease in the Trichinella spiralis adult. Exp Parasitol. 2018;195:8–18. https://doi.org/10.1016/j.exppara.2018.09.009.
Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. https://doi.org/10.1080/20013078.2018.1535750.
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18:116. https://doi.org/10.1186/s12943-019-1041-z.
Tzelos T, Matthews JB, Buck AH, Simbari F, Frew D, Inglis NF, et al. A preliminary proteomic characterisation of extracellular vesicles released by the ovine parasitic nematode. Teladorsagia circumcincta Vet Parasitol. 2016;221:84–92. https://doi.org/10.1016/j.vetpar.2016.03.008.
Samoil V, Dagenais M, Ganapathy V, Aldridge J, Glebov A, Jardim A, et al. Vesicle-based secretion in schistosomes: Analysis of protein and microRNA (miRNA) content of exosome-like vesicles derived from Schistosoma mansoni. Sci Rep. 2018;8:3286. https://doi.org/10.1038/s41598-018-21587-4.
Rooney J, Northcote HM, Williams TL, Cortés A, Cantacessi C, Morphew RM. Parasitic helminths and the host microbiome—A missing “extracellular vesicle-sized” link? Trends Parasitol. 2022;38:737–47. https://doi.org/10.1016/j.pt.2022.06.003.
Kifle DW, Pearson MS, Becker L, Pickering D, Loukas A, Sotillo J. Proteomic analysis of two populations of Schistosoma mansoni-derived extracellular vesicles: 15k pellet and 120k pellet vesicles. Mol Biochem Parasitol. 2020;236:111264. https://doi.org/10.1016/j.molbiopara.2020.111264.
Gao X, Yang Y, Liu X, Xu F, Wang Y, Liu L, et al. Extracellular vesicles from Trichinella spiralis: Proteomic analysis and protective immunity. PLoS Negl Trop Dis. 2022;16:e0010528. https://doi.org/10.1371/journal.pntd.0010528.
Liu Y, Xu N, Li Y, Tang B, Yang H, Gao W, et al. Recombinant cystatin-like protein-based competition ELISA for Trichinella spiralis antibody test in multihost sera. PLoS Negl Trop Dis. 2021;15:e0009723. https://doi.org/10.1371/journal.pntd.0009723.
Li F, Cui J, Wang ZQ, Jiang P. Sensitivity and optimization of artificial digestion in the inspection of meat for Trichinella spiralis. Foodborne Pathog Dis. 2010;7:879–85. https://doi.org/10.1089/fpd.2009.0445.
Protein Structure Prediction Server (PSIPRED). http://bioinf.cs.ucl.ac.uk/psipred Accessed 28 Apr 2023.
Pfam. http://pfam-legacy.xfam.org/ Accessed 28 Apr 2023.
SWISS-MODEL. https://swissmodel.expasy.org/ Accessed 28 Apr 2023.
ProtParam. https://www.expasy.org/resources/protparam Accessed 28 Apr 2023.
Gene Ontology Resource. http://geneontology.org/ Accessed 20 Mar 2023.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. https://doi.org/10.1093/molbev/msy096.
Bruschi F, Gómez-Morales MA, Hill DE. International Commission on Trichinellosis: Recommendations on the use of serological tests for the detection of Trichinella infection in animals and humans. Food Waterborne Parasitol. 2019;14:e00032. https://doi.org/10.1016/j.fawpar.2018.e00032.
Wang ZQ, Shi YL, Liu RD, Jiang P, Guan YY, Chen YD, et al. New insights on serodiagnosis of trichinellosis during window period: early diagnostic antigens from Trichinella spiralis intestinal worms. Infect Dis Poverty. 2017;6:41. https://doi.org/10.1186/s40249-017-0252-z.
Ofir-Birin Y, Regev-Rudzki N. Extracellular vesicles in parasite survival. Science. 2019;363:817–8. https://doi.org/10.1126/science.aau4666.
Greening DW, Xu R, Ale A, Hagemeyer CE, Chen W. Extracellular vesicles as next generation immunotherapeutics. Semin Cancer Biol. 2023;90:73–100. https://doi.org/10.1016/j.semcancer.2023.02.002.
Marcilla A, Martin-Jaular L, Trelis M, de Menezes-Neto A, Osuna A, Bernal D, et al. Extracellular vesicles in parasitic diseases. J Extracell Vesicles. 2014;3:25040. https://doi.org/10.3402/jev.v3.25040.
Hansen EP, Fromm B, Andersen SD, Marcilla A, Andersen KL, Borup A, et al. Exploration of extracellular vesicles from Ascaris suum provides evidence of parasite-host cross talk. J Extracell Vesicles. 2019;8:1578116. https://doi.org/10.1080/20013078.2019.1578116.
Nowacki FC, Swain MT, Klychnikov OI, Niazi U, Ivens A, Quintana JF, et al. Protein and small non-coding RNA-enriched extracellular vesicles are released by the pathogenic blood fluke Schistosoma mansoni. J Extracell Vesicles. 2015;4:28665. https://doi.org/10.3402/jev.v4.28665.
Wu Z, Wang L, Li J, Wang L, Wu Z, Sun X. Extracellular vesicle-mediated communication within host–parasite interactions. Front Immunol. 2018;9:3066. https://doi.org/10.3389/fimmu.2018.03066.
Song YY, Zhang Y, Ren HN, Sun GG, Qi X, Yang F, et al. Characterization of a serine protease inhibitor from Trichinella spiralis and its participation in larval invasion of host’s intestinal epithelial cells. Parasit Vectors. 2018;11:499. https://doi.org/10.1186/s13071-018-3074-3.
Wang J, Jin X, Li C, Chen X, Li Y, Liu M, et al. In vitro knockdown of TsDNase II-7 suppresses Trichinella spiralis invasion into the host’s intestinal epithelial cells. PLoS Negl Trop Dis. 2023;17:e0011323. https://doi.org/10.1371/journal.pntd.0011323.
Page MJ, Di Cera E. Serine peptidases: classification, structure and function. Cell Mol Life Sci. 2008;65:1220–36. https://doi.org/10.1007/s00018-008-7565-9.
Xue Y, Zhang B, Wang N, Huang HB, Quan Y, Lu HN, et al. Oral vaccination of mice with Trichinella spiralis putative serine protease and murine interleukin-4 DNA delivered by invasive Lactiplantibacillus plantarum elicits protective immunity. Front Microbiol. 2022;13:859243. https://doi.org/10.3389/fmicb.2022.859243.
Long SR, Liu RD, Kumar DV, Wang ZQ, Su CW. Immune protection of a helminth protein in the DSS-induced colitis model in mice. Front Immunol. 2021;12:664998. https://doi.org/10.3389/fimmu.2021.664998.
Li C, Bai X, Liu X, Zhang Y, Liu L, Zhang L, et al. Disruption of epithelial barrier of caco-2 cell monolayers by excretory secretory products of Trichinella spiralis might be related to serine protease. Front Microbiol. 2021;12:634185. https://doi.org/10.3389/fmicb.2021.634185.
Xu D, Tang B, Yang Y, Cai X, Jia W, Luo X, et al. Vaccination with a DNase II recombinant protein against Trichinella spiralis infection in pigs. Vet Parasitol. 2021;297:109069. https://doi.org/10.1016/j.vetpar.2020.109069.
Xu D, Bai X, Xu J, Wang X, Dong Z, Shi W, et al. The immune protection induced by a serine protease from the Trichinella spiralis adult against Trichinella spiralis infection in pigs. PLoS Negl Trop Dis. 2021;15:e0009408. https://doi.org/10.1371/journal.pntd.0009408.
Acknowledgements
We thank Yang Wang and Lixiao Zhang for their help in the use of flow detection equipment, Yi Xin for his help in the operation of scanning electron microscopy and Hongju Dong for his help in the operation of nanoparticle size and zeta potentiometer.
Funding
This study was supported by the National Key Research and Development Program of China (2021YFC2600202-LXL), the National Natural Science Foundation of China (NSFC32230104-LMY, 82201959-JXM) and Jilin University Young Teachers and Students Cross-disciplinary Cultivation Project (2022-JCXK-22).
Author information
Authors and Affiliations
Contributions
Methodology: CL, FX, HW. Conceptualization: CL, XB. Investigation: CL, YZ, RW. Writing—original draft: CL, XJ. Writing—review and editing: ML, YY, XY. Funding acquisition: XL, ML, XJ, CL. The authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All animal experiments were in strict accordance with the Regulations on the Administration of Laboratory Animals issued by the Ministry of Science and Technology, PRC, and the Animal Welfare Regulations AWR of the US Public Health Service. Animal protocols were reviewed and approved by the Ethics Committee of Jilin University, affiliated with the Provincial Animal Health Committee, Jilin Province, China (Ethical Clearance No. 201803022).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, C., Li, C., Xu, F. et al. Identification of antigens in the Trichinella spiralis extracellular vesicles for serological detection of early stage infection in swine. Parasites Vectors 16, 387 (2023). https://doi.org/10.1186/s13071-023-06013-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-023-06013-7