Abstract
Background
The two most abundant sand fly species on the Honduran Pacific coast are Lutzomyia (Lutzomyia) longipalpis and Pintomyia (Pifanomyia) evansi. Both species are known vectors of Leishmania (Leishmania) infantum, the etiological agent of visceral leishmaniasis (VL) in the Americas. Although VL and non-ulcerative cutaneous leishmaniasis (NUCL) are endemic on the Pacific versant of the Central American Pacific, the latter is the most frequent manifestation of leishmaniasis there. We evaluated the circulation of Leishmania spp. in the sand fly species on El Tigre Island, an endemic area of NUCL.
Results
We collected 222 specimens of six sand fly species. Lu. longipalpis (180 specimens; 81%) and Pif. (Pi.) evansi (35 specimens; 16%) were the most abundant species. L. (L.) infantum DNA was detected in nine of the 96 specimens analyzed; seven of these specimens were identified as Lu. longipalpis, and the remaining two were Pi. evansi, with an infection rate of 9.4% and 2.7%, respectively.
Conclusion
We present the first record of L. (L.) infantum DNA in Pi. evansi from a NUCL endemic region of Central America. Our results suggest that Pi. evansi could be a secondary vector of L. (L.) infantum in the transmission cycle of leishmaniasis. The detection of natural infections of L. (L.) infantum in sand flies in this region contributes to an understanding of the epidemiology of leishmaniasis in Honduras.
Graphical Abstract

Similar content being viewed by others
Background
Leishmaniasis is a vector-borne parasitic disease caused by species of the genus Leishmania (Kinetoplastida: Trypanosomatidae). Leishmania is widespread mainly in tropical and subtropical regions of 98 countries throughout Europe, Africa, Asia, and America [1]. More than 1,000 sand fly species have been identified worldwide, of which 530 species are present in the Americas [2, 3], and at least 30 species are considered Leishmania vectors [4]. In Honduras, 29 sand fly species have been reported [2, 5,6,7]. Members of the Lutzomyia longipalpis species complex are the main vectors of Leishmania (Leishmania) infantum; however, other sand fly species, including Lutzomyia (Lutzomyia) cruzi and Pintomyia (Pifanomyia) evansi, play a role as vectors of this parasite in some endemic areas of visceral leishmaniasis (VL) in South and Central America [8, 9]. Lu. longipalpis s.l. is the most abundant sand fly species in the area endemic for VL and non-ulcerative cutaneous leishmaniasis (NUCL) in the southern region of Honduras [6, 10], and is the only species reported as a vector of L. (L.) infantum [11]. Nevertheless, the role of Pi. evansi in VL transmission dynamics in Honduras is unknown, although evidence of its vector competence has been reported in South America [12]. For this reason, the aim of this study was to evaluate the DNA circulation of L. (L.) infantum in Lu. longipalpis and Pi. (Pif.) evansi on a Mesoamerican Pacific island.
Methods
Study area and sand fly collection
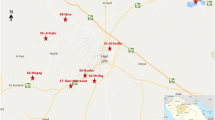
This study was carried out in Amapala municipality (13°17′26.082″N, 87°39′5.543″W), Valle department, an area covering 80.7 km2. The municipality comprises two islands, Zacate Grande and El Tigre, located in the Gulf of Fonseca in southern Honduras. Sand flies were sampled for five consecutive nights in May 2018 from five localities: Las Pelonas (13°16′58.332″N, 87°37′4.799″W), Punta Honda (13°16′21.576″N, 87°36′56.879″W), Tigüilotada (13°15′45.720″N, 87°36′57.527″W), Islitas (13°15′39.456″N, 87°37′24.276″W) and Playa Grande (13°16′4.183″N, 87°39′31.104″W). The captures were carried out from 6:00 p.m. until 6:00 a.m., using automatic CDC miniature light traps (model 512; John W. Hock, Gainesville, FL) in neighborhoods where active cases of NUCL were evident. The traps were installed in peridomiciliary environments, near decomposing organic matter or next to latrines. The specimens were separated and processed 1 day after capture.
Taxonomic identification of the sand fly species
The taxonomy of the phlebotomine sand flies was conducted by analyzing their morphological characteristics. In adherence to the identification procedures outlined by Mejía et al. [6], the sand flies were first mounted and the specific species identified in accordance with Young and Duncan [5]. Finally, Galati et al. [13] were the primary academic resource used during the classification stage of the genera and the species.
Genomic material extraction and polymerase chain reaction
Genomic DNA was extracted from gut tissues dissected from individual female sand flies using the Chelex 100 Kit (Bio-Rad, Hercules, CA). As an internal control for the DNA extraction, the cacophony IVS6 gene present in the sand fly genome was amplified [14]. For the detection of the genus Leishmania, we used the primers Leish1: 5ʹ-AACTTTTCTCTGGTCCTCCGGGTAG-3ʹ and Leish2: 5′-ACCCCCAGTTTCCCGCC-3ʹ to amplify an approximate 120-base pair (bp) product [15]. Amplifications were performed using a commercial kit (Master Mix 2X; Promega). Each reaction was performed by adding 4 µl of target DNA and 0.6 µmol/L of each primer to obtain a final volume of 20 µl. The polymerase chain reaction (PCR) reactions were done in an Applied Biosystem 2770 Thermal Cycler (ThermoFisher Scientific, USA), under the following conditions: initial denaturation cycle at 94 °C for 5 min, followed by 35 cycles at 94 °C for 15 s, 60 °C for 20 s and 72 °C for 60 s, and final extension at 72 °C for 10 min. The amplification products were analyzed by electrophoresis in 1.5% agarose gel.
To characterize the Leishmania species, PCR-restriction fragment length polymorphism (RFLP) was performed, which amplified a specific region of the hsp70 gene [16]. The primers used were hsp70 sense (5' GGACGAGATCGAGCGCATGGT 3') and hsp70 antisense (5' TCCTTCGACGCCTCCTGGTTG 3'). The reaction mixture was prepared in a final volume of 20 µl with 10 µl Master Mix 2X (Promega), 4 µl of target DNA and 0.6 µmol/L of each primer. The PCR reactions were done under the following conditions: initial denaturation at 94 °C for 5 min, followed by 37 cycles at 94 °C for 30 s, 61 °C for 1 min and 72 °C for 3 min, and a final extension cycle at 72 °C for 10 min. The amplification products were analyzed by electrophoresis in 2% agarose gel. To perform the restriction of the PCR products, the restriction enzyme HaeIII (Promega) was used, 5 µl of amplified DNA was added to the reaction and the mixture incubated at 37 °C for 3 h. The species profiles of each sample and reference controls were observed in 4% of agarose gel subjected to electrophoresis for 3.5 h.
Results and discussion
A total of 222 sand fly specimens were collected, which were predominately males (66%) (Table 1). Six species were identified by using morphological characters. The most predominant sand fly species collected was Lu. (Lu.) longipalpis, followed by Pi (Pif.) evansi. The other species that were captured included Micropygomyia (Micropygomyia) cayennensis cayennensis, Micropygomyia (Coquillettimyia) chiapanensis, Dampfomyia (Coromyia) beltrani and Lutzomyia (Tricholateralis) gomezi. Lu. (Lu.) longipalpis has been previously studied in the region and was incriminated as the vector of L. (L.) infantum [11]. The behavioral characteristics of this species in the study area were described by Carrasco et al. [7]. Recently, Mejía et al. [6] expounded on various aspects of sand flies’ feeding preferences within the Pacific Honduran area. These authors observed a predominance of Pi. (Pif.) evansi and Lu. (Lu.) longipalpis, but did not detect the presence of L. (L.) infantum in these species [6]. These sand flies were also predominant in other NUCL-endemic areas of Central America (i.e., Costa Rica and Nicaragua) [17, 18]
Thirty-seven of the 96 analyzed female specimens were positive for the genus Leishmania according to the PCR results (Fig. 1). L. (L.) infantum DNA was revealed in nine specimens: seven Lu. (Lu.) longipalpis and two Pi. (Pif.) evansi. The L. (L.) infantum infection rate was 9.4% for Lu. (Lu.) longipalpis and 2.7% for Pi. (Pif.) evansi. All of the samples analyzed produced an amplified product of 220 bp, corresponding to a Lutzomyia spp. constitutive gene (cacophony), which confirmed the integrity of the insect DNA preparation and the absence of PCR inhibitors [14]. We used a method for the detection of Leishmania spp. described in Francino et al. [15]. However, one limitation of this method is the small size of the PCR product (120 bp), which makes sequencing unlikely. Therefore, the method described by Graça et al. [16] was used to differentiate Leishmania species. This method amplifies a Leishmania 234-bp hsp70 fragment and shows similar sensitivity to the PCR-internal transcribed spacer 1 (>70%) method used to detect Leishmania DNA; however, associating this 234-bp hps70 with a RFLP protocol may give researchers the advantage of being able to distinguish this Leishmania species by electrophoresis [19].
Polymerase chain reaction (PCR) to determine Leishmania spp. infection using Leish1-Leish2 primers to target conserved DNA regions of the kinetoplast DNA from Leishmania spp. [120 base pairs (bp)]. Lane M Molecular weight marker (100-bp DNA ladder). Lanes 1–16 Female sand fly DNA [lanes 1-10 Lutzomyia (Lutzomyia) longipalpis, positive female; lanes 11, 13–16 Pintomyia (Pifanomyia) evansi, positive female]. Lane 17 PCR positive control [DNA extracted from a mixture of the male insect pool containing Leishmania (Leishmania) infantum DNA]. Lane 18 Amplification reaction without added DNA (PCR negative control)
Our study is the first to report the presence of L. (L.) infantum DNA in Pi. evansi females in Central America. In two studies in Colombia, in endemic areas of VL, the natural infection rate of L. (L.) infantum in Pi. evansi was found to be 0.10% [12] and 0.34% [19]. The natural infection rates of Lu. longipalpis were between 0.5% and 1.1% according to direct observations of dissected intestines [11, 20] and from PCR of dissected intestines [21,22,23]. We report a 9.4% infection rate of L. (L.) infantum in Lu. longipalpis, which is in agreement with other research [22, 23]. Although the detection of Leishmania DNA in sand flies does not indicate the ability of these species to transmit this parasite, we evidenced contact between Pi. evansi and Lu. longipalpis with the natural host of L. (L.) infantum in the study area, in the Amapala municipality. Considering that the vector competence of Pi. evansi has been previously described [12], it is probable that both Lu. longipalpis and Pi. evansi transmit L. (L.) infantum in southern Honduras.
Conclusion
We present for the first time evidence of the presence of L. (L.) infantum DNA in Pi. evansi in a NUCL endemic region of Central America. Considering the natural infection of Lu. longipalpis by L. (L.) infantum, our results indicate that Pi. evansi might be a secondary vector of this parasite, and might be involved in the transmission cycle of leishmaniasis. The detection of natural infections of L. (L.) infantum in sand flies in this region contributes to our understanding of the epidemiology of leishmaniasis in Honduras.
Availability of data and materials
Data supporting the conclusions of this article are included within the article. Specimens of Lutzomyia analyzed during the study are available from the corresponding author upon reasonable request.
Abbreviations
- VL:
-
Visceral leishmaniasis
- NUCL:
-
Non-ulcerative cutaneous leishmaniasis
- PCR:
-
Polymerase chain reaction
- bp:
-
Base pairs
- RFLP:
-
Restriction fragment length polymorphism
References
Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, et al. WHO Leishmaniasis Control Team. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE. 2012;7:e35671.
Shimabukuro PHF, De Andrade AJ, Galati EAB. Checklist of American sand flies (Diptera, Psychodidae, Phlebotominae): genera, species, and their distribution. ZooKeys. 2017;660:67–106.
Galati EAB, Galvis-Ovallos F, Lawyer P, Léger N, Depaquit J. An illustrated guide for characters and terminology used in descriptions of Phlebotominae (Diptera, Psychodidae). Parasite. 2017;24:26.
Beati L, Caceres AG, Lee JA, Munstermann LE. Systematic relationships among Lutzomyia sand flies (Diptera: Psychodidae) of Peru and Colombia based on the analysis of 12S and 28S ribosomal DNA sequences. Int J Parasitol. 2004;34(2):225–34.
Young DG, Duncan MA. Guide to the identification and geographic distribution of Lutzomyia sand flies in Mexico, the West Indies, Central and South America (Diptera: Psychodidae) Gainesville. Mem Am Entomol Inst. 1994;54:1–881.
Mejia A, Matamoros G, Fontecha G, Sosa-Ochoa W. Bionomic aspects of Lutzomyia evansi and Lutzomyia longipalpis, proven vectors of Leishmania infantum in an endemic area of non-ulcerative cutaneous leishmaniasis in Honduras. Parasites Vectors. 2018;11(1):15.
Carrasco J, Morrison A, Ponce C. Behaviour of Lutzomyia longipalpis in an area of southern Honduras endemic for visceral/atypical cutaneous leishmaniasis. Ann Trop Med Parasitol. 1998;92(8):869–76.
Lainson R, Rangel EF. Lutzomyia longipalpis and the eco-epidemiology of American visceral leishmaniasis with particular reference to Brazil: a review. Mem Inst Oswaldo Cruz. 2005;100(8):811–27.
Chaves LF, Anez N. Nestedness patterns of sand fly (Diptera: Psychodidae) species in a Neotropical semi-arid environment. Acta Trop. 2016;153:7–13.
Sosa-Ochoa W, Morales Cortedano X, Argüello A, Zuniga C, Henríquez J, Mejía R, et al. Ecoepidemiología de la Leishmaniasis cutánea no ulcerada en Honduras. Rev Cienc Tecnol UNAH. 2014;14:115–28.
Ponce C, Ponce E, Morrison A, Cruz A, Kreutzer R, McMahon-Pratt D, et al. Leishmania donovani chagasi: new clinical variant of cutaneous leishmaniasis in Honduras. Lancet. 1991;337(8733):67–70.
Travi BL, Vélez ID, Brutus L, Segura I, Jaramillo C, Montoya J. Lutzomyia evansi, an alternate vector of Leishmania chagasi in a Colombian focus of visceral leishmaniasis. Trans R Soc Trop Med Hyg. 1990;84(5):676–7.
Galati EAB.Morfologia e terminologia de Phlebotominae (Diptera: Psychodidae). Classificação e identificação de táxons das Américas. Vol I. Apostila da Disciplina Bioecologia e Identificação de Phlebotominae do Programa de Pós-Graduação em Saúde Pública. Faculdade de Saúde Pública da Universidade de São Paulo. 2019. https://www.fsp.usp.br/egalati/wpcontent/uploads/2020/02/Apostila_Vol_I_2019.pdf. Accessed 18 May 2019.
Pita-Pereira D, Souza GD, Zwetsch A, Alves CR, Britto C, Rangel EF. First report of Lutzomyia (Nyssomyia) neivai (Diptera: Psychodidae: Phlebotominae) naturally infected by Leishmania (Viannia) braziliensis in a periurban area of south Brazil using a multiplex polymerase chain reaction assay. Am J Trop Med Hyg. 2009;80(4):593–5.
Francino O, Altet L, Sánchez-Robert E, Rodriguez A, Solano-Gallego L, Alberola J, et al. Advantages of real-time PCR assay for diagnosis and monitoring of canine leishmaniosis. Vet Parasitol. 2006;137:214–21.
Graça GC, Volpini AC, Romero GAS, Oliveira Neto MP, Hueb M, Porrozzi R, et al. Development and validation of PCR-based assays for diagnosis of American cutaneous leishmaniasis and identification of the parasite species. Mem Inst Oswaldo Cruz. 2012;107:664–74.
Zeledón R, Murillo J, Gutiérrez H. Observaciones sobre la ecología de Lutzomyia longipalpis (Lutz & Neiva, 1912) y posibilidades de existencia de leishmaniasis visceral en Costa Rica. Mem Inst Oswaldo Cruz. 1984;79(4):455–9.
Raymond RW, McHugh CP, Kerr SF. Sand flies of Nicaragua: a checklist and reports of new collections. Mem Inst Oswaldo Cruz. 2010;105(7):889–94.
Travi BL, Montoya J, Gallego J, Jaramillo C, Llano R, Velez ID. Bionomics of Lutzomyia evansi (Diptera: Psychodidae) vector of visceral leishmaniasis in northern Columbia. J Med Entomol. 1996;33(3):278–85.
Silva JGD, Werneck GL, Cruz MSP, Costa CHN, Mendonça IL. Infecção natural de Lutzomyia longipalpis por Leishmania sp. em Teresina, Piauí Brasil. Cad Saúde Públ. 2007;23(7):1715–20.
Flórez M, Martínez JP, Gutiérrez R, Luna KP, Serrano VH, Ferro C, et al. Lutzomyia longipalpis (Diptera: Psychodidae) en un foco suburbano de leishmaniosis visceral en el Cañón del Chicamocha en Santander Colombia. Biomédica. 2006;26(Supl1):109–20.
Michalsky EM, Guedes KS, França-Silva JC, Dias CLF, Barata RA, Dias ES. Infecção natural de Lutzomyia (Lutzomyia) longipalpis (Diptera: Psychodidae) por Leishmania infantum chagasi em flebotomíneos capturados no município de Janaúba, Estado de Minas Gerais Brasil. Rev Soc Bras Med Trop. 2011;44(1):58–62.
Mota TF, de Sousa OMF, Silva YJ, Borja LS, Leite BMM, Solcà MDS, et al. Natural infection by Leishmania infantum in the Lutzomyia longipalpis population of an endemic coastal area to visceral leishmaniasis in Brazil is not associated with bioclimatic factors. PLoS Negl Trop Dis. 2016;13:e0007626.
Acknowledgements
We thank the Ministry of Health of Honduras for allowing us to further our research in the NUCL endemic area of Amapala municipality and for assisting us in this endeavor. We would also like to thank Jessica Cardenas for her outstanding technical support.
Funding
This study was financially supported by the Research Directorate of the National Autonomous University of Honduras.
Author information
Authors and Affiliations
Contributions
Conceptualization: MDL, FG, and WSO. Methodology: WSO, FG, JV, YL, GVAF, CMSP, CZ, and GS. Formal analysis: WSO, FG, and MDL. Preparation of the first draft: WSO, FG, and MDL. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Sosa-Ochoa, W., Varela Amador, J., Lozano-Sardaneta, Y. et al. Detection of Leishmania infantum DNA in Pintomyia evansi and Lutzomyia longipalpis in Honduras. Parasites Vectors 13, 593 (2020). https://doi.org/10.1186/s13071-020-04462-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04462-y