Abstract
Background
Cutaneous leishmaniasis is a disease transmitted by sand fly bites. This disease is highly prevalent in Syria where Leishmania major and Leishmania tropica are the known aetiological agents. In 2011, more than 58,000 cases were reported in the country by the Ministry of Health. The central region of the country harbors 20 % of the reported cases. However, the epidemiology of the disease in this area is not well understood. An epidemiological survey was conducted in 2010 to identity the circulating parasite and the sand fly vector in the central provinces of Edlib and Hama.
Methods
Sand fly specimens were collected using CDC light traps and identified morphologically. Total DNA was extracted from the abdomens of female specimens and from Giemsa-stained skin lesion smears of 80 patients. Leishmania parasites were first identified by sequencing the ITS1 gene amplicons. Then polymorphism analysis was performed using the RFLP technique.
Results
A total of 2142 sand flies were collected. They belonged to eight species, among which Phlebotomus sergenti and Phlebotomus papatasi were the most predominant.
L. tropica ITS1 gene was amplified from two pools of P. sergenti specimens and from skin smears of cutaneous leishmaniasis patients. This suggests that P. sergenti is the potential vector species in the study area. The digestion profiles of the obtained amplicons by TaqI restriction enzyme were identical for all analysed L. tropica parasites. Moreover, L. infantum ITS1 gene was amplified from two pools of Phlebotomus tobbi in the relatively humid zone of Edlib.
Conclusions
L. tropica is confirmed to be the aetiological agent of cutaneous leishmaniasis cases in the central provinces. RFLP technique failed to show any genetic heterogeneity in the ITS1 gene among the tested parasites. The molecular detection of this parasite in human skin smears and in P. sergenti supports the vector status of this species in the study area. The detection of L. infantum in P. tobbi specimens indicates a potential circulation of this parasite in the humid zone of Edlib. Further epidemiological studies are needed to evaluate the burden of this visceral parasite in the study region.
Similar content being viewed by others
Background
Cutaneous leishmaniasis (CL) is a disease caused by Leishmania parasites and transmitted to humans by sand fly bites in more than 70 countries around the world, in climates ranging from tropical to desert. Syria is considered one of the most endemic countries for CL [1]. Syrian health authority records show that the vast majority (90 %) of reported CL cases in the country are caused by Leishmania tropica, whereas the remaining cases are caused by the zoonotic parasite L. major [2]. Visceral leishmaniasis due to Leishmania infantum is also spread in Syria. The incidence for the period 2004–2008 was 14 cases/year, mainly reported from Edlib province. Dogs are suspected to be the main reservoir [2]. Published data from the Syrian Ministry of Health show that, despite the implementation of several control programmes, the incidence of CL has increased during the last fifteen years from 12,027 cases in 1997 to 58,156 cases in 2011 [3]. Since 2011, the ongoing Syrian war and the resulting massive population displacement have led to a significant increase of the incidence of CL in the country and in neighbouring Turkey, Jordan and Lebanon [4, 5]. In 2013, more than 1000 cases of CL (mainly caused by L. tropica) were recorded in Lebanon among Syrian refugees [4].
The northern province of Aleppo accounts for 56 % of CL, followed by the central provinces of Edlib and Hama which together harbor 20 % of all declared cases. Several studies on leishmaniasis have been conducted in the Aleppo area [6–10]. The causative agent was typed as L. tropica ZMON-76 [11]. However, the epidemiology of the disease in the central provinces remains poorly studied. Records from Hama province suggest that L. tropica is the main circulating parasite [12]. During the summer of 2010, we conducted an entomological survey in the provinces of Hama and Edlib in order to identify the potential vectors of CL and establish an inventory of existing sand fly species. In addition, we analysed Giemsa-stained smears of skin lesions belonging to CL patients residing in these provinces to determine the species identity and polymorphism of the circulating Leishmania parasites. Classically, polymerase chain reaction (PCR) based methods performed on various targets such as microsatellite, kinetoplast DNA, or ribosomal RNA have been used for identification of Leishmania isolates in various foci [13–15]. The internal transcribed spacers 1 (ITS1) gene, a ribosomal RNA gene, is considered a primary target for this purpose as shown in many studies conducted in endemic areas in the Old and New World [16–22]. In this study, sequence and restriction fragment length polymorphism (RFLP) analyses were performed on the ITS1 amplicons in order to identify the circulating species and to detect potential genetic polymorphism.
Methods
Description of the study area
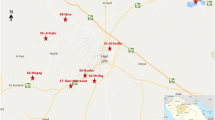
The provinces of Hama (8883 km2) and Edlib (5933 Km2) are located in the central region of Syria (Fig. 1), which is characterized by an arid climate with an annual average rate of precipitation lower than 200 mm. The western side of this region is influenced by the Mediterranean climate and, therefore, is obviously more humid. Agriculture is the main economic resource for the two million residents of this area.
Sample collection
Sand flies were captured during July and August 2010, in 11 geographical locations in the province of Hama and 17 locations in the province of Edlib (Fig. 1), from where human CL cases were reported in 2010 by the Syrian Ministry of Health. CDC miniature light traps (John Hock Co., Gainesville, FL, USA) were placed in indoor (bedrooms, animal shelters) and outdoor habitats (private gardens). Collected specimens were preserved in 90 % ethanol. In addition, Giemsa-stained skin smear slides of patients with confirmed CL were provided to us by the Ministry of Health authorities. These smears were performed on skin lesions of patients seeking diagnosis and treatment at local primary health care centers in Hama and Edlib provinces. All tested smears were confirmed positive by observing Leishmania amastigotes microscopically.
Sand fly identification
Male specimens were cleared in Marc-Andrée solution [23] and then mounted on slides in chloral gum medium [24]. Female specimens were dissected; their head and genitalia were cleared and then mounted, whereas their abdomen and thorax were preserved in ethanol for further molecular analysis. Female and male specimens were morphologically identified by observing head and genital structures under the microscope using local and regional identification keys [25, 26].
DNA extraction
Abdomens of female sand flies were pooled (up to 10 per pool) according to species and capture locality. These samples were then ground in microtubes (1.5 mL). Slides of skin lesion smears were individually scraped using sterile blade scalpels. The scraping product was collected in microtubes. Total DNA was extracted from both types of materials using the extraction kit QIAamp DNA Mini Kit (Qiagen, Valencia, CA).
PCR amplification and sequencing of ITS1 gene
DNA extracts were screened for Leishmania DNA by conventional PCR technique. A 339-bp fragment of the internal transcribed spacer region (ITS1) of the small subunit ribosomal DNA was amplified using specific primers: L5.8S: 5′-TGATACCACTTATCGCACTT-3′ and LITSR: 5′-CTGGATCATTTTCCGATG-3′ [17]. The PCR mix comprised 2.5 U Taq polymerase (Promega, Madison, WI), 0.6 pM of each primer, 1.5 mM MgCl2, 0.1 mM dNTPs, and 5 μL DNA template in a final volume of 50 μL. Cycling conditions were as follow: 95 °C, 120 s and (94 °C, 30 s; 53 °C, 60 s; 72 °C, 60 s) repeated 37 times.
Amplicons were visualized after electrophoresis in 1.5 % agarose gel in TBE (1X) buffer. DNA extracts from L. infantum parasite of the reference strain MHOM/FR/78/LEM 75 and male sand fly specimen were used respectively as positive and negative controls. Amplicons were purified using the Qiagen PCR Clean up Kit (Qiagen, Valencia, CA). Fifty fmol of purified DNA were sequenced in the forward direction with an ABI Prism 3130 Genetic Analyzer, Big Dye Terminator technology (Applied Biosystems, Foster City, CA, USA) using 1.6 pmol of each primer. Sequences were manually edited and used to query GenBank for species identity.
RFLP analysis of amplified ITS1
In order to detect potential genetic polymorphism among the identified Leishmania parasites, 10 μL of non-purified ITS1 amplicons were digested with Taq I restriction enzyme as indicated by the manufacturer (New England Biolabs, Ipswich, MA, USA). The digested products were run on 1.5 % electrophoresis gel agar at 100 V in TBE buffer and visualized under ultraviolet light. Bands were stained by adding GelRed dye (Phenix research, USA) to the gel preparation.
Results
Identification of collected sand fly specimens
The total number of sand flies captured was 2142, including 603 in Hama province and 1539 in Edlib province. Their identification revealed eight species, of which seven belonged to the Phlebotomus genus and one to the Sergentomyia genus (Tables 1 and 2).
The dominant species among collected specimens was Phlebotomus sergenti in both provinces, followed by Phlebotomus papatasi. These species are known vectors of anthroponotic and zoonotic forms of CL respectively [27–32]. Larroussius subgenus is dominantly represented by Phlebotomus gallilaeus. Specimens of this species were mainly collected from the locality of Khamrieh in the humid area west of Edlib, near the Turkish border. Phlebotomus tobbi is another Larroussius member found in the study area. This species is spread mainly in countries of the mid-northern to eastern sides of the Mediterranean basin. Moreover, P. tobbi is the vector of L. infantum in Cyprus [33] and a suspected vector of this parasite in several countries, including Turkey and Iran [34, 35].
Parasite identification
The presence of ITS1 gene of Leishmania parasite was investigated by PCR on 1172 specimens of female sand flies, divided into 216 pools, as well as on skin smears of 80 patients of CL (40 from each province). Amplification results identified two positive pools of P. sergenti, one collected in the locality of Deir Charki (Edlib), the other in Smakh (Hama) (Fig. 2). In total, 489 female specimens of this species were tested in the study zone, including 254 from Edlib province (of which 14 were from Deir Charki) and 235 from Hama province (16 from Smakh). We also identified two Leishmania positive pools of P. tobbi, which were collected in the localities of El Jebb and El Safsafeh in Edlib. In this province, a total of 85 female specimens of P.tobbi were collected, of which only 6 were captured in El Jebb and 19 in El Safsafeh. On the other hand, ITS1 gene amplicons were obtained from 70 skin smears (34 from Edlib and 36 from Hama) out of the 80 tested samples.
All the obtained amplicons were sequenced. Their analysis revealed that the gene segments amplified from the two pools of P. sergenti and those from all analysed skin smears from both Syrian provinces are identical and belong to L. tropica. These sequences perfectly matched with ITS1 sequences of L. tropica isolate MHOM/AF/88/KK27 from Afghanistan and MHOM/IR/2012/Savodjbolagh isolate from Iran [GenBank: GQ913688.1, GenBank: KC574386.1]. Moreover, analysis of ITS1 gene sequences amplified from the two pools of P. tobbi from Edlib showed that the parasite belongs to L. infantum and is identical to the Iranian strain ITOB/IR/2009/Bilesavar [GenBank: HQ535858.1], which was also isolated from P. tobbi.
RFLP results
After species identification, all PCR products of L. tropica from the skin smears of CL patients and from P. sergenti pools were digested by Taq1 restriction enzyme in order to detect potential genetic polymorphism. Digestion products showed that all isolates displayed two DNA bands of approximately 140 and 190 bp (Fig. 3).
Discussion
The objectives of the current study were to identify the sand fly vectors and Leishmania species responsible for the CL cases in the central provinces of Hama and Edlib. Sequences of the ITS1 gene amplified from skin lesions of 70 patients showed that L. tropica is the causative agent of CL in both provinces. This species is also the main agent of CL in the highly endemic Aleppo province, northern Syria [9, 36] and in several foci in neighbouring countries [37, 38]. PCR-RFLP analysis of the ITS1 gene is commonly used to detect intraspecific genetic diversity among L. tropica species [21, 22]. This method is usually used as an alternative to sequence comparison by alignment. RFLP results revealed that all analysed strains from the study area had identical digestion patterns of the ITS1-gene, characterized by two DNA bands. This suggests the absence of mutations on the restriction site of the Taq1 enzyme. The absence of other polymorphic sites on the ITS1-gene was confirmed by sequence alignment (data not shown). Nevertheless, the existence of genetic diversity on other gene markers, such as Kinetoplast DNA minicircle, remains a possibility and requires further investigation. Our findings corroborate the observation of Pratlong and collaborators (2009) who identified only one L. tropica zymodeme (MON-76) among 51 tested samples [39]. Whereas, in Iran, Doudi and collaborators (2012) [22] were able to detect one dominant L. tropica isolate characterized by a specific digestion pattern using Taq 1 enzyme, in addition to two other types of isolates with different digestion patterns, reflecting the existence of a genetic polymorphism at the level of the ITS1 gene. Moreover, our results suggest that the sand fly vector responsible for L. tropica transmission in the study area is P. sergenti. Two pools of sand flies belonging to this species were positive for Leishmania ITS1 gene. PCR-RFLP analysis of the amplified gene produced the same digestion pattern as that observed for all L. tropica isolates from skin lesions. Besides, P. sergenti is the dominant sand fly (43 %) in our collections. This species is known to be the vector of L. tropica in several countries [27–30]. L. tropica is an anthroponotic parasite; nevertheless, zoonotic transmission has been documented in certain foci in the Middle East. The rock hyrax (Procavia capensis) is the main reservoir of L. tropica in certain Israeli and Palestinian foci [40]. In addition, dogs (and other Canidae) are suspected to be involved in the transmission of this Leishmania species in Morocco, Iran and Israel [41–43]. The high endemicity of L. tropica in the central provinces in Syria suggests an anthroponotic transmission pattern of this parasite. However, the involvement of dogs as reservoirs should be investigated. It is worth noting the absence of L. major, the agent of zoonotic CL, among the tested skin smears, despite the important spread of its specific vector, P. papatasi. The rodent reservoir of this species (Psammomys obesus) does not occur in the central provinces, while it is found mainly in the desert area, east of Damascus [44].
L. infantum seems to circulate in the study area, as it was detected in two pools of female specimens of P. tobbi collected in the humid area of Edlib. L. infantum is the known agent of visceral leishmaniasis in the Mediterranean basin. Few cases of VL are reported annually in Syria. Recent reports showed an incidence of VL of 14 cases per year, based on a 2–4-fold underreporting ratio [1]. The involvement of P. tobbi in the transmission of visceral Leishmania parasites was also recorded during a study conducted in the province of Lattakia that borders the western side of the study zone [45]. Nevertheless, L. infantum was also proven to be a causative agent of CL in the Mediterranean basin [46–50]. In the neighbouring Anatolia (South of Turkey), Svobodova and collaborators (2009) identified L. infantum as the causative agent of cutaneous lesions [33]. These authors incriminated P. tobbi as the vector of this dermotropic form. We expect a similar epidemiological situation in the humid zones west of Syria. Analysis of skin smears of patients originating from these zones is required to confirm the circulation of dermotropic L. infantum.
Conclusions
Using PCR-ITS1 method, L. tropica was identified as the parasitic agent responsible for the high number of CL cases in the central provinces of Syria. RFLP analysis of the ITS1 gene amplicons obtained from skin lesions and from the sand fly vector did not reveal any polymorphism among the circulating parasites. Entomological data showed that P. sergenti and P. papatasi are the most dominant species in our collections. The detection, for the first time in Syria, of L. tropica ITS1 gene in two pools of P. sergenti, supports the vector status of this species. The amplification of L. infantum ITS1 gene from two pools of P. tobbi in the humid part of Edlib suggests an active circulation of this parasite. Further studies in humans, vectors and potential reservoirs are needed to determine the burden of L. infantum in the study area and in other parts of Syria.
References
Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, et al. Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7(5), e35671.
World Health Organization. Leishmaniasis information resources. http://www.who.int/leishmaniasis/resources/SYRIAN_ARAB_REPUBLIC.pdf
National reports for the Ministry of Health, Syrian Arab Republic. http://www.moh.gov.sy
Sharara SL, Kanj SS. War and Infectious Diseases: Challenges of the Syrian Civil War. PLoS Pathog. 2014;10(11), e1004438.
Salman IS, Vural A, Unver A, Saçar S. Cutaneous leishmaniasis cases in Nizip, Turkey after the Syrian civil war. Mikrobiyol Bul. 2014;48(1):106–13.
Douba M, Mowakeh A, Wali A. Current status of cutaneous leishmaniasis in Aleppo, Syrian Arab Republic. Bull World Health Organ. 1997;75(3):253–9.
Douba MD, Abbas O, Wali A, Nassany J, Aouf A, Tibbi MS, et al. Chronic cutaneous leishmaniasis, a great mimicker with various clinical presentations: 12 years experience from Aleppo. J Eur Acad Dermatol Venereol. 2012;26(10):1224–9.
Tayeh A, Jalouk L, Cairncross S. Twenty years of cutaneous leishmaniasis in Aleppo, Syria. Trans R Soc Trop Med Hyg. 1997;91(6):657–9.
Maroli M, Jalouk L, Al Ahmed M, Bianchi R, Bongiorno G, Khoury C, et al. Aspects of the bionomics of Phlebotomus sergenti sandflies from an endemic area of anthroponotic cutaneous leishmaniasis in Aleppo Governorate. Syria Med Vet Entomol. 2009;23(2):148–54.
Hayani K, Dandashli A, Weisshaar E. Cutaneous leishmaniasis in Syria: clinical features, current status and the effects of war. Acta Derm Venereol. 2015;95(1):62–6.2.
Belazzoug S, Pratlong F, Rioux JA. Un nouveau zymodème de Leishmania tropica, agent du Bouton d’Alep (Syrie). Archives de l’lnstitut Pasteur, Algérie. 1988;56:95–9.
Al-Nahhas S, Kaldas R: Characterization of Leishmania species isolated from cutaneous human samples from Central Region of Syria by RFLP Analysis. ISRN Parasitology. 2013, Article ID 308726, 5 pages.
Al-Jawabreh A1, Diezmann S, Müller M, Wirth T, Schnur LF, Strelkova MV, et al. Identification of geographically distributed sub-populations of Leishmania (Leishmania) major by microsatellite analysis. BMC Evol Bio. 2008;8:183.
Singh S, Sivakumar R. Recent advances in the diagnosis of leishmaniasis. J Postg Med. 2003;49:55–60.
Smith DF, Searle S, Ready PD, Gramiccia M, Ben-Ismail R. A kinetoplast DNA probe diagnostic for Leishmania major: sequence homologies between regions of Leishmania minicircles. Mol Biochem Parasitol. 1989;37(2):213–23.
Berzunza-Cruz M, Cabrera N, Crippa-Rossi M, Sosa Cabrera T, Pérez-Montfort R, Becker I. Polymorphism analysis of the internal transcribed spacer and small subunit of ribosomal RNA genes of Leishmania mexicana. Parasitol Res. 2002;88:918–25.
Cupolillo E, Brahim LR, Toaldo CB, de Oliveira-Neto MP, de Brito ME, Falqueto A, et al. Genetic polymorphism and molecular epidemiology of Leishmania (Viannia) braziliensis from different hosts and geographic areas in Brazil. J Clin Microbiol. 2003;41:3126–32.
Doudi M, Hejazi SH, Razavi MR, Narimani M, Khanjani S, Eslami G. Comparative molecular epidemiology of Leishmania major and Leishmania tropica by PCR-RFLP technique in hyper endemic cities of Isfahan and Bam. Iran Med Sci Monit. 2010;16:CR530–5.
El Tai NO, Osman OF, El Fari M, Presber W, Schönian G. Genetic heterogeneity of ribosomal internal transcribed spacer in clinical samples of Leishmania donovani spotted on filter paper as revealed by single-strand conformation polymorphisms and sequencing. Trans R Soc Trop Med Hyg. 2000;94(5):575–9.
Mirzaie F, Eslami G, Yosefi MH, Pestehchian N. Molecular identification of Leishmania isolates obtained from patients suspected as having cutaneous leishmaniasis referred to reference laboratories from Yazd province in central Iran. Adv Biomed Res. 2013;2:92.
Schonian G, Schnur L, el Fari M, Oskam L, Kolesnikov AA, Sokolowska-Kohler W, et al. Genetic heterogeneity in the species Leishmania tropica revealed by different PCR-based methods. Trans R Soc Trop Med Hyg. 2001;95(2):217–24.
Doudi M, Ghasemi F, Setorki M. Genetic polymorphism analysis of Leishmania tropica isolated from three endemic regions (Bam, Kermanshah and Mashhad) in Iran by PCR-RFLP technique and based on ITS1 sequences. Afr J Microbiol Res. 2012;6(12):2970–5.
Abonnenc E. Les phlébotomes de la région éthiopienne (Diptera: Psychodidae). Sér Ent Méd Parasitol: Cah. ORSTOM; 1972. p. 55. 239.
Madulo-Leblond G: Les phlébotomes (Diptera: Phlebotomidae) des Iles Ioniennes. PhD thesis Champagne-Ardenne University, Faculty of Pharmacy, Reims, France; 1983.
Léger N, Pesson B, Madulo-Leblond G, Abonnenc E. Differentiation of females of the subgenus Larroussius Nitzulescu 1931 (Diptera-Phlebotomidae) of the Mediterranean region. Ann Parasitol Hum Comp. 1983;58(6):611–23.
Haddad N, Léger N, Sadek R. Sandflies of Lebanon: faunistic inventory. Parasite. 2003;10(2):99–110.
Ajaoud M, Es-sette N, Hamdi S, El-Idrissi AL, Riyad M, Lemrani M. Detection and molecular typing of Leishmania tropica from Phlebotomus sergenti and lesions of cutaneous leishmaniasis in an emerging focus of Morocco. Parasit Vectors. 2013;6:217.
Tabbabi A, Bousslimi N, Rhim A, Aoun K, Bouratbine A. First report on natural infection of Phlebotomus sergenti with Leishmania promastigotes in the cutaneous leishmaniasis focus in southeastern Tunisia. Am J Trop Med Hyg. 2011;85(4):646–7.
Dvorak V, Votypka J, Aytekin AM, Alten B, Volf P. Intraspecific variability of natural populations of Phlebotomus sergenti, the main vector of Leishmania tropica. J Vector Ecol. 2011;36 Suppl 1:49–57.
Oshaghi MA, Rasolian M, Shirzadi MR, Mohtarami F, Doosti S. First report on isolation of Leishmania tropica from sandflies of a classical urban cutaneous leishmaniasis focus in southern Iran. Exp Parasitol. 2010;126(4):445–50.
Yavar R, Abedin S, Reza AM, Ali OM, Sina R, Mehdi M, et al. Phlebotomus papatasi and Meriones libycus as the vector and reservoir host of cutaneous leishmaniasis in Qomrood district, Qom province, central Iran. Asian Pac J Trop Med. 2011;4(2):97–100.
Hamarsheh O. Distribution of Leishmania major zymodemes in relation to populations of Phlebotomus papatasi sand flies. Parasit Vectors. 2011;25(4):9.
Léger N, Depaquit J, Ferté H, Rioux JA, Gantier JC, Gramiccia M, et al. Phlebotomine sandflies (Diptera-Psychodidae) of the isle of Cyprus. II--Isolation and typing of Leishmania (Leishmania infantum Nicolle, 1908 (zymodeme MON 1) from Phlebotomus (Larroussius) tobbi Adler and Theodor, 1930. Parasite. 2000;7(2):143–6.
Svobodová M, Alten B, Zídková L, Dvorák V, Hlavacková J, Mysková J, et al. Cutaneous leishmaniasis caused by Leishmania infantum transmitted by Phlebotomus tobbi. Int J Parasitol. 2009;39(2):251–6.
Oshaghi MA, Rassi Y, Hazratian T, Fallah E, Rafizadeh S. Natural infection of wild caught Phlebotomus tobbi to Leishmania infantum in East Azerbaijan province, northwestern Iran. J Vector Borne Dis. 2013;50(1):24–9.
Abazid N, Jones C, Davies CR. Knowledge, attitudes and practices about leishmaniasis among cutaneous leishmaniasis patients in Aleppo, Syrian Arab Republic. East Mediterr Health J. 2012;18(1):7–14.
Gürel MS, Yeşilova Y, Olgen MK, Ozbel Y. Cutaneous leishmaniasis in Turkey. Turkiye Parazitol Derg. 2012;36(2):121–9.
Azmi K, Schönian G, Nasereddin A, Schnur LF, Sawalha S, Hamarsheh O, et al. Epidemiological and clinical features of cutaneous leishmaniases in Jenin District, Palestine, including characterization of the causative agents in clinical samples. Trans R Soc Trop Med Hyg. 2012;106(9):554–62.
Pratlong F, Dereure J, Ravel C, Lami P, Balard Y, Serres G, et al. Geographical distribution and epidemiological features of Old World cutaneous leishmaniasis foci, based on the isoenzyme analysis of 1048 strains. Trop Med Int Health. 2009;14(9):1071–85.
Talmi-Frank D, Jaffe CL, Nasereddin A, Warburg A, King R, Svobodova M, et al. Leishmania tropica in rock hyraxes (Procavia capensis) in a focus of human cutaneous leishmaniasis. Am J Trop Med Hyg. 2010;82:814–8.
Dereure J, Rioux JA, Gallego M, Perieres J, Pratlong F, Mahjour J, et al. Leishmania tropica in Morocco: infection in dogs. Trans R Soc Trop Med Hyg. 1991;85:595.
Mohebali M, Malmasi A, Hajjaran H, Jamshidi S, Akhoundi B, Rezaei M, et al. Disseminated Leishmaniasis Caused by Leishmania tropica in a Puppy from Karaj. Central Iran Iran J Parasitol. 2011;6:69–73.
Baneth G, Zivotofsky D, Nachum-Biala Y, Yasur-Landau D, Botero AM. Mucocutaneous Leishmania tropica infection in a dog from a human cutaneous leishmaniasis focus. Parasit Vectors. 2014;7:118.
Rioux JA, Ashford RW, Khiami A. Ecoepidemiology of leishmaniases in Syria. 3. Leishmania major infection in Psammomys obesus provides clues to life history of the rodent and possible control measures. Ann Parasitol Hum Comp. 1992;67(6):163–5.
Rioux JA, Leger N, Haddad N, Jalouk L, Dereure J, Al-Khiami A, et al. Natural infestation of Phlebotomus tobbi (Diptera, Physholidae) by Leishmania donovani (Kinetoplastida, Trypanosomatidae) in Syria. Parassitologia. 1998;40(1):148.
Rioux JA, Lanotte G, Maazoun R, Perello R, Pratlong F. Leishmania infantum Nicolle, 1908, the agent of the autochthonous oriental sore. About the biochemical identification of 2 strains isolated in the eastern Pyrenees. C R Seances Acad Sci D. 1980;291:701–3.
Maroli M, Gramiccia M, Gradoni L. Natural infection of sandfly Phlebotomus perfiliewi with Leishmania infantum in a cutaneous leishmaniasis focus of the Abruzzi region, Italy. Trans R Soc Trop Med Hyg. 1987;81:596–8.
Gramiccia M, Gradoni L, Pozio E. Leishmania infantum sensu lato as an agent of cutaneous leishmaniasis in Abruzzi region (Italy). Trans R Soc Trop Med Hyg. 1987;81:235–7.
Del Giudice P, Marty P, Lacour JP, Perrin C, Pratlong F, Haas H, et al. Cutaneous leishmaniasis due to Leishmania infantum. Case reports and literature review. Arch Dermat. 1998;134:193–8.
Serin MS, Daglioglu K, Bagirova M, Allahverdiyev A, Uzun S, Vural Z, et al. Rapid diagnosis and genotyping of Leishmania isolates from cutaneous and visceral leishmaniasis by microcapillary cultivation and polymerase chain reaction restriction fragment length polymorphism of miniexon region. Diagn Microbiol Infect Dis. 2005;53:209–14.
Acknowledgments
This work was funded by the Department of Defense Global Emerging Infections System (GEIS), ID number C0303_10_ N3_OC HFHSC, the Doctorate School of Science and Technology of the Lebanese University, the Syrian Ministry of Health. The views expressed herein are those of the authors and do not necessarily reflect the official policy or position of the U.S. Department of the Navy, U.S. Department of Defense, or U.S. Government. A co-author was an employee of the U.S. Government, and this work was prepared as part of his official duties. Title 17 U.S.C. §105 provides that “Copyright protection under this title is not available for any work of the United States Government.” Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
NH was involved in the study design, conducted field work (Sand fly collection), performed molecular study, contributed to the sand fly identification and drafted the manuscript. HS identified the sand flies and performed molecular work. AA organized and participated in field work (samples collection), JV was involved in the optimization of the molecular work protocol. SA initiated the study, provided local contacts, contributed to the study design, samples collection and manuscript reviewing. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Haddad, N., Saliba, H., Altawil, A. et al. Cutaneous leishmaniasis in the central provinces of Hama and Edlib in Syria: Vector identification and parasite typing. Parasites Vectors 8, 524 (2015). https://doi.org/10.1186/s13071-015-1147-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-015-1147-0






