Abstract
Background
Indonesia is home to many species of non-human primates (NHPs). Deforestation, which is still ongoing in Indonesia, has substantially reduced the habitat of NHPs in the republic. This has led to an intensification of interactions between NHPs and humans, which opens up the possibility of pathogen spillover. The aim of the present study was to determine the prevalence of malarial parasite infections in NHPs in five provinces of Indonesia in 2022. Species of the genus Anopheles that can potentially transmit malarial pathogens to humans were also investigated.
Methods
An epidemiological survey was conducted by capturing NHPs in traps installed in several localities in the five provinces, including in the surroundings of a wildlife sanctuary. Blood samples were drawn aseptically after the NHPs had been anesthetized; the animals were released after examination. Blood smears were prepared on glass slides, and dried blood spot tests on filter paper. Infections with Plasmodium spp. were determined morphologically from the blood smears, which were stained with Giemsa solution, and molecularly through polymerase chain reaction and DNA sequencing using rplU oligonucleotides. The NHPs were identified to species level by using the mitochondrial cytochrome c oxidase subunit I gene and the internal transcribed spacer 2 gene as barcoding DNA markers. Mosquito surveillance included the collection of larvae from breeding sites and that of adults through the human landing catch (HLC) method together with light traps.
Results
Analysis of the DNA extracted from the dried blood spot tests of the 110 captured NHPs revealed that 50% were positive for Plasmodium, namely Plasmodium cynomolgi, Plasmodium coatneyi, Plasmodium inui, Plasmodium knowlesi and Plasmodium sp. Prevalence determined by microscopic examination of the blood smears was 42%. Species of the primate genus Macaca and family Hylobatidae were identified by molecular analysis. The most common mosquito breeding sites were ditches, puddles and natural ponds. Some of the Anopheles letifer captured through HLC carried sporozoites of malaria parasites that can cause the disease in primates.
Conclusions
The prevalence of malaria in the NHPs was high. Anopheles letifer, a potential vector of zoonotic malaria, was identified following its collection in Central Kalimantan by the HLC method. In sum, the potential for the transmission of zoonotic malaria in several regions of Indonesia is immense.
Graphical Abstract

Similar content being viewed by others
Background
Malaria, which is one of the oldest infectious diseases, remains a public health problem in Indonesia despite a significant reduction in its incidence in the western parts of the archipelago. Malaria is caused by species of the apicomplexan parasite genus Plasmodium, which are transmitted between their vertebrate hosts by mosquito vectors. In Southeast Asia, at least 13 Plasmodium species are able to infect non-human primates (NHPs), and seven of them can naturally infect macaques [1,2,3]. Infection of their natural hosts by any one of these seven Plasmodium spp., i.e., Plasmodium cynomolgi, Plasmodium brasilianum, Plasmodium eylesi, Plasmodium knowlesi, Plasmodium inui, Plasmodium schwetzi, and Plasmodium simium, usually results in very low parasitemia and causes mild or asymptomatic disease [4]. Plasmodium falciparum, P. vivax, P. malariae, and P. ovale are commonly found in humans, but P. knowlesi, which causes zoonotic malaria throughout Southeast Asia, also infects macaques naturally [5, 6]. Thirty-eight species of NHPs have been identified in Indonesia [7,8,9]; nine of these are macaque species endemic to the country and include the Mentawai pig-tailed macaque (Macaca pagensis), the black macaque (Macaca nigra), the Moor macaque (Macaca maura), Heck’s macaque (Macaca hecki), the booted macaque (Macaca ochreata), the Tonkean macaque (Macaca tonkeana), the Buton macaque (Macaca brunnescens), and the Gorontalo macaque (Macaca nigrescens) [10, 11]. Is it considered prudent to monitor wild macaques for the Plasmodium spp. that they harbor, and the mosquito vectors of these parasites, to better understand the epidemiology of the disease so that feasible, effective measures can be proposed to reduce the risk of zoonotic malaria infection.
Within the last three decades, deforestation for agriculture, mining, and resettlement has substantially reduced the habitat of NHPs in the western part of Indonesia, including the islands of Sumatra, Java, Kalimantan (the Indonesian part of Borneo), Sulawesi, Bali, and the Lesser Sunda Islands [12]. This has led to the intensification of interactions between NHPs and humans in many parts of Indonesia and thus opened up the possibility of pathogen spillover from NHPs, and other wild animals, to humans, as evidenced by reports of cases of zoonotic malaria in Sumatra and Kalimantan [13,14,15]. The aim of the present study was to determine the prevalence of malarial parasite infections in NHPs in five provinces of Indonesia in 2022. The wild NHPs were captured using traps installed in several localities, including in the surroundings of a wildlife sanctuary that borders settlements, and were released after examination. Settlers who live adjacent to the wildlife sanctuary were investigated for zoonotic malaria infections. Mosquito vectors of malaria were monitored in the five provinces.
Methods
Study sites
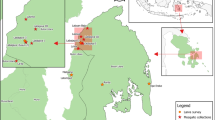
Sampling of NHPs and mosquito vectors was conducted in the following five provinces of Indonesia: Central Java (Cikakak wildlife sanctuary and Kalisalak village in Banyumas Regency); North Sumatra (forest adjacent to Aek Batang Paya village, Sipirok Subdistrict, South Tapanuli Regency); Aceh (Iboih village, Sukakarya Subdistrict, Sabang Municipality; the village is adjacent to Weh Island Natural Park); West Sumatra (Mount Meru, Lubuk Begalung subdistrict and Lubuk Minturun, Koto Tangah district, Padang City Regency); and Central Kalimantan (Nyaru Menteng Arboretum, Taman Wisata Bukit Tangkiling Nature Park, and a settlement near the urban forest of Palangkaraya) (Fig. 1).
Collection of NHPs
Traps were installed in NHP study sites from August to November 2022 and monitored every day for the presence of trapped NHPs. The NHPs were anesthetized by intramuscular injection with Zoletil (4 mg/kg body weight) (Virbac). Sex, morphological characteristics, weight, and body temperature were recorded for each animal. Blood (3-ml samples) was drawn and then transferred into tubes containing ethylenediaminetetraacetic acid. Thin and thick blood smears were prepared on glass slides, and three drops of blood were collected on Whatman 3 MM filter papers for DNA extraction and molecular analysis.
Mass blood survey
Mass blood surveys were conducted in Aceh and Central Java. Residents of the village adjacent to the wildlife sanctuary were asked if they would like to voluntarily participate in the study after they had received information about it. Blood was obtained from the participants by aseptic finger prick and the drops collected on Whatman filter paper for dried blood spot testing and on a glass slide for the preparation of thin and thick blood smears, in accordance with standard procedures of the World Health Organization [16].
Microscopic analysis
Thick and thin blood films of the NHPs and human participants were stained with Giemsa and examined under light microscopy using a 100x objective lens with immersion oil for the presence of malaria parasites. At least 200 microscopic fields of the thick smear were examined to determine parasite densities; if no parasites were found the sample was considered negative.
Mosquito breeding site survey
Water bodies in the study areas were surveyed for the presence of mosquito larvae using a standardized World Health Organization protocol [17]. Larvae were recorded at various sampling locations to identify and evaluate breeding habitats. The different types of water bodies were characterized according to depth, water clarity, and vegetation, and the geographic coordinates recorded using the Open Data Kit (ODK) Collect application. The mosquito larvae in each water body were counted and collected using a dipper and pipette. The salinity and pH of the water were recorded during sampling.
Collection of adult mosquitoes and DNA extraction
Mosquito species diversity was determined using the human landing catch (HLC) method. Residents that voluntarily participated in the study were trained in the HLC method. Two collectors were assigned per house, one positioned indoors and one outdoors (on the veranda). The volunteers collected the mosquitoes that landed on their exposed lower legs by using a mouth aspirator. Collections were conducted for 50 min every hour from 1800 to 0600 hours. The mosquitoes were immediately killed in the field with chloroform vapor and transported in a tube containing silica gel. The adult mosquitoes were identified to species based on their morphology by the use of illustrated keys [18]. A randomly selected subset of morphologically identified samples was also molecularly identified by sequencing the internal transcribed spacer 2 (ITS2) and cytochrome oxidase subunit I (COI) genes [19,20,21,22,23]. These samples were chosen to represent all of the morphologically identified specimens and the locations in which they were collected. The mosquito samples were ground with a Teflon pestle and then transferred to 1.5-ml Eppendorf microtubes containing 50 µl of water and 100 µl saponin. Mosquito DNA was extracted using a Chelex-100 ion exchanger (Bio-Rad Laboratories, Hercules, CA) according to a published procedure [24]. The DNA was either used immediately for polymerase chain reaction (PCR) or stored at -20 °C for later analysis.
DNA extraction for NHPs and parasites
Parasite DNA and NHP DNA was extracted from the NHP blood samples using a QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). The DNA was either used immediately for PCR amplification or stored at − 20 °C for later analysis.
Molecular analyses
Extracted genomic DNA was amplified by PCR targeting the ITS2 and COI genes for species identification of the mosquitoes, and only COI for the NHP samples. The forward and reverse gene-specific oligonucleotides for ITS2 were ITS2A (5ʹ-TGTGAACTGCAGGACACAT-3ʹ) and ITS2B (5ʹ-TATGCTTAAATTCAGGGGGT-3ʹ) [24], respectively; the universal primers used for COI were LCO1490 (5'-GGTCAACAAATCATAAAGATATTGG-3') and HCO2198 (5'-TAAACTTCAGGGTGACCAAAA AATCA-3ʹ) [25]. The PCR conditions for COI amplification were as follows: incubation at 95 °C for 1 min, followed by 35 cycles at 95 °C for 15 s, 53 °C for 15 s, and 72 °C for 60 s, with final extension at 72 °C for 5 min. For Plasmodium sp. detection, a semi-nested PCR assay was used targeting the small subunit ribosomal RNA genes, using primers rplU1 (5ʹ-TCAAAGATTAAGCCATGCAAGTGA-3ʹ) and rplU5 (5ʹ-CCTGTTGTTGCCTTAAACTCC-3ʹ), and a second PCR using rplU1 and rplU4 (5ʹ-TACCCGTCATAGCCATGTTAGGCCAATACC-3ʹ) [22]. The PCR conditions were as described in [26]. All the amplicons were visualized by using gel electrophoresis with 1–2% agarose gels. The consensus sequences of these small subunit ribosomal RNA genes were compared to those in the NCBI nr database by using BLAST to confirm species identification.
Results
In the North Sumatra site, the NHP habitat partly comprised a plantation. In the other sites, the NHPs lived in their natural habitat but also interacted with settlers in the surroundings. A total of 110 NHPs were captured during a 20-day period in the traps that had been installed in the five study areas. The baseline characteristics of the NHPs are shown in Table 1. Sixty-one of the NHPs were male and 49 were female. Body weight of the NHPs ranged from 3.4 to 5.5 kg, and the average body temperature from 36.7 to 39.5 °C. The NHPs were either macaques (family Cercopithecidae) and gibbon (family Hylobatidae). Most of the macaques that were trapped were long-tailed macaques, and a few were pig-tailed macaques. The distributions of the NHPs sampled in this study are summarized in Table 2.
Molecular identification of the NHP species
PCR amplification of the COI gene was successful for 93 of the captured NHPs and unsuccessful for 17 of them. Amplicon sequencing confirmed that these 93 NHPs included individuals of the two species of macaque identified morphologically, namely Macaca fascicularis and Macaca nemestrina. Two species of the family Hylobatidae were also identified molecularly, Hoolock hoolock and Hylobates albibarbis. Variation in the DNA sequence of the COI gene of M. fascicularis was noted between some of the study sites.
Prevalence of Plasmodium spp. infections in NHPs
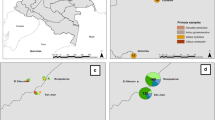
Of the total 110 NHPs captured in the five provinces, Plasmodium infections were morphologically diagnosed in 47 individuals (Table 3). However, molecular analysis using the ribosomal DNA or mitochondrial COI genes indicated that 55 of the NHPs were infected with Plasmodium spp. (Table 3), namely Plasmodium inui, Plasmodium cynomolgi, Plasmodium coatneyi, Plasmodium knowlesi, and other Plasmodium spp. (Table 4; Figs. 2, 3, 4). The highest rate of Plasmodium infection in the macaques was in Iboih village, Aceh. No P. knowlesi infection was found among the macaques captured in Aceh or North Sumatra. The most prevalent species were P. inui and P. cynomolgi, followed by P. coatneyi and then P. knowlesi (Table 4). Plasmodium sp. was identified in 16 of the NHP positive for malaria. Two species of the genus Hylobates identified in this study were not infected with any species of Plasmodium.
Morphology of the primate malaria species
Figures 2, 3 and 4 show morphological characteristics of different stages of the Plasmodium species found in this study. Several blood stages of P. cynomolgi are shown in Fig. 2. The infected red blood cells are distinctly enlarged. Plasmodium coatneyi and P. knowlesi are difficult to distinguish morphologically in thick and thin blood smears. Plasmodium inui, which can cause malaria in NHPs, clearly shows a pigmented trophozoite stage in the red blood cells.
Mosquito breeding sites
Fourteen types of mosquito breeding sites were found (Table 5; Fig. 5), of which ditches and puddles were the most common. The types of mosquito breeding sites that contained Anopheles larvae included artificial water containers, coconut shells, ditches, hoof prints, man-made ponds, mangroves, natural ponds, paddy fields, puddles, springs, stream margins, swamps, tire tracks, and wells. The pH of the breeding sites ranged from 2.5 to 9.6, and the salinity from 0 to 23 p.p.m.
Collection of adult mosquitoes
Species of the genus Anopheles and other species of mosquito were collected in Central Java, North Sumatra, Aceh, and Central Kalimantan (Table 6). Mosquito collection was limited to larvae only in West Sumatra due to time constraints. Many species of mosquito in addition to Anopheles spp. were collected by HLC. Mosquito diversity was highest in Central Kalimantan, where the predominant species collected was An. letifer, and lowest in Central Java. The predominant species collected in North Sumatra was Anopheles kochi and in Aceh Anopheles dirus.
Morphological and molecular identification of the mosquitoes
Based on the morphology of the adult mosquitoes, six species of Anopheles could be identified: Anopheles dirus, Anopheles kochi, Anopheles montanus, Anopheles nigerrimus, Anopheles letifer, and Anopheles umbrosus (Table 7). The other identified mosquito species were Armigeres subalbatus, Culex quinquefasciatus, Culex vishnui and Mansonia sp. The molecular analysis confirmed the presence of An. dirus, An. kochi, An. sinensis, An. sundaicus and An. vagus.
Vector incrimination for zoonotic malaria
The PCR amplification of samples from 399 adult Anopheles collected through HLC revealed that two were positive for Plasmodium DNA. These positive samples were identified as An. letifer, captured in the Nyaru Menteng Arboretum in Central Kalimantan (Table 8).
Mass blood survey
Limited mass blood surveys for zoonotic malaria infection were conducted in areas of Aceh and Central Java where there is intense interaction between human and NHPs. In Aceh, 36 residents of Iboih village participated in the blood survey; in Central Java, 123 residents participated in the survey. No cases of zoonotic malaria were found in either study area.
Discussion
There was a high prevalence of Plasmodium infection within the macaque populations examined in the five provinces, with the highest prevalence in Sabang, Aceh. The most common Plasmodium species identified in the five provinces were P. inui and P. cynomolgi. Plasmodium knowlesi, the most common cause of zoonotic malaria, was only found in West Sumatra, Central Kalimantan, and Central Java. Our findings are not consistent with reports of zoonotic malaria in Acen [15]. In Sabang, Aceh, cases of zoonotic malaria were reported to have been caused by P. knowlesi [15, 27], but we found no P. knowlesi infection among the macaques examined in the present study. In addition, Plasmodium cynomolgi, Plasmodium inui, and Plasmodium fieldi were reported to have caused zoonotic malaria in NHP in South Sumatra, Bintan Island, West Java, and Lampung [28,29,30]. The discrepancies between these and our findings indicate the importance of regular monitoring for the detection of zoonotic malaria in these areas and the implementation of mitigation efforts to prevent its spread. The lowest prevalence, which was found in North Sumatra, may be attributable to the fact that fewer NHPs were captured there. It is particularly noteworthy that although human–macaque interactions are relatively intense on the fringes of Cikakak sanctuary, Central Java, no cases of zoonotic malaria cases have ever been reported in the province. This is probably due to the absence from this region of Anopheles vectors that are capable of transmitting primate malaria to humans. Vectors of zoonotic malaria thus far identified are mainly of theAnopheles leucosphyrus group, with a few other species, such as Anopheles kochi and Anopheles letifer, also capable of transmitting the disease [31,32,33,34,35].
Macaca fascicularis was the most common NHP species trapped in the study areas, and the molecular analysis revealed high DNA sequence variation between individuals from different study sites. This latter finding corroborates the identification of different subspecies of M. fascicularis, from Sumatra, Kalimantan, Java, Bali, Sumba, and Timor [10]. Macaca fascicularis live in primary and secondary forest in lowland areas and in highland areas at > 1000 m above sea level. In highland areas, they are typically found in secondary forest or agricultural areas [10]. In the present study, most of the M. fascicularis were trapped in Central Java, West Sumatra and Aceh, which may be explained by the fact that domesticated, captive macaques predominate in North Sumatra and Central Kalimantan. This, in turn, may explain the absence or lower prevalences of Plasmodium spp. infection in these latter two areas. Based on information from villagers in North Sumatra and Central Java, M. fascicularis often scavenge for food in settlements. NHPs in areas popular with tourists in Iboih, Aceh, and Cikakak, Central Java are often fed by the visitors, which causing changes in the natural behavior of these animals and encourages them to spend more time around settlements in search of food. Environmental changes such as deforestation and urban expansion are also linked to an increase in human-macaque interactions, which increase the potential for zoonotic transmission of malaria [36].
Several Anopheles species, including An. dirus collected in Sabang and An. kochi collected in North Sumatra, were identified following their capture through HLC, but only An. letifer, caught in Central Kalimantan, was infected with Plasmodium sp. The DNA of this parasite was found in the head-thoracal parts of the mosquito, which indicated the presence of sporozoites. It is highly likely that An. dirus plays an important role in zoonotic malaria transmission in Sabang, the area in which it was found in the present study, as it has been reported to transmit the disease in many other sites in Southeast Asia [37]. Further surveillance is required to elucidate the role of An. kochi in the transmission of zoonotic malaria in North Sumatra. In Central Kalimantan, and particularly in Nyaru Menteng Arboretum, where orangutans (Pongo pygmaeus) and macaques are protected, An. letifer, which bites the NHPs and settlers that live in the area, has been confirmed as a zoonotic malaria vector. The role of An. letifer as a vector of zoonotic malaria has also been described for Sarawak [38]. The very low number of individuals of Anopheles species sampled during the present study, with the exception of An. kochi, may partly explain the low sporozoite rates reported here.
The intensity of NHP–human interactions in the five areas surveyed in this study indicate the potential for zoonotic malaria transmission where the mosquito vectors of this disease are present. At present, mosquito vector surveillance and mass blood surveys of the settlers who live adjacent to the NHPs in these areas are inadequate. We propose that regular vector surveillance should be mandatory in these areas, as should mass blood surveys, particularly for the settlers who live around the forest habitats of the NHPs and the visitors to them, as they are the groups most at risk of zoonotic malaria infection.
Conclusions
The increased potential for spillover infection of malaria carried by NHPs due to the ongoing reduction in their habitat and consequent increase in their interactions with humans should be monitored by regular vector surveillance and mass blood surveys. A further study is underway in four localities in Indonesia to determine the factors that may contribute to the transmission of zoonotic malaria.
Availability of data and materials
All of the datasets generated and analyzed during this study are included in the manuscript.
References
Jeyaprakasam NK, Liew JWK, Low VL, Wan-Sulaiman WY, Vythilingam I. Plasmodium knowlesi infecting humans in Southeast Asia: what’s next? PLoS Negl Trop Dis. 2020;14:e0008900. https://doi.org/10.1371/journal.pntd.0008900.
Lee KS, Vythilingam I. Plasmodium knowlesi: emergent human malaria in Southeast Asia. In: Parasites and their vectors. Vienna: Springer Vienna; 2013. p. 5–31.
Collins WE. Major animal models in malaria research: simians. In: Malaria: principles and practice of malariology. Edinburgh: Churchill Livingstone; 1988.
Yap NJ, Hossain H, Nada-Raja T, Ngui R, Muslim A, Hoh B, et al. Natural human infections with Plasmodium cynomolgi, P. inui, and 4 other Simian malaria parasites. Malaysia Emerg Infect Dis. 2021;27:2187–91. https://doi.org/10.3201/eid2708.204502.
Sato S. Plasmodium—a brief introduction to the parasites causing human malaria and their basic biology. J Physiol Anthropol. 2021;401:1–13. https://doi.org/10.1186/S40101-020-00251-9.
Subbarao SK. Centenary celebrations article: Plasmodium knowlesi: from macaque monkeys to humans in Southeast Asia and the risk of its spread in India. J Parasit Dis. 2011;35:87–93. https://doi.org/10.1007/S12639-011-0085-9.
Groves C. The what, why and how of primate taxonomy. Int J Primatol. 2004;25:1105–26. https://doi.org/10.1023/B:IJOP.0000043354.36778.55.
Mittermeier RA, Raylands AB, Wilson DE. Handbook of the mammals of the world. 3rd ed. Barcelona: Lynx Editions; 2016.
Roos C, Ramesh B, Supriatna J, Fellowes JR, Groves CP, Nash SD, et al. An updated taxonomy and conservation status review of Asian primates. UCN/SSC Primate Specialist Group. 2014;4:1.
Supriatna J. Field guide to the primates of Indonesia. Cham: Springer International Publishing; 2022.
Mustari AH. Manual Identifikasi dan Bio-Ekologi Spesies Kunci di Sulawesi. PT Penerbit IPB Press, 2020.
Ministry of Environment and Forestry, Republic of Indonesia. Deforestasi Indonesia Turun, Terendah Dalam Sejarah - Kementerian LHK. https://www.menlhk.go.id/site/single_post/3640/deforestasi-indonesia-turun-terendah-dalam-sejarah (Accessed Jan 29 2023).
Said IB, Kouakou YI, Omorou R, Bienvenu A, Ahmed K, Culleton R, et al. Systematic review of Plasmodium knowlesi in Indonesia: a risk of emergence in the context of capital relocation to Borneo? Parasit Vectors. 2022;15:1–9. https://doi.org/10.1186/S13071-022-05375-8.
Setiadi W, Sudoyo H, Trimarsanto H, Sihite BA, Saragih RJ, Juliawaty R, et al. A zoonotic human infection with simian malaria, Plasmodium knowlesi, in Central Kalimantan. Indonesia Malar J. 2016;15:218. https://doi.org/10.1186/S12936-016-1272-Z.
Herdiana H, Irnawati I, Coutrier FN, Munthe A, Mardiati M, Yuniarti T, et al. Two clusters of Plasmodium knowlesi cases in a malaria elimination area, Sabang municipality. Aceh Indonesia Malar J. 2018;17:186. https://doi.org/10.1186/s12936-018-2334-1.
World Health Organization. Basic malaria microscopy. World Health Organization, Geneva PP - Geneva, 2010, https://apps.who.int/iris/handle/10665/44208.
World Health Organization. Manual on practical entomology in malaria / prepared by the World Health Organization Division of Malaria and Other Parasitic Diseases. World Health Organization, Geneva PP - Geneva, https://apps.who.int/iris/handle/10665/42481.
Atmosoedjono S, Bangs M. Illustrated key to the Anopheles of Indonesia. Jakarta: Ministry of Health, 1989.
Lobo NF, Laurent BS, Sikaala CH, Hamainza B, Chanda J, Chunula D, et al. Unexpected diversity of Anopheles species in Eastern Zambia: implications for evaluating vector behavior and interventions using molecular tools. Sci Rep. 2015;5:17952. https://doi.org/10.1038/srep17952.
Laurent BS, Cooke M, Krishnankutty SM, Asih P, Mueller JD, Kahindi S, et al. Molecular characterization reveals diverse and unknown malaria vectors in the Western Kenyan highlands. Am Soc Trop Med Hyg. 2016;94:327–35. https://doi.org/10.4269/ajtmh.15-0562.
Davidson J, Wahid I, Sudirman R, Small ST, Baskin RN, Burton TA, et al. Molecular analysis reveals a high diversity of Anopheles species in Karama, West Sulawesi, Indonesia. Parasit Vectors. 2020;13:379. https://doi.org/10.1186/s13071-020-04252-6.
Beebe NW, Ellis JT, Cooper RD, Saul A. DNA sequence analysis of the ribosomal DNA ITS2 region for the Anopheles punctulatus group of mosquitoes. Insect Mol Biol. 1999;8:381–90. https://doi.org/10.1046/J.1365-2583.1999.83127.X.
Beebe NW. DNA barcoding mosquitoes: advice for potential prospectors. Parasitol. 2018;145:622–33. https://doi.org/10.1017/S0031182018000343.
White TJ, Bruns T, Lee J, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics in PCR protocols. Amsterdam: Elsevier; 1990.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9.
Jiram AI, Hisam S, Reuben H, Husin SZ, Roslan A, Ismail WRW. Submicroscopic evidence of the simian malaria parasite, Plasmodium knowlesi, in an Orang Asli community. Southeast Asian J Trop Med Public Health. 2016;47:4.
Ekawati LL, Johnson KC, Jacobson JO, Cueto CA, Zarlinda I, Elyazar IRFA, et al. Bennett, defining malaria risks among forest workers in Aceh, Indonesia: a formative assessment. Malar J. 2020;19:144. https://doi.org/10.1186/s12936.
Herdiana H, Cotter C, Coutrier FN, Zarlinda I, Zelman BW, Tirta YK, et al. Malaria risk factor assessment using active and passive surveillance data from Aceh Besar, Indonesia, a low endemic, malaria elimination setting with Plasmodium knowlesi, Plasmodium vivax, and Plasmodium falciparum. Malar J. 2016;15:468. https://doi.org/10.1186/s12936-016-1523-z.
Zhang X, Kadir KA, Quintanilla-Zarinan LF, Villano J, Houghton P, Du H, et al. Distribution and prevalence of malaria parasites among long-tailed macaques (Macaca fascicularis) in regional populations across Southeast Asia. Malar J. 2016;15:8. https://doi.org/10.1186/S12936-016-1494-0.
Siregar JE, Faust CL, Murdiyarso LS, Rosmanah L, Saepuloh U, Dobson AP, et al. Non-invasive surveillance for Plasmodium in reservoir macaque species. Malar J. 2015;14:404. https://doi.org/10.1186/s12936-015-0857-2.
Obsomer V, Defourny P, Coosemans M. The Anopheles dirus complex: spatial distribution and environmental drivers. Malar J. 2007;6:26. https://doi.org/10.1186/1475-2875-6-26.
Klein TA, Harrison BA, Dixon SV, Burge JR. Comparative susceptibility of Southeast Asian Anopheles mosquitoes to the simian malaria parasite Plasmodium cynomolgi. J Am Mosq Control Assoc. 1991;7:481–7.
Marchand R. Co-infections of Plasmodium knowlesi, P. falciparum, and P. vivax among humans and Anopheles dirus mosquitoes in southern Vietnam. Emerg Infect Dis. 2011;17:1232–9. https://doi.org/10.3201/eid1707.101551.
De Ang JX, Yaman K, Kadir KA, Matusop A, Singh B. New vectors that are early feeders for Plasmodium knowlesi and other simian malaria parasites in Sarawak Malaysian Borneo. Sci Rep. 2021;111:1–12. https://doi.org/10.1038/S41598-021-86107-3.
Laurent BS, Burton TA, Zubaidah S, Miller HC, Asih PB, Baharuddin A, et al. Host attraction and biting behavior of Anopheles mosquitoes in South Halmahera, Indonesia. Malar J. 2017;16:310. https://doi.org/10.1186/s12936-017-1950-5.
Tananchai C, Pattanakul M, Nararak J, Sinou V, Manguin S, Chareonviriyaphap T. Diversity and biting patterns of Anopheles species in a malaria endemic area, Umphang valley, Tak province, western Thailand. Acta Trop. 2019;190:183–92. https://doi.org/10.1016/J.ACTATROPICA.2018.11.
Van de Straat B, Sebayang B, Grigg MJ, Staunton K, Garjito TA, Vythilingam I, et al. Zoonotic malaria transmission and land use change in Southeast Asia: what is known about the vectors. Malar J. 2022;21:109. https://doi.org/10.1186/s12936-022-04129-2.
Figtree M, Lee R, Bain L, Kennedy T, Mackertich S, Urban M, et al. Plasmodium knowlesi in human Indonesian Borneo. Emerg Menginfeksi Dis. 2010;16:672–4. https://doi.org/10.3201/idul1604.091624.
Acknowledgements
We wish to thank Lembaga Penelitian dan Pengabdian Pada Masyarakat, the University of Hasanuddin, the University of Andalas, Direktorat Riset dan Pengembangan, the University of Indonesia, and the National Research and Innovation Agency Indonesia, for their continued support and encouragement. The authors are grateful to Balai Konservasi Sumber Daya Alam (Ministry of Environment and Forestry, Indonesia) for their assistance in sample collection in the five study sites; Meyby Lempang Eka Putri for her assistance in sampling in Central Java; and Mahfur Zurahman and Kurnia Ilham for their assistance in sampling in Sumatra Barat. These activities comprised part of the doctoral program undertaken by DHP at the University of Indonesia.
Funding
The NHP sample collection and several of the molecular assays were supported by funding from Riset Kolaborasi Indonesia 2022, Ministry of Education, Culture, Research, and Technology, Republic of Indonesia. The collection of mosquito samples and their analysis were supported by the National Research and Innovation Agency Indonesia. The funders had no role in the design of the study or in the collection and interpretation of the data.
Author information
Authors and Affiliations
Contributions
DHP, H, DAS, PBSA, and DS designed the study, supervised the data collection, and contributed to the writing of the manuscript. DHP, H, IER, LP, WS, NI, R, SW, YY, I, HA, and PBSA performed the data collection, laboratory work, and data analysis. DHP, PBSA and DS drafted the manuscript. All the authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved in 2022 by the Ethics Committee for Medical Research, Faculty of Medicine, University of Hasanuddin, Makassar, Indonesia. The ethical approval identifiers for the human subjects are 368/UN4.6.4.5.31/PP36/2022 and 371/UN4.6.4.5.31/PP36/2022, and that for the animals is 367/UN4.6.4.5.31/PP36/2022.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Permana, D.H., Hasmiwati, Suryandari, D.A. et al. The potential for zoonotic malaria transmission in five areas of Indonesia inhabited by non-human primates. Parasites Vectors 16, 267 (2023). https://doi.org/10.1186/s13071-023-05880-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-023-05880-4