Abstract
Background
The genus Huffmanela Moravec, 1987 (Nematoda, Trichosomoididae, Huffmanelinae), represents a group of nematodes that infect both marine and freshwater fish, and the main gross feature of infection with different species of the genus is the presence of noticeable dark spots or tracks within the parasitized tissues. The purpose of this study was to describe morphologically and morphometrically the eggs of a new marine species of Huffmanela (Huffmanela persica sp. nov.), which was found in the form of black spots in the ovary and the tunica serosa of the stomach of the daggertooth pike conger (Muraenesox cinereus). The new species differs from Huffmanela hamo, another species reported from musculature of this host in Japan, in egg metrics, eggshell features and targeted organ. Molecular identification and pathological examination of the lesions caused by the new species are also reported.
Methods
Nematode eggs with varying degrees of development were separated from the infected tissues (ovary and tunica serosa of stomach) and investigated using light and scanning electron microscopy. Different species-specific markers (small subunit ribosomal DNA, 18S; large subunit ribosomal DNA, 28S; internal transcribed spacer, ITS) were used for molecular identification and phylogenetic study of the new species. Infected tissues were fixed in buffered formalin for pathological investigations.
Results
The fully developed eggs of H. persica sp. nov. are distinguished from those previously described from this host on the basis of their measurements (size, 54–68 × 31–43 µm; polar plugs, 6.4–9.7 × 8.4–12 µm; shell thickness, 3.5–6.1 µm) and a delicate but ornate uterine layer (UL) covering the entire eggshell including the polar plugs. Histopathological examination revealed a fibro-granulomatous inflammation in the ovary and the serosal layer of the stomach of infected fish. Maximum-likelihood (ML) phylogenetic analysis recovered a sister relationship between the new species of marine origin and Huffmanela species previously collected from freshwater hosts.
Conclusions
The present study is the first to report the molecular characterization and phylogenetic position of a teleost-associated marine species of the genus Huffmanela. A comprehensive list of nominal and innominate populations of Huffmanela is also provided.
Graphical abstract

Similar content being viewed by others
Background
Huffmanellosis, which is known to occur in a variety of fish species inhabiting marine and freshwater environments, is a parasitic infection caused by members of the genus Huffmanela Moravec, 1987 (Nematoda, Trichinelloidea, Trichosomoididae, Huffmanelinae) [1]. To date, 23 nominal species and approximately 12 innominate species of the genus Huffmanela have been described or reported, most of which were distinguished based on morphology of their remarkable eggs (eggs generally are regarded as syntypes) without collecting the rarely found adult worms [2]. Many previously recognized congeneric species were collected from marine fishes distributed across global ocean ecosystems. Infections due to freshwater species of Huffmanela were, however, only reported in certain fish belonging to Centrarchidae and Poeciliidae in the USA and Tetraodontidae in Brazil [3,4,5,6]. The amphipod crustacean Hyalella serves as the experimental intermediate host in the life cycle of freshwater populations of Huffmanela (Huffmanela huffmani, Moravec, 1987) [7, 8]. There is lack of information regarding the life cycle and biology of marine species of Huffmanela, but closely related amphipods of marine origin probably act as their intermediate hosts [9]. In general, eggs are ingested by the intermediate host, and then by a fish as the definitive host, where the larval nematodes mature to the adult forms. After mating, the adult females lay numerous eggs within the species-specific tissues, and shortly afterwards the adult nematodes begin to disappear [3, 10]. Eggs continue to develop and appear mostly as grossly visible dark spots in the infected sites [11, 12]. The presence of dark discolored spots or tracks originated from eggs deposited in fish tissue as the result of movements of adult female worm is pathognomonic of Huffmanela infection [13]. Extra-intestinal infections with eggs and adult forms of the histozoic parasite Huffmanela have been recorded in a diverse range of teleost and elasmobranch fish, mainly in skin, swim bladder, serous membrane, mesentery, muscle, gonads and bone [14, 15].
Generally, huffmanellosis is not a severe problem in wild fish. Nonetheless, infections observed in external organs and musculature (flesh) could negatively impact the marketability of affected fish and result in their rejection by final consumers [16]. In addition, infection diagnosed in the swim bladder was found to reduce the physiological efficiency of this specific organ [5]. Eggs associated with Huffmanela species have also been detected in stool samples collected from humans; however, there is no report on the zoonotic potential of Huffmanela species from a public health perspective [17,18,19].
In the present study, we describe a new species of Huffmanela (Huffmanela persica sp. nov.) from the daggertooth pike conger (Muraenesox cinereus) according to morphometric and morphological examinations of the parasite eggs using light and scanning electron microscopy, provide for the first time to our knowledge the molecular sequence data associated with three different genetic markers (18S, 28S and ITS) in a teleost-associated marine species of Huffmanela and report the pathological lesions caused by naturally occurring infection with the nematode in the host tissues.
Methods
Sample collection
During January 2021, a total of 20 freshly caught fish (M. cinereus) with average length 86.65 ± 7.74 cm (mean ± SD) were purchased from local fishermen off the coast of Zir Ahak, Bushehr, Iran (28°17' N, 51°13' E). Gross and stereomicroscopic examinations of fish and their organs were respectively performed to detect any parasitic infection. While no adult specimen of H. persica sp. nov. was recovered from the internal organs, the ovary and the serous coat (tunica serosa) of the stomach were found to be infected by discernible dark egg clusters deposited by the female nematode. Infected tissues containing eggs were collected and divided into two portions. The first portion was stored in 10% neutral buffered formalin for morphological and pathological examinations. The second portion was preserved in 70% ethanol for molecular and SEM analyses [15]. Simultaneously, eggs were scraped from unfixed infected tissues with a scalpel, wet mounted and cover slipped on glass slides, and subjected to gentle pressure before being fixed in formalin for measurement of the size of nematode larvae [10, 20].
Morphological and morphometric analyses
To investigate the morphology and morphometry of the eggs, egg clusters within the infected tissues were separated, cleared in glycerol and mounted in glycerin jelly (for temporary mounting) or Canada balsam (for permanent mounting). All measurements were performed using cellSens imaging software integrated with a digital camera (Olympus SC50 CMOS) installed on a compound microscope (Olympus BX-53). Egg measurements and morphological characteristics considered in the present study (see Fig. 1) were according to those previously described [15, 21,22,23,24]. To describe the architecture of the eggs of H. persica sp. nov., the novel anatomical and terminological framework proposed by Bond and Huffman, 2023, was adopted [25]. Measurements are given in micrometers as range (minimum–maximum) followed by mean ± standard deviation (SD) in parentheses. Line drawings were made with the aid of a drawing tube.
Representation of an advanced egg of Huffmanela persica sp. nov. with measurements considered in this study. Total length with protruding polar plug and UL (black line); total length without protruding polar plug and UL (red line); total width with UL (yellow line); total width without UL (light blue line); shell thickness with UL (green line); shell thickness without UL (white line); polar plug width (dark blue line)
Scanning electron microscopy
Nematode eggs fixed in 70% ethanol (stored at 4 °C) were carefully separated, transferred into 70% acetone overnight and dehydrated in an ascending series of acetone ranging from 70 to 90% at 10-min intervals followed by anhydrous acetone three times for 30 min [15]. The samples were subsequently treated with a mixture (1:1 v/v) of anhydrous acetone and hexamethyldisilazane (HMDS, Sigma-Aldrich, Germany) and immersed in HMDS (as the final desiccation step) three times for 60 min each. After several hours of air drying, they were mounted on metal stubs using double-sided adhesive tape, sputter coated with a thin layer (4 nm) of gold in a sample coater (Balzers SCD 050) and examined with a tabletop scanning electron microscope (Hitachi TM-1000, operated at accelerating voltage of 15 kV) equipped with a high-sensitive semiconductor BSE detector.
Preparation of histopathological sections
Infected tissues fixed in formalin were rinsed in water, dehydrated in ascending grades of ethanol, cleared in xylene and impregnated in paraffin. The specimens embedded in paraffin were micro-sectioned at 4 μm thickness using a microtome (Microm HM 360 microtome, MICROM Laborgeräte GmbH, Walldorf, Germany), double stained with hematoxylin and eosin (H&E) and mounted on glass slides [26]. The histological sections were examined using light microscopy and the images were taken with optical magnifications 5×, 10× and 100× (Leica DM 2500).
DNA barcoding
Egg aggregations were removed from the infected tissues preserved in 70% ethanol, transferred into 2-ml Eppendorf tubes, rinsed in nuclease-free water and collected by centrifugation at 9660×g for 1 min. Genomic DNA from the nematode eggs was extracted using the DNeasy Blood and Tissue kit following the manufacturer’s instructions (Qiagen GmbH, Hilden, Germany). Briefly, 180 μl of the lysis buffer ATL and 40 μl of proteinase K (20 mg/ml) were added to each tube containing collected eggs, and the tubes were vortexed for 15 s and incubated overnight on an Eppendorf Thermomixer R (Hamburg, Germany) at 56 °C. Thereafter, 200 μl of the lysis buffer AL was added to each tube followed by incubation at 56 °C for 10 min. The unlysed chitinous material from the eggs was precipitated by centrifugation at 9660×g for 30 s, and the supernatant containing the DNA was transferred to a new 2-ml Eppendorf tube. A total of 200 μl of pure ethanol was added, and the mixture was pipetted into a DNeasy Mini spin column placed in a 2-ml collection tube. After two washing steps using the wash buffers AW1 and AW2, the genomic DNA was eluted in a 1.5-ml tube by adding 35 μl of the buffer AE. Total DNA was also extracted from uninfected host tissue and included in the polymerase chain reaction (PCR) reactions as a negative control. Concentration and purity of the extracted DNA were measured using a NanoDrop 2000 (Thermo Scientific, USA). The concentration of purified DNA from the eggs was found to be 1050 ng/µl and subsequently adjusted at 100 ng/µl before being used for PCR reactions. The 260/280 and 260/230 absorbance ratios were 2.11 and 2.18, respectively. Three different markers were used for molecular identification of the parasite. The 18S rDNA gene was partially amplified by PCR using the primers Nem_18S_F and Nem_18S_R as described previously [27]. PCR amplification of the 28S rDNA was performed using the primer sets D2A and D3B and 28SF and 28SR [28, 29]. The entire ITS region comprising the ITS1, 5.8S gene and the ITS2 was amplified by PCR using the primers NC5 and NC2 [30]. The primer sequences used are listed in Table 1. PCR reactions were performed in 50 µl reaction mixture composed of 25 µl DreamTaq Green PCR master mix (Thermo Scientific), 2 µl 10 pmol/ul forward and reverse primers, 1 µl 25 mM MgCl2 (Thermo Scientific), 15 µl nuclease-free water and 5 µl 100 ng/µl DNA. PCR cycling conditions used in this study were those optimized in previous studies [27,28,29,30]. PCR products were verified on 1% agarose gel, excised from the gel and purified using a MinElute Gel Extraction Kit as per the manufacturer’s protocol (Qiagen GmbH, Hilden, Germany). As purified PCR product amplified from the entire ITS region (ITS1, 5.8S and ITS2) was not successfully sequenced, it was cloned into the pCR™4-TOPO® TA vector using a commercially available kit (TOPO® TA Cloning® Kit, Invitrogen, USA). The resultant construct was transformed into competent cells (Invitrogen™ One Shot™ TOP10 Chemically Competent E. coli), and transformants were plated on Luria–Bertani (LB) agar supplemented with 50 µg/ml kanamycin. Positive clones were then analyzed by colony PCR screening, and plasmid DNA was isolated using the QIAprep Spin Miniprep Kit according to the protocol included in the manual. Purified DNA and plasmid samples were sequenced in both orientations using the same primers used in PCR reactions. Barcode sequencing was carried out using an automated sequencer (ABI 3730 XL) at LGC Biosearch™ Technologies (LGC Genomics GmbH, Berlin, Germany). To extract consensus sequences related to each gene, the obtained sequence files were aligned using the software MEGA X, checked by the program Chromas version 2.6.6 and assembled manually [31].
Phylogenetic analysis
Standard nucleotide BLAST (blastn) was used to compare the target sequences (18S, 28S and ITS) with sequence data previously registered in GenBank repository. All available sequences of different species of genera belonging to the family Trichosomoididae Hall, 1916, and sequence data representing species of important genera within the other known families of the superfamily Trichinelloidea Ward, 1907 (Capillariidae Railliet, 1915; Trichinellidae Ward, 1907; Trichuridae Ransom, 1911; and Cystoopsidae Skryabin, 1923), were retrieved from GenBank and included in phylogenetic analysis (Table 2) [5, 32, 33]. Likewise, only sequences with approximately similar length of those newly generated in this study were selected. A phylogenetic tree was constructed based on the 18S rDNA dataset obtained (as 28S rDNA and ITS sequences related to Huffmanela and its closely related taxa have not been yet deposited in the GenBank sequence database) by maximum likelihood (ML) method using MEGA X software [34]. Multiple alignments were performed using ClustalW implemented in the same software with default parameters. Alignments were then trimmed (at both ends) to the shortest sequence length and best-fit nucleotide substitution model was selected [35, 36]. ML analysis was carried out following the substitution model K2 + G (Kimura 2-parameter with gamma distribution) with 1000 bootstrap replications.
Results
Huffmanela persica sp. nov.
Taxonomic summary
Type definitive host: Daggertooth pike conger, Muraenesox cinereus (Forsskål), Anguilliformes, Muraenesocidae, date of collection, February 2021.
Type locality: Persian Gulf off the coast of Zir Ahak, Bushehr, Iran (28°17' N, 51°13' E).
Site of infection: Ovary, tunica serosa of stomach (Fig. 2A). Conspicuous masses of eggs of H. persica sp. nov. were observed in the ovaries of all 13 fish infected. The egg aggregations were simultaneously found in the serosae of stomach of 3 out of 13 fish infected by the nematode. All fish examined were also infected by larval (viscera) and adult (stomach and intestine) forms of anisakids and raphidascaridids. Adult forms (male and female) of H. persica sp. nov. were not observed.
Macroscopic and microscopic appearance of fully developed eggs of Huffmanela persica sp. nov. A Grossly visible lesions of eggs previously deposited by adult forms of H. persica sp. nov. in the form of dark spots of various size within the infected tissues (ovary and serosa of stomach) of Muraenesox cinereus. B Wet mount prepared from infected ovary illustrating variously oriented advanced eggs of H. persica sp. nov. within the egg clusters
Prevalence: 65% (13 infected out of 20).
Etymology: The specific name persica (Persian) refers to the country of its occurrence (Iran).
Molecular characterization: Nucleotide sequences of the 18S rDNA (OQ418445), 28S rDNA (OQ428648) and ITS rDNA (OQ428649) of the new species have been deposited in GenBank.
Deposited specimens: Syntype eggs collected from the host (M. cinereus) were deposited in the Helminthological Collection, Institute of Parasitology, Biology Centre of the Czech Academy of Sciences, České Budějovice, Czech Republic (catalog no. N-1277).
ZooBank registration:
To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature [37], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:84ABB846-5AF2-4008-97F6-573A68DFD1BB. The LSID for the new species name Huffmanela persica is urn:lsid:zoobank.org:act: urn:lsid:zoobank.org:act:E12D7B01-E298-4BEE-AD69-8C075F84B33E.
Description of eggs
Grossly visible eggs of H. persica sp. nov. were mostly observed in the form of black aggregates with different dimensions which were randomly distributed in the infected tissues (Fig. 2B). Scalpel scrapings obtained from fresh and fixed egg clusters showed thousands of variously oriented eggs with untinted, brown or black eggshells depending on the stage of development of each egg. Four stages of egg development (from less developed to fully developed eggs) are reported herein based upon morphology and morphometry of eggs as well as embryonic stages of development observed in eggs. These include stage I (meiotic stage), stage II (early mitotic embryonated stage), stage III (late embryonated stage) and stage IV (vermiform larvated stage).
Stage I eggs
Stage I eggs (Figs. 3A, a; 4A–D; 5H) were evidently recently laid eggs in probably meiosis I with a distinctly spherical nucleus mainly located centrally in the granular cytoplasm and no developed or two poorly developed plugs at poles. Eggs in stage I spherical shaped (occasionally shrunken and wrinkled), with light amber rigid eggshell walls and no discernable layering. Egg morphometrics (n = 15) recorded in stage I was as follows: total length (with uterine layer, UL) 34.14–44.32 (38.12 ± 2.97); total length (without UL) 30.51–39.51 (34.06 ± 2.76); total width (with UL) 32.65–41.25 (35.50 ± 2.06); total width (without UL) 30.45–37.20 (32.60 ± 1.72); shell thickness (with UL) 3.27–6.07 (4.74 ± 0.81); shell thickness (without UL) 1.54–2.99 (2.47 ± 0.44); nucleus length 8.2–10.71 (9.24 ± 0.87); nucleus width 7.57–10.08 (8.53 ± 0.74).
General morphology and surface ornamentation pattern of eggs of Huffmanela persica sp. nov. in various stages of development. A–H Photomicrographs and a–h corresponding line drawings of individual eggs at different stages of development (scale bars: 20 µm). A, a Eggs in stage I at very early stage, probably meiosis I, with a spherical nucleus in granular cytoplasm and incompletely developed chitinous layer and polar plugs (note early appearance of superficial projections of UL already apparent). B, b Eggs in stage II at later stage of development (probably meiosis II); chitin deposition appears to be complete. C, c Two-celled mitotic stage of early embryonic development (embryonated). D, d Later multicellular stage of embryonic development; chitinous layer still uniformly translucent with no apparent division into outer and inner chitinous layers. E, e Eggs in stage III with bean-like embryo and chitinous layer appearing two-layered under bright-field (light) microscopy with darker inner layer. F, f Tadpole-like embryos with UL appearing to have been partially dislodged from chitinous layer. G, g Eggs in stage IV with darker-brown shell; embryo now vermiform (larvated) and in-folded three times (pretzel stage). H, h Later stage IV egg with chitinous layer very dark brown; larva nearing final development and folded 5–6 times. I–T Photomicrographs of less developed (I–L), moderately developed (M–P) and fully developed (Q–T) eggs, where the first set of images (I, M, Q) represents overview of these variously advanced eggs, and the second (J, N, R), third (K, O, S) and fourth (L, P, T) series of images focus on the pattern of their surface ornamentation by adjusting the focal plane. Black and yellow arrows represent illusions of superficial ridges (well demonstrated in less developed eggs; occasionally appearing as interconnecting ridges, blue arrowhead) and sculptures on the egg surface, respectively. Green arrowheads exhibit irregular protuberances on the eggshell surface. Red arrowheads indicate an illusory spinous appearance in fully developed eggs
Scanning electron micrographs of poorly developed (A–D) and fully developed (E–I) eggs of Huffmanela persica sp. nov. (separated from infected ovary). Less developed eggs spherical shaped (white arrow shows a shrunken and wrinkled egg) and with no evidently developed polar plugs. Fully developed eggs oblong and containing two plugs at poles. Eggs completely surrounded by a UL bearing uniformly based mammiform mounds adorned with tendril-like vermiform appendage emerging from the apex, occasionally adjoined to that of a neighboring mound (green arrowheads). Note the illusory appearance of serrated-like ridges (occasionally in the form of illusory interconnecting ridges, black arrows) in side views which is caused by overlapping of the mound bases (well observed in less developed eggs, red arrowheads). Outer surface of eggshell with irregular protuberances (yellow arrowheads)
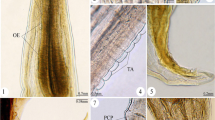
Photomicrographs from histological sections of infected tissues of a daggertooth pike conger eel. A, B Histological sections of the stomach infected by eggs of Huffmanela persica sp. nov. at various stages of development as well as degenerated encysted metazoans (⁎). C, D Sections of the tunica serosa of the stomach parasitized with completely developed eggs. E Sections of the ovarian lamellae infected by both clusters of immature (➔) and developing (➤) eggs, representing immature and previtellogenic oocytes embedded within a loose fibro-granulomatous infiltration containing histiocytes and eosinophilic granular leukocytes (⁎). F Magnified view of histological section of infected ovary showing a fibro-granulomatous infiltrate surrounding clusters of eggs at different stages of development (mainly highly developed eggs, ➤). G High magnification (100×) view of a fully developed egg of H. persica (cross-sectional view) showing larva in-folded within the eggshell and a protruding polar plug at either end. H Less developed eggs with a nearly central nucleus (⁎) and thin eggshell layer and I fully developed eggs containing twisted larvae of H. persica cut on varying planes of section
Stage II eggs
Stage II represents various stages of early embryonic development within the eggshell from single-cell (probably meiosis II) stage to multiple cell mitotic stages (Fig. 3B, b; C, c; D, d). Eggs in the single-cell stage (Fig. 3B, b) were distinguished by having a spherical nucleus present in the granular cytoplasm and two newly developed polar plugs. Eggs in the two-cell stage (Fig. 3C, c) had a symmetric pattern as the result of the first cell division. Eggs containing multi-cell stages (Fig. 3D, d) showed further development of embryo, where the inner space of the rigid eggshell wall was partly or completely occupied by a multicellular mass. Eggs in stage II were elliptical or oblong, light yellow to light brown, with polar plugs (apparently two-layered) slightly or markedly protruding and no distinct shell layers. Egg morphometrics (n = 40) obtained in stage II: total length (inclusive of protruding polar plugs and UL) 46.79–65.84 (58.26 ± 4.83); total length (exclusive of protruding part of polar plugs and UL) 43.24–61.85 (54.08 ± 4.85); total width (with UL) 31.51–40.80 (35.43 ± 2.23); total width (without UL) 30.35–39.52 (33.57 ± 2.35); shell thickness (with UL) 3.43–8.44 (5.20 ± 1.11); shell thickness (without UL) 2.70–7.57 (4.28 ± 1.14); polar plug width 7.52–15.76 (9.78 ± 1.65).
Stage III eggs
Eggs in stage III (Fig. 3E, e; F, f) demonstrating higher development of embryo were characterized by possessing spatially differentiated forms (bean-like or tadpole-like embryos; Fig. 3E, e; F, f), two easily distinguishable apparent shell layers and mild to moderate degrees of embryonic bending within the eggshell. Eggs in stage III elliptical or oblong in shape, brown in color, containing two protruding polar plugs, with bilayer shell inclusive of a thin and light (hyaline) inner layer alongside with a thick and dark outer layer (this layering is an optical illusion under light microscopy caused by the birefringent properties of chitin; see Bond and Huffman, 2023) [25]. Morphometric measurements of eggs (n = 27) in stage III are summarized as follows: total length (with protruding polar plugs and UL) 59.39–70.98 (62.44 ± 2.60); total length (without protruding part of polar plugs and UL) 54.75–61.67 (57.39 ± 1.94); total width (with UL) 32.46–44.75 (34.95 ± 3.03); total width (without UL) 30.62–43.30 (32.88 ± 3.05); shell thickness (with UL) 3.81–5.73 (4.90 ± 0.52); shell thickness (without UL) 2.92–5.02 (3.77 ± 0.46); polar plug width 8.18–11.09 (9.39 ± 0.68).
Stage IV eggs
Stage IV eggs (Figs. 3G, g; H, h; 4E–I; 5G, I) were differentiated by having brown to dark-brown color, oblong or elliptical shape, varying degrees of larval formation within egg from threefold stage (pretzel, Fig. 3G, g) to five–sixfold stage (fully developed larva, Fig. 3H, h), dark outer layer much thicker in width and two distinctly protruding polar plugs. Morphometric data obtained from stage IV eggs (n = 71) were as follows: total length (with protruding polar plugs and UL) 60.37–75.13 (66.62 ± 3.04); total length (without protruding part of polar plugs and UL) 54.37–67.86 (62.03 ± 2.94); total width (with UL) 32.54–44.63 (36.46 ± 2.65); total width (without UL) 30.96–43.39 (34.41 ± 2.78); shell thickness (with UL) 4.48–7.17 (5.63 ± 0.53); shell thickness (without UL) 3.51–6.11 (4.63 ± 0.55); polar plug width 8.43–12.00 (10.29 ± 0.79); larval width in fully developed eggs 6.09–9.16 (7.91 ± 0.61); larval length (n = 17) in fully developed eggs 196.27–231.49 (209.51 ± 10.64).
Egg surface ornamentations
The rigid eggshell wall (or its integral outermost layer, Pellicula Ovi) at all stages adorned with irregular protuberances on the surface (Fig. 4B–D, H, I). Eggs thoroughly surrounded by a thin transparent UL decorated with closely and regularly spaced mammiform mounds (superficial projections, Figs. 3 I–T, 4B–D, F, G) appearing to form pointed serrated-like ridges (Fig. 3J, N, R) (occasionally appearing as interconnecting ridges; Figs. 3J, 4D) on the egg surface (the appearance of ridges is most likely a geometric illusion; see Figs. 1–3, 7, 8 of Žďárská et al. 2001, Fig. 30 of Bond, 2020 and Fig. 24 of Bond and Huffman, 2023) [25], resulting in manifestation of a fine superficial sculpture on the surface of eggs examined with light microscope (Fig. 3K, O, S). UL mammiform mounds predominantly with a tendril-like vermiform appendage extending from the tip, sometimes adjoined to that of a neighboring mound (Fig. 4A–F).
Pathological lesions
The serosal surface of the stomach in infected fish contained eggs of H. persica sp. nov. at various stages of development (Fig. 5A–D) and degenerated encapsulated metazoans (Fig. 5A, B; more likely larval forms of Anisakis and less likely adult forms of Huffmanela as the females are usually extremely long, are rarely found encapsulated and are not much thicker than the eggs), all of which were enclosed in a fibro-granulomatous infiltrate. Ovigerous lamellae of the ovary were infected with nests of developing eggs from the nematode H. persica. The eggs were located within both clusters of immature and previtellogenic oocytes and the connective tissue of the lamellae and encased by a fibro-granulomatous infiltrate containing both histiocytes and eosinophilic granular leukocytes (Fig. 5E, F).
Gene characterization and phylogenetic analysis
Sequencing of purified PCR products of 18S, 28S and ITS regions from H. persica sp. nov. resulted in DNA fragments with different lengths (855 bp, 1264 bp and 1127 bp, respectively). The BLAST analysis revealed that 18S rDNA gene sequence of H. persica sp. nov. shared 93–96% identity (ID) and 83–96% query coverage (QC) with Huffmanela species collected from freshwater bony fish including H. cf. huffmani of Bullard et al., 2022 (ON838248, 93.12% ID and 96% QC), H. huffmani (ON838249, 94.62% ID and 88% QC), H. huffmani (ON838251, 96.08% ID and 83% QC) and H. cf. huffmani (ON838247, 95.80% ID and 83% QC) and 86–89% ID and 81–95% QC with those isolated from marine cartilaginous fish including Huffmanela sp. (ON838247, 86.28% ID and 95% QC) and Huffmanela markgracei Ruiz and Bullard, 2013 (ON838250, 89.38% ID and 81% QC). According to the ML tree generated from 18S rDNA sequences (Fig. 6), H. persica was not grouped with previously reported species of Huffmanela. However, a sister relationship was recovered between the new species of marine origin with freshwater species of Huffmanela (H. huffmani and H. cf. huffmani), all of which are known to infect teleost fish. A sister group relationship was also found between Huffmanela species infecting teleost fish (H. huffmani, H. cf. huffmani and H. persica) with those parasitizing elasmobranch fish (H. markgracei and Huffmanela sp.). Phylogenetic analysis based on 18S rDNA dataset strongly corroborated monophyly of the subfamily Huffmanelinae (Fig. 6).
Maximum likelihood (ML) phylogram reconstructed using the 18S rDNA dataset of the new species Huffmanela persica sp. nov. and other related species within the families Trichosomoididae, Trichinellidae, Trichuridae and Capillariidae. ML analysis was performed using the substitution model K2 + G with 1000 bootstrap replications
Discussion
The description and examination of developing eggs of Huffmanela provide valuable information regarding the taxonomy and biodiversity of the genus [38, 39]. However, differential comparison of various species of the genus Huffmanela based on the eggs has been suggested to be principally performed according to parasitological analyses of fully developed eggs with consideration of host-parasite interactions [15, 39]. Accordingly, a combination of diagnostic characteristics has been previously applied for differentiation of Huffmanela species. These include morphometric analyses (egg measurements), general morphology (color, shape, structure, surface ornamentation), host species, preferred site of infection in the host’s body (specific tissues infected by the parasite) and geographical locality. Huffmanela persica sp. nov. can be principally distinguished from the other accepted and unnamed species of Huffmanela by having a range of features including different size of egg proper, distinct ornamentation of UL and rarely observed eggshell adornment. The dimensions (considering both length and width) of advanced eggs (54–68 × 31–43 µm) of the new species closely or partly resemble those of H. huffmani (54–60 × 30–33 µm) [1], H. cf. huffmani (54–66 × 27–33 µm) [5], H. markgracei plectropomi Justine, 2011 (64–76 × 29–35 µm) [40], H. hamo Justine and Iwaki, 2014 (65–78 × 33–38 µm) [12], Huffmanela moraveci Carballo and Navone, 2007 (50–57 × 23–31 µm) [41], H. longa Justine, 2007 (58–72 × 22–34 µm) [2], H. balista Justine, 2007 (63–78 × 32–41 µm) [2], H. mexicana Moravec and Fajer-Avila, 2000 (63–66 × 30–33 µm) [22], H. selachii Al-Sabi et al., 2022 (63–90 × 30–56 µm) [42] and Huffmanela sp. of Attia et al., 2021b (62–75 × 30–36 µm) [43]. On the other hand, as observed in H. persica sp. nov. in the form of protuberance and mammiform mounds, distinctively shaped ornamentations associated with eggshell or UL have been previously described for Huffmanela schouteni Moravec and Campbell, 1991 (UL with protuberances) [44], H. banningi Van Banning, 1980 (UL apparently spinose) [20], H. huffmani [1], H. japonica Moravec et al., 1998 (eggshell with protuberances) [18], H. canadensis Moravec et al., 2005 (eggshell with transverse ridges) [45], H. lata Justine, 2005 (eggshell apparently spinose) [39], H. balista (eggshell with longitudinal ridges) [2], H. moraveci (eggshell spinose) [41], H. markgracei (shell with transverse ridges) [46], H. oleumimica Ruiz et al., 2013 (eggshell spinous) [15], Huffmanela sp. (eggshell with protuberance) [43], H. cf. huffmani (UL with spines) [5] and H. psittacus Carvalho et al., 2022 (shell with longitudinal, oblique and transverse ridges) [6]. Of the abovementioned species, H. persica sp. nov. can be easily differentiated from H. schouteni, H. banningi, H. oleumimica, H. markgracei, H. lata and H. psittacus by having dissimilar size of eggs (Table 3). Likewise, the eggs of H. persica sp. nov. are wider compared to those of H. japonica and H. moraveci. The eggs of new species are remarkably different from those of Huffmanela plectropomi, H. hamo, H. longa, H. mexicana, H. japonica, H. canadensis, H. moraveci, H. ballista, H. selachii and Huffmanela sp. [43] by possessing unique surface structures (eggshell with protuberances and UL with mammiform mounds adorned with a tendril-like vermiform appendage in H. persica sp. nov. versus smooth eggshell in H. hamo, H. longa, H. mexicana and H. selachii; eggshell with transverse ridges in H. canadensis; eggshell with longitudinal ridges in H. balista; absence of UL in H. plectropomi, H. hamo, H. canadensis and H. selachii; smooth UL in H. japonica, H. moraveci, H. balista and Huffmanela sp. of Attia et al., 2021b). Egg measurements of H. huffmani and H. cf. huffmani [5] closely overlap those of the new species. Nevertheless, these species of freshwater origin can be separated from the marine species H. persica sp. nov. depending upon the characteristics such as eggshell without protuberances (versus eggshell with protuberances) and distinct egg UL with papilla-like spines (versus UL with mammiform mounds). Huffmanela huffmani and H. cf. huffmani were respectively described from freshwater fish within the families Centrarchidae and Poeciliidae in the USA [3, 5].
Muraenesox cinereus is an oceanodromous fish species widely distributed in the Western Indo-Pacific realm including the Persian Gulf [47,48,49]. Previous reports are available on the infection of marine teleost and elasmobranch fish collected from the Persian Gulf region with nominal (H. selachii) [42] and unnamed species of Huffmanela (Huffmanela sp. of Attia et al., 2021a, Huffmanela sp. of Attia et al., 2021b, and Huffmanela sp. of Al-Hasson et al., 2019) [43, 50, 51]. However, H. persica sp. nov. is distinguishable from these species on the basis of egg size, egg overlays, infected tissues and host fish (Table 3). Justine and Iwaki (2014) described the eggs of H. hamo from the somatic musculature of the anguilliform fish M. cinereus (off Japan), which as previously stated is morphologically different from the new species [12]. In this study, however, eggs of H. persica sp. nov. were detected in the ovary and serosa of stomach of the same species, suggesting that tissues parasitized by different populations of Huffmanela are species specific in this genus [11, 45]. The biology of larval and adult forms of the daggertooth pike conger is largely unknown. No food material has ever been detected in the intestine of any anguillid leptocephali inhabiting natural waters. As concluded by Hulet (1978), the gut is poorly differentiated in anguilliform leptocephali, and protoplasmic juices from organisms punctured by the projecting teeth could be absorbed through the epithelium of the esophagus and stomach in larval stages [52]. In a previous study, no captive-reared larvae of pike conger were found to strike at foods such as Chlorella, rotifers, Protozoa, fertilized eggs of sea urchin and phytoplankton [53]. However, pike conger of larger size is an active predator feeding on the bathypelagic-pelagic, demersal and small bottom living organisms including fishes, crustaceans and cephalopods [54,55,56]. The stomach content of pike congers collected from Porto-Novo, West Africa, was found to be predominantly composed of mackerel, Clupeids, Caranx species, squids and flying fish [57]. Another study also revealed that Engraulis japonicus (Engraulidae) was the main preferred prey of pike congers collected from the coastal waters off Goseong, South Korea [58]. Unfortunately, there is a dearth of studies on habitat use, feeding habits and seasonal dietary preferences of pike conger and its preferred prey in the Persian Gulf, and therefore further studies are required to determine whether intermediate or paratenic hosts are involved in the life cycle of H. persica sp. nov.
Masses of eggs of H. persica sp. nov. were observed in the ovary and the tunica serosa of the stomach in infected fish. Similarly, tissues with a serous membrane were infected with eggs of H. schouteni (intestinal serosa) [44], H. longa (mesentery) [2], H. plectropomi (mesentery near swim bladder) [40], H. cf. huffmani (visceral peritoneum) [5] and Huffmanela sp. (mesentery) [51]. Infection with eggs of H. cf. huffmani was also detected in the gonad (testes) of platyfish (Xiphophorus variatus) [5]. In this study, eggs of H. persica were found to be surrounded by epithelioid cells and eosinophilic granulocytes, implicating the inflammatory response resulted from infection with the parasite. Immunopathological responses induced by intercellular and intracellular infection with eggs or worms of different species of Huffmanela have been previously reported in various fish hosts, reflecting the physical and chemical damage caused by the parasite to the infected tissues. These include granulomatous inflammation (characterized mainly by infiltration of mononuclear phagocytes), inter-cellular edema (spongiosis), cellular adaptive response (hypertrophic, atrophic and hyperplastic changes), degenerative and necrotic changes and degenerative and dystrophic calcification [5, 6, 10, 16, 43, 59,60,61,62,63].
The superfamily Trichinelloidea currently comprises five families including Capillariidae, Trichinellidae, Trichuridae, Cystoopsidae and Trichosomoididae [33, 38]. According to Moravec (2001), the family Trichosomoididae includes three subfamilies, Anatrichosomatinae Smith and Chitwood, 1954, Huffmanelinae Moravec, 1987, and Trichosomoidinae Hall, 1916 [38]. Anatrichosomatinae and Huffmanelinae contain only one genus, namely Anatrichosoma (Smith and Chitwood, 1954) and Huffmanela, whereas Trichosomoidinae represents two genera, Trichosomoides (Railliet, 1895) and Trichuroides (Ricci, 1949). Hodda (2022) recently listed the genus Paratrichosoma Ashford and Muller, 1978, in the family Trichosomoididae [33]; however, the genus was previously suggested to be the synonymy of Capillaria and placed in the family Capillariidae [24, 38, 64, 65]. In this study, phylogenetic analysis based on the 18S rDNA sequences recovered a sister relationship between Huffmanela and Trichinella (Railliet, 1895). Sister group relationship between these two genera was also supported by Bayesian phylogenetic analysis in a previous study [5]. Phylogenetic relationships among species of the genus Huffmanela revealed two distinct clades with maximum support. The first clade, Chondrichthyes (elasmobranch)-associated clade, includes Huffmanela species (H. markgracei and Huffmanela sp.) collected from marine elasmobranch fish, whereas the second clade, Osteichthyes (teleost)-associated clade, represents Huffmanela species infecting marine (H. persica) and freshwater (H. huffmani and H. cf. huffmani) teleost fish. However, further nucleotide sequences representing the nuclear and mitochondrial rRNA genes from marine species of Huffmanela infecting teleost fish are required to elucidate whether freshwater species of Huffmanela are derived from marine ancestors [66]. If freshwater species of Huffmanela presented a monophyletic group that is nested within a group of marine taxa, it may support the hypothesis that the genus Huffmanela might have a marine origin [5, 67, 68].
Conclusions
To the best of our knowledge, the present study provides the molecular sequences from the 28S and ITS regions of nuclear rDNA in a species belonging to the genus Huffmanela for the first time. The phylogenetic position of a marine species of Huffmanela was demonstrated for the first time based on analysis of the 18S rDNA sequence data obtained from the new species and those previously described from freshwater species of Huffmanela.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request. Nucleotide sequences of the 18S rDNA (OQ418445), 28S rDNA (OQ428648) and ITS rDNA (OQ428649) of the new species have been deposited in GenBank.
Abbreviations
- 18S:
-
Small subunit ribosomal DNA
- 28S:
-
Large subunit ribosomal DNA
- ID:
-
Identity
- ITS:
-
Internal transcribed spacer
- HMDS:
-
Hexamethyldisilazane
- LSID:
-
Life Science Identifier
- ML:
-
Maximum likelihood
- PCR:
-
Polymerase chain reaction
- QC:
-
Query coverage
- rDNA:
-
Ribosomal DNA
- UL:
-
Uterine layer
References
Moravec F. Revision of capillariid nematodes (subfamily Capillariinae) parasitic in fishes. Praha: Academia; 1987. p. 141.
Justine JL. Huffmanela spp. (Nematoda, Trichosomoididae) parasites in coral reef fishes off New Caledonia, with descriptions of H. balista n. sp. and H. longa n. sp. Zootaxa. 2007;1628:23–41.
Huffman DG, Moravec F. First description of adult Huffmanela huffmani Moravec, 1987 (Nematoda: Trichosomoididae) from the swimbladder of centrarchid fishes of the upper San Marcos River, central Texas. Folia Parasitol. 1988;35:227–34.
Johnson K. Parasites of the Redbreast Sunfish (Lepomis auritus) in the South Concho River of West Central Texas (Doctoral dissertation). 2019;32 p.
Bullard SA, Moravec F, Ksepka SP, Warren MB, Dutton HR, Huffman DG, et al. Huffmanela cf. huffmani (Nematoda: Trichosomoididae) infecting swim bladder, peritoneum, and gonad of variable platyfish, Xiphophorus variatus (Cyprinodontiformes: Poeciliidae) and eastern mosquitofish, Gambusia holbrooki (Poeciliidae) in Florida; taxonomy, phylogenetic analysis, and pathological changes. Parasitol Res. 2022;121:2307–23.
Carvalho EL, Santana RL, Corrêa GC, Sindeaux Neto JL, Pinheiro RH, et al. A new species of Huffmanela (Nematoda: Trichosomoididae) parasitizing Colomesus psittacus (Tetraodontiformes) from Marajó Island, Pará State, Brazil. Rev Bras Parasitol Vet. 2022;22:31.
Bond A. Investigations into Huffmanela (Nematoda Trichosomoididae): New Populations, Life Cycles, and Eggshell Ultrastructure. 2020;70 p.
Worsham ML, Bond A, Gibson JR, Huffman DG. Biogeography of selected spring endemics in texas interglacial-drought refugia with unexpected insights from a spring-dependent nematode parasite. Hydrobiol. 2023;2:97–133.
Worsham ML, Huffman DG, Moravec F, Gibson JR. The life cycle of Huffmanela huffmani Moravec, 1987 (Nematoda: Trichosomoididae), an endemic marine-relict parasite of Centrarchidae from a Central Texas spring. Folia Parasitol. 2016;63:1.
Bullard SA, Ruiz CF, McElwain A, Murray MJ, Borucinska JD, Benz GW. Huffmanela cf. carcharhini (Nematoda: Trichosomoididae: Huffmanelinae) from skin of a sandbar shark, Carcharhinus plumbeus, in the Pacific Ocean. J Parasitol. 2012;98:333–40.
Moravec F, Garibaldi F. Huffmanela paronai sp. n.(Nematoda: Trichosomoididae), a new parasite from the skin of swordfish Xiphias gladius in the Ligurian Sea (Western Mediterranean). Folia Parasitol. 2000;47:309–14.
Justine JL, Iwaki T. Huffmanela hamo sp. N. (Nematoda: Trichosomoididae: Huffmanelinae) from the dagger-tooth pike conger Muraenesox cinereus off Japan. Folia Parasitol. 2014;61:267.
Gillett-Kaufman JL, Wilson F. Fish Nematode Huffmanela spp. (Enoplea: Trichinellida: Trichosomoididae): EENY744/IN1276, 1/2020. EDIS. 2021;2021:4–4.
Žďárská Z, Huffman DG, Moravec F, Nebesářová J. Egg shell ultrastructure of the fish nematode Huffmanela huffmani (Trichosomoididae). Folia Parasitol. 2001;48:231–4.
Ruiz CF, Ray CL, Cook M, Grace MA, Bullard SA. A New Species of Trichosomoididae (Nematoda) from Skin of Red Snapper, Lutjanus campechanus (Perciformes: Lutjanidae), on the Texas-Louisiana Shelf, Northern Gulf of Mexico. J Parasitol. 2013;99:318–26.
Ramos P, Carvalho R, Rosa F, Alexandre-Pires G, Seixas F, Esteves A, et al. Huffmanela lusitana sp. N.(Nematoda: Trichosomoididae) infecting pouting, Trisopterus luscus (Teleostei: Gadidae) off the Atlantic coast of Portugal. Int J Parasitol Parasites Wildl. 2019;9:266–73.
Gállego J, Riera C, Portús M. Huffmanela sp. eggs (Nematoda: Trichosomoididae), as a human spurious parasite in a child from Barcelona (Spain). Folia Parasitol. 1993;40:208–10.
Moravec F, Koudela B, Ogawa K, Nagasawa K. Two new Huffmanela species, H. japonica n. sp. and H. shikokuensis n. sp. (Nematoda: Trichosomoididae), from marine fishes in Japan. J Parasitol. 1998;84:589–93.
Moravec F, Garibaldi F. First record of Huffmanela schouteni (Nematoda: Trichosomoididae), a histozoic parasite of flyingfishes, in Europe. Dis Aquat Org. 2003;57:173–5.
Van Banning P. The occurrence of black spots in the tongue sole, Cynoglossus browni Chabanaud, due to nematode eggs (Capillaria spinosa) previously described in the shark Carcharhinus mttberti Müller & Henle. J Fish Biol. 1980;17:305–9.
Wharton D. Nematode egg-shells. Parasitology. 1980;81:447–63.
Moravec F, Fajer-Avila E. Huffmanela mexicana n. sp. (Nematoda: Trichosomoididae) from the marine fish Sphoeroides annulatus in Mexico. J Parasitol. 2000;86:1229–31.
Schierenberg E. Embryological variation during nematode development. WormBook. 2006. https://doi.org/10.1895/wormbook.1.55.1.
Borba VH, Martin C, Machado-Silva JR, Xavier SC, de Mello FL, Iñiguez AM. Machine learning approach to support taxonomic species discrimination based on helminth collections data. Parasit Vectors. 2021;14:1–5.
Bond AT, Huffman DG. Nematode eggshells: a new anatomical and terminological framework, with a critical review of relevant literature and suggested guidelines for the interpretation and reporting of eggshell imagery. Parasite. 2023;30:6.
Mirzazadeh A, Grafl B, Abbasnia M, Emadi-Jamali S, Abdi-Hachesoo B, Schachner A, et al. Reduced performance due to adenoviral gizzard erosion in 16-day-old commercial broiler chickens in Iran, confirmed experimentally. Front Vet Sci. 2021;8:635186.
Floyd RM, Rogers AD, Lambshead PJ, Smith CR. Nematode-specific PCR primers for the 18S small subunit rRNA gene. Mol Ecol Notes. 2005;5:611–2.
Nadler SA, Hudspeth DS. Ribosomal DNA and phylogeny of the Ascaridoidea (Nemata: Secernentea): implications for morphological evolution and classification. Mol Phylogenet Evol. 1998;10:221–36.
Pereira FB, Tavares LE, Scholz T, Luque JL. A morphological and molecular study of two species of Raphidascaroides Yamaguti, 1941 (Nematoda: Anisakidae), parasites of doradid catfish (Siluriformes) in South America, with a description of R. moraveci n. sp. Syst Parasitol. 2015;91:49–61.
Zhu X, Gasser RB, Podolska M, Chilton NB. Characterisation of anisakid nematodes with zoonotic potential by nuclear ribosomal DNA sequences. Int J Parasitol. 1998;28:1911–21.
Amor N, Farjallah S, Merella P, Said K, Ben SB. Molecular characterization of Hysterothylacium aduncum (Nematoda: Raphidascaridae) from different fish caught off the Tunisian coast based on nuclear ribosomal DNA sequences. Parasitol Res. 2011;109:1429–37.
Smythe AB, Holovachov O, Kocot KM. Improved phylogenomic sampling of free-living nematodes enhances resolution of higher-level nematode phylogeny. BMC Evol Biol. 2019;19:1–5.
Hodda M. Phylum Nematoda: a classification, catalogue and index of valid genera, with a census of valid species. Zootaxa. 2022;5114:1–289.
Pereira FB, Luque JL. Morphological and molecular characterization of cucullanid nematodes including Cucullanus opisthoporus n. sp. in freshwater fish from the Brazilian Amazon. J Helminthol. 2017;91:739–51.
Soliman H, Lewisch E, El-Matbouli M. Identification of new genogroups in Austrian carp edema virus isolates. Dis Aquat Org. 2019;136:193–7.
Sudhagar A, El-Matbouli M, Kumar G. Identification and expression profiling of toll-like receptors of brown trout (Salmo trutta) during proliferative kidney disease. Int J Mol Sci. 2020;21:3755.
International Commission on Zoological Nomenclature (ICZN). Amendment of articles 8, 9, 10, 21 and 78 of the international code of zoological nomenclature to expand and refine methods of publication. Bull Zool Nomencl. 2012;69:161–9.
Moravec F. Trichinelloid nematodes parasitic in cold-blooded vertebrates. Academia: Praha; 2001. p. 429.
Justine JL. Huffmanela lata n. sp. (Nematoda: Trichosomoididae: Huffmanelinae) from the shark Carcharhinus amblyrhynchos (Elasmobranchii: Carcharhinidae) off New Caledonia. Syst Parasitol. 2005;61:181–4.
Justine JL. Huffmanela plectropomi n. sp. (Nematoda: Trichosomoididae: Huffmanelinae) from the coralgrouper Plectropomus leopardus (Lacépède) off New Caledonia. Syst Parasitol. 2011;79:139–43.
Carballo MC, Navone GT. A new Huffmanela species (Nematoda: Trichosomoididae) parasitizing atherinid fishes in north Patagonian gulfs, Argentina. J Parasitol. 2007;93:377–82.
Al-Sabi MN, Ibrahim MM, Al-Hizab F, Al-Jabr OA, Al-Shubaythi S, Huffman DG. Huffmanela selachii n. sp. (Nematoda: Trichosomoididae: Huffmanelinae): a new species infecting the skin of the great hammerhead shark (Sphyrna mokarran) and the blacktip reef shark (Carcharhinus melanopterus) in the Arabian Gulf, off-shore Saudi Arabia. Saudi J Biol Sci. 2022;29:103430.
Attia MM, Mahmoud MA, Ibrahim MM. Morphological and pathological appraisal of Huffmanela sp. (Nematoda: Trichosomoididae) infecting orange-spotted grouper (Epinephelus coioides, Hamilton, 1822) at Jubail Province, Saudi Arabia: a case report. J Parasit Dis. 2021;45:980–5.
Moravec F, Campbell BG. A new Huffmanela species, H. schouteni sp. n. (Nematoda: Trichosomoididae) from flying fishes in Curaçao. Folia Parasitol. 1991;38:29–32.
Moravec F, Conboy GA, Speare DJ. A new trichosomoidid from the skin of Sebastes spp. (Pisces) from British Columbia Canada. J Parasitol. 2005;91:411–4.
Ruiz CF, Bullard SA. Huffmanela markgracei sp. n. (Nematoda: Trichosomoididae) from buccal cavity of Atlantic sharpnose shark, Rhizoprionodon terraenovae (Carcharhiniformes: Carcharhinidae), in the northwestern Gulf of Mexico off Texas. Folia Parasitol. 2013;60:353.
Owfi F, Fatemi SM, Motallebi AA, Coad B. Systematic review of Anguilliformes order in Iranian museums from the Persian Gulf and Oman Sea, Iran. J Fish Sci. 2014;13:407–26.
Ji HS, Kim JK, Oh TY, Choi KH, Choi JH, Seo YI, et al. Larval distribution pattern of Muraenesox cinereus (Anguilliformes: Muraenesocidae) leptocephali in waters adjacent to Korea. Ocean Sci J. 2015;50:537–45.
Eagderi S, Fricke R, Esmaeili HR, Jalili P. Annotated checklist of the fishes of the Persian Gulf: diversity and conservation status. Iran J Ichthyol. 2019;6:1–71.
Attia MM, Ibrahim MM, Mahmoud MA, Al-Sabi MN. Huffmanela sp. (Nematoda: Trichosomoididae: Huffmanelinae) encountered in the whitecheek shark (Carcharhinus dussumieri) in the Arabian Gulf. Helminthologia. 2021;58:281–91.
Al-Hasson HA, Al-Niaeem KS, Al-Azizz SA. Occurrence of six larval nematode species from marine fishes of Iraq. Biol Appl Environ Res. 2019;3:127–41.
Hulet WH. Structure and functional development of the eel leptocephalus Ariosoma balearicum (De La Roche, 1809). Philos Trans R Soc. 1978;282:107–38.
Umezawa A, Otake T, Hirokawa J, Tsukamoto K, Okiyama M. Development of the eggs and larvae of the pike eel, Muraenesox cinereus. J Ichthyol. 1991;38:35–40.
Ali AM, Algurabi MA, Nasibulina BM, Kurochkina TF, Bakhshalizadeh S. New records of some fishes from Hadhramout coast, Gulf of Aden, Yemen. Iran J Ichthyol. 2021;8:189–203.
Irmak E, Özden U. After four decades-Occurrence of the daggertooth pike conger, Muraenesox cinereus (Actinopterygii: Anguilliformes: Muraenesocidae), in the Mediterranean Sea. Acta Ichthyol Piscat. 2021;51:245.
McCosker J, Smith DG, Tighe K, Torres AG, Leander NJS. Muraenesox cinereus. IUCN Red List Threat Species. 2021. https://doi.org/10.2305/IUCN.UK.2021-1.RLTS.T199344A2585390.en.
Devadoss P, Pillai PK. Observations on the food and feeding habits of the eel, Muraenesox cinereus (Forskal) from Porto Novo. Indian J Fish. 1979;26:244–6.
An YS, Park JM, Kim HJ, Baeck GW. Feeding habits of daggertooth pike conger Muraenesox cinereus in the coastal water off Goseong, Korea. Korean J Fish Aquat Sci. 2012;45:76–81.
Conboy GA, Speare DJ. Dermal nematodosis in commercially captured rockfish (Sebastes spp.) from coastal British Columbia, Canada. J Comp Pathol. 2002;127:211–3.
Esteves A, Seixas F, Carvalho S, Nazário N, Mendes M, Martins C. Huffmanela sp. (Nematoda: Trichosomoididae) muscular parasite from Trisopterus luscus captured off the Portuguese coast. Dis Aquat Org. 2009;84:251–5.
Dill JA, Field CL, Camus AC. Pathology in Practice. J Am Vet Med Assoc. 2016;249:275–7.
Esteves A, Oliveira I, Ramos P, Carvalho A, Nazário N, Seixas F. Huffmanela spp. (Nematoda, Trichosomoididae) from Microchirus azevia: tissue location and correspondence of host muscle discoloration with parasite burden. J Fish Aquat Sci. 2016;11:304–10.
Eissa IA, Gadallah AO, Hashim M, Noureldin EA, Bayoumy EM, Haridy M. First record of the nematode, Huffmanela sp. infecting the broomtail wrasse (Cheilinus lunulatus) from Egypt. J Parasit Dis. 2021;45:228–35.
Spratt DM. Description of capillariid nematodes (Trichinelloidea: Capillariidae) parasitic in Australian marsupials and rodents. Zootaxa. 2006;1348:1–82.
Anderson RC, Chabaud AG, Willmott S. Keys to the nematode parasites of vertebrates: archival volume. Wallingford: Cabi; 2009. p. 48.
Chan AH, Chaisiri K, Saralamba S, Morand S, Thaenkham U. Assessing the suitability of mitochondrial and nuclear DNA genetic markers for molecular systematics and species identification of helminths. Parasit Vectors. 2021;14:1–3.
Conway KW, Kim D, Rüber L, Pérez HS, Hastings PA. Molecular systematics of the New World clingfish genus Gobiesox (Teleostei: Gobiesocidae) and the origin of a freshwater clade. Mol Phylogenet Evol. 2017;112:138–47.
Bullard SA, Roberts JR, Warren MB, Dutton HR, Whelan NV, Ruiz CF, et al. Neotropical turtle blood flukes: two new genera and species from the Amazon river basin with a key to genera and comments on a marine-derived parasite lineage in South America. J Parasitol. 2019;105:497–523.
MacCallum GA. Eggs of a new species of nematoid worm from a shark. Proc US Natl Mus. 1925;67:1.
Schouten H, Suriel-Smeets RM, Kibbelaar MA. The simultaneous occurrence of ova resembling Dicrocoelium dendriticum or Capillaria hepática in the stools of inhabitants of Curaçao. Trop geogr med. 1968;20:271–5.
Grabda J, Ślósarczyk W. Parasites of marine fishes from New Zealand. Acta Ichthyol Piscat. 1981;11:85–103.
Cox MK, Huffman DG, Moravec F. Observations on the distribution and biology of Huffmanela huffmani (Nematoda: Trichosomoididae). Folia Parasitol. 2004;51:50.
Worsham, M. 2015. Huffmanela huffmani: Life cycle, natural history, and biogeography. 119 p.
Justine JL. Three new species of Huffmanela Moravec, 1987 (Nematoda: Trichosomoididae) from the gills of marine fish off New Caledonia. Syst Parasitol. 2004;59:29–37.
MacLean RA, Fatzinger MH, Woolard KD, Harms CA. Clearance of a dermal Huffmanela sp. in a sandbar shark (Carcharhinus plumbeus) using levamisole. Dis Aquat Org. 2006;73:83–8.
Moravec F, Justine JL. Some trichinelloid nematodes from marine fishes off New Caledonia, including description of Pseudocapillaria novaecaledoniensis sp. Nov. (Capillariidae). Acta Parasitologica. 2010;55:71–80.
Tamaru M, Yamaki S, Jimenez LA, Sato H. Morphological and molecular genetic characterization of three Capillaria spp. (Capillaria anatis, Capillaria pudendotecta, and Capillaria madseni) and Baruscapillaria obsignata (Nematoda: Trichuridae: Capillariinae) in avians. Parasitol Res. 2015;114:4011–22.
Sakaguchi S, Yunus M, Sugi S, Sato H. Integrated taxonomic approaches to seven species of capillariid nematodes (Nematoda: Trichocephalida: Trichinelloidea) in poultry from Japan and Indonesia, with special reference to their 18S rDNA phylogenetic relationships. Parasitol Res. 2020;119:957–72.
Honisch M, Krone O. Phylogenetic relationships of Spiruromorpha from birds of prey based on 18S rDNA. J Helminthol. 2008;82:129–33.
Leis E, Easy R, Cone D. Report of the potential fish pathogen Pseudocapillaria (Pseudocapillaria) tomentosa (Dujardin, 1843) (Nematoda) from red Shiner (Cyprinella lutrensis) shipped from Missouri to Wisconsin. Comp Parasitol. 2016;83:275–8.
Zarlenga DS, Rosenthal BM, La Rosa G, Pozio E, Hoberg EP. Post-Miocene expansion, colonization, and host switching drove speciation among extant nematodes of the archaic genus Trichinella. Proc Natl Acad Sci. 2006;103:7354–9.
Krivokapich SJ. Detection and molecular, biological, morphological and evolutionary study of the first autochthonous Trichinella species found in the Neotropical region. PhD thesis (In Argentinian). 2017;165 p. https://hdl.handle.net/20.500.12110/tesis_n6276_Krivokapich.
Aguinaldo AM, Turbeville JM, Linford LS, Rivera MC, Garey JR, Raff RA, et al. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–93.
Callejón R, Nadler S, De Rojas M, Zurita A, Petrášová J, Cutillas C. Molecular characterization and phylogeny of whipworm nematodes inferred from DNA sequences of cox1 mtDNA and 18S rDNA. Parasitol Res. 2013;112:3933–49.
Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–5.
Acknowledgements
The scanning electron microscopy was performed by the Electron Microscopy Facility at Vienna BioCenter Core Facilities (VBCF), member of the Vienna BioCenter (VBC), Austria. The authors thank Thomas Heuser and Nicole Drexler, Vienna BioCenter Core Facilities, Vienna, Austria, for their excellent technical support. The authors sincerely appreciate Prof. František Moravec (Institute of Parasitology, Biology Centre of the Czech Academy of Sciences, Branišovská 31, 37005 České Budějovice, Czech Republic) and the two anonymous reviewers for their insightful suggestions and helpful comments on different versions of this paper. The first author thanks Prof. Rahim Peyghan and Dr. Zahra Tulaby Dezfuly for their valuable assistance in a preliminary investigation performed at Department of Clinical Sciences, Faculty of Veterinary Medicine, Shahid Chamran University of Ahvaz, Iran. The authors express their appreciation to Dr. Amin Mirzazadeh (Boehringer Ingelheim, RCV GmbH and Co KG, Vienna, Austria), whose assistance contributed greatly to this research.
Funding
Open Access funding for this article was provided by the University of Veterinary Medicine Vienna (Vetmeduni Vienna). This research was funded by the University of Veterinary Medicine, Vienna, Austria.
Author information
Authors and Affiliations
Contributions
ME and RG designed the research. RG collected the samples, carried out the experiments and drafted the manuscript. HS, MS and GK participated in molecular experiments. DG and GK contributed to pathological examination. ME, MF and MS investigated the study. ME, MF, MS and HS provided critical comments and edited the final version of manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ghanei-Motlagh, R., Fast, M.D., Groman, D. et al. Description, molecular identification and pathological lesions of Huffmanela persica sp. nov. (Nematoda: Trichosomoididae: Huffmanelinae) from the daggertooth pike conger Muraenesox cinereus. Parasites Vectors 16, 182 (2023). https://doi.org/10.1186/s13071-023-05772-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-023-05772-7