Abstract
Equine theileriosis, a tick-transmitted disease caused by the hemoprotozoan parasites Theileria equi and Theileria haneyi, affects equids throughout tropical and subtropical regions of the world. It is a significant regulatory concern in non-endemic countries, where testing for equine theileriosis is required prior to horse import to prevent parasite entry. Within endemic areas, infection causes significant morbidity and mortality, leading to economic losses. No vaccine for equine theileriosis is available, and current drug treatment protocols are inconsistent and associated with significant side effects. Recent work has revealed substantial genetic variability among equine theileriosis organisms, and analysis of ribosomal DNA from affected animals around the world indicates that the organisms can be grouped into five distinct clades. As these diverse parasites are capable of infecting a wide range of both tick and mammalian hosts, movement of different equine Theileria species between endemic countries, and eventually into non-endemic countries, is a significant concern. Furthermore, the substantial genetic variability of these organisms will likely render currently utilized importation diagnostic tests unable to detect all equine Theileria spp. To this end, more complete characterization of these diverse parasites is critical to the continued global control of equine theileriosis. This review discusses current knowledge of equine Theileria spp. in this context, and highlights new opportunities and challenges for workers in this field.

Similar content being viewed by others
Background
Equine theileriosis, caused by Theileria equi and Theileria haneyi, is common throughout tropical and subtropical regions of the world, and greatly constrains international movement of horses [1]. To prevent disease entry, testing of horses for T. equi is required prior to import into non-endemic countries. Furthermore, in equine theileriosis-endemic countries, in which over 90% of the world’s horses reside [2], economic losses due to morbidity and mortality are significant [3], and prevention of disease via vaccination is highly desirable, but not yet possible. Through improved surveillance and molecular technology, our understanding of the diversity of equine Theileria spp. parasites is rapidly expanding and is critical to the continued development of effective diagnostic and preventive strategies. Herein, we review current research regarding the genetic complexity of equine theileriosis, highlighting knowledge gaps and new research opportunities.
The genus Theileria
The phylum Apicomplexa is a diverse and ancient one; some studies suggest it is approximately 800 million years old [4, 5]. The best-known members are mosquito-transmitted malaria parasites in the genus Plasmodium. Apicomplexans are defined by an apical complex of organelles, including rhoptries, micronemes and microspheres (also known as dense granules and spherical bodies). In many of the most economically and medically important genera, such as Theileria, Toxoplasma, Plasmodium and Babesia, the apical complex is necessary for invasion of mammalian host cells and subsequent intracellular establishment of the parasite [6,7,8,9].
Apicomplexan parasites in the genus Theileria are transmitted by ticks and can be divided into transforming and non-transforming types. The transforming Theileria, which infect and immortalize mammalian host leukocytes, resulting in a “cancer-like” phenotype, include the highly pathogenic T. parva, T. annulata and T. lestoquardi. Other host leukocyte-transforming species, such as T. taurotragi, a parasite of Eland, bushbuck and cattle, are less pathogenic. Transforming Theileria spp. undergo schizogony in concert with transformed host cell proliferation, which subsequently gives rise to an intraerythrocytic stage and a sexual cycle in the tick vector. In these parasitic infections, clinical disease is secondary to host cell transformation and the resultant inflammatory immune response [10, 11].
In contrast, non-transforming Theileria species, including the equine parasites T. equi and T. haneyi, and bovine parasites T. orientalis and T. mutans, have only a transient nucleated cell stage and persist as intra-erythrocytic piroplasms [12,13,14,15]. In these infections, clinical disease is secondary to red blood cell destruction and resultant anemia. Due to the prominent intraerythrocytic stage, transplacental transmission [16, 17], as well as iatrogenic transmission via contaminated needles and/or blood products [18,19,20], are common means of transmission in non-transforming Theileria species.
Equine Theileria species
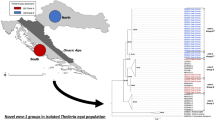
The 18S ribosomal RNA gene sequences of equid-infective Theileria species separate into five clades (Fig. 1 ), which are discussed in detail later in this review. For the purposes of this review, parasites in rDNA clade A comprise T. equi (sensu stricto) which are the best characterized in terms of biology and economic importance, while those in clades B–E, (including T. haneyi in clade C) are designated T. equi (sensu lato).
18S rDNA sequences placed in GenBank and identified as Theileria equi but detected within various host species were gathered and aligned in CLC Genomics Workbench v.10.1.1 (Qiagen), along with outgroup 18S rDNA sequences from other piroplasmids as identified in the figure. All sequences were trimmed to identical length of 401 bp, and span the 4th hypervariable region of full-length 18S rDNA. Aligned sequences were compared in a maximum likelihood phylogeny in the same program, with a Kimura 80 nucleotide substitution model, with neighbor-joining used in constructing initial trees to enable enhanced evolutionary inference. Five hundred replicates were performed in a bootstrap analysis with the values for nodes above a cut-off of 50 indicated on the tree for major branches. The tree provides a representative sample of ribosomal sequences falling into distinct clades across different infected host species and is not is intended to provide an exhaustive depiction of the relationships of all equid Theileria isolates
Comparative genomics studies [12, 21] indicate that T. equi and T. haneyi have features intermediate between Theileria and Babesia and may represent a distinct taxon, closer to Theileria than Babesia. Similar to all other Theileria spp., T. equi undergoes schizogony in leukocytes prior to the development of intraerythrocytic stages [22], unlike Babesia spp., which infect only mammalian erythrocytes. An intraleukocytic life-cycle stage has not yet been demonstrated for T. haneyi, and research is underway to determine whether one exists.
Ultrastructural analysis of Theileria spp. that have so far been studied shows that the mammalian cell entry involves ‘zippering’ of closely juxtaposed parasite and host membranes, but does not require orientation dependent binding of the apical complex and release of the organelle contents, as in Babesia spp. and species of most other apicomplexan genera [6, 7]. Furthermore, in Theileria, the rhoptries and other apical organelles are involved in establishment within the host cell, but not in the entry process as in most Apicomplexa. The role of apical complex organelles in T. equi and T. haneyi host cell entry is currently unknown; however, unlike T. parva and T. annulata, T. equi sporozoites and merozoites possess morphological structures similar to micronemes [23] and may therefore utilize a different mechanism of host cell invasion. One important mechanism of protective immunity to transforming Theileria, including T. parva, depends both on the development of a CD8+ T-cell response to schizont infected host leukocytes and an antibody response to sporozoite antigens [24, 25]. While non-transforming Theileria also have a brief period of schizogony in host leukocytes [22], the existence and significance of a cell-mediated immune response to this parasite stage remains uncharacterized. Unlike the better-known transforming Theileria species, but similar to Babesia, transmission of T. equi in ticks occurs both transtadially and intrastadially [1].
In contrast to other Theileria, T. equi-type parasites infect multiple families of mammals, with reports of infection in dogs [26, 27], camels [28], tapirs [29], waterbuck, cattle, goats [30], and sheep [31] in addition to equids. Clinical disease has been observed in tapirs and dogs, but not in camels. Most other transforming and non-transforming Theileria species parasitize only a single mammalian family and frequently only one to three species within that family [32]. Theileria equi is also promiscuous in the range of ixodid tick vectors utilized, with at least 14 phylogenetically divergent tick species in four different genera implicated in natural and experimental transmission [2]. The ability to survive in many species of mammals and arthropod vectors may be a consequence of the larger complement of genes in T. equi, whose 11.7 megabase (Mb) genome contains 1985 predicted protein-encoding genes that are absent in Babesia bovis and host cell-transforming Theileria species [12]. Theileria haneyi has an approximately 10 Mb genome, intermediate in size between host cell-transforming Theileria and T. equi. This species has a minimum of 878 genes that are absent in T. parva and T. annulata [33].
Novel features of the T. equi genome include major expansions of the multicopy ATP binding cassette transporter protein family and the type II secretory protein family [21], which may contribute to the ability of the parasite to survive in multiple host and vector tissues. Many of the additional genes in T. equi and T. haneyi are located in paralogous multicopy gene families that exhibit no significant sequence identity to proteins in the public databases, although some contain conserved domains. Improved understanding of the molecular basis of promiscuous host and vector preferences therefore awaits a system for genetic manipulation and phenotyping of these parasites. Equid infective Theileria spp. possess FAINT (frequently-associated in Theileria) domains in multiple open reading frames (ORFS). FAINT domains, whose function is unknown, are present in the virtual proteomes of T. parva, T annulata and T. orientalis, but are absent in those of Babesia species [34, 35].
The EMA gene family of T. equi and T. haneyi
An important feature common to T. equi and the newly discovered T. haneyi is a multicopy gene family encoding an immunodominant family of proteins believed to be involved in host erythrocyte invasion and/or exit. These are known as equine merozoite antigen (EMA) proteins, of which there are nine family members (EMA1-9) in both species that are thought to have evolved by gene duplication [21]. By contrast, the genomes of T. parva, T. annulata and T. orientalis each contain only single-copy genes that exhibit synteny with EMA5 in the genomes of the two equine Theileria species [33]. EMA5 is therefore likely orthologous to the single copy homologues in other Theileria species. However, whether EMA5 remains functionally equivalent, given the expansion of the EMA family in equid-infective Theileria, is unknown. Three copies of the family in T. equi, including EMA1, are absent from equivalent positions in the genome of T. haneyi according to syntenic analysis. However, the genome of T. haneyi contains three additional homologous genes with significant identity to the EMA family, but in different genomic locations. These copies are believed to have evolved more recently, and preserve the number of EMA genes [12]. Whether a complement of nine EMA genes is more widely conserved within the complex of Theileria species that are infective to equids and the possible functional divergence of the paralogous copies awaits further analysis. However, the ratio of non-synonymous to synonymous mutations in the EMA family indicates selection for conservation at certain residues, suggesting functional importance [33]. In T. equi, immune response research has focused almost entirely on the humoral immune response to EMA family antigens [1, 33, 36]. The situation is similar in T. orientalis, where almost all immune response data is centered on the immunodominant EMA orthologue, major piroplasm surface protein (MPSP) [37,38,39]. While infected animals develop robust humoral immune responses to these antigens, the responses are largely non-protective. Wise et al. [33] recently showed that the conservation of the EMA family present within the genomes of T. equi and T. haneyi, is probably not a response to immune pressure from the host. Thus, significant work is still required to obtain an understanding of the nature of protective immunity to non-transforming Theileria and its role in parasite evolution and genome diversification.
Phylogeny of equine Theileria species
Parasites that have been classified under the umbrella term ‘Theileria equi’ infect zebra, domestic horses and donkeys [40, 41]. The 18S rRNA gene sequences of equine Theileria fall into either four or five clades based on phylogenetic analysis of hypervariable region 4, depending on how a cluster is defined [42,43,44,45]. Using a maximum likelihood algorithm, following initial implementation of a neighbor-joining approach, we identify five clades in our analysis with clades C and D being relatively similar (Fig. 1, clades A–E). Theileria sequences were selected for inclusion in this analysis from the NCBI nucleotide database by searching for ‘Theileria equi 18S’ and then choosing sequence derived from hypervariable region 4 based on diversity of the mammalian host species listed as the source of the sample. Redundant sequences from the same host and from the same locality were reduced to one representative. Selection was done without regard for whether the sequence was from a published journal article and because accession numbers are provided in the tree, no further references were added to the citations. The ribosomal RNA gene clade containing T. parva and T. annulata is also shown in Fig. 1 in order to provide a scale illustrating the distinctness of the equine-infective Theileria clusters. Co-infection of horses with equine Theileria spp. from different clades was recently documented experimentally [46] and, importantly, in horses and donkeys from the field in South Africa [42] and the Gambia [47], as well as wild zebra from South Africa [42]. Interestingly, Theileria spp. diversity was significantly greater in zebra, with clades A, B, C and D all detected in South African mountain zebra (Equus zebra) and several in Burchell’s zebra (Equus quagga), suggesting that zebra could be an ancestral mammalian host of parasites causing equine theileriosis [42].
Clade A contains Theileria equi (sensu stricto), which causes infections of variable severity, but is capable of inducing severe anemia and resultant mortality in domestic equids. In most instances, however, the incalculable cost of subclinical morbidity and reduced performance of animals used for transport, traction and racing far surpasses the impact of disease mortality [1]. The parasite, which is endemic throughout most of Africa, mainland Europe, the Middle East and Latin America [2], has recently been detected in the UK, previously thought to be disease-free [47]. Diagnosis of T. equi (sensu stricto), which is implemented using a competitive ELISA based on the antigen EMA1, is important for regulation of the global movement of horses [1]. Thus, genetic variation from T. equi (sensu stricto), especially at the EMA1 locus, can significantly hinder detection of such organisms with the currently available diagnostic assays. For instance, due to its complete lack of the EMA1 gene, T. haneyi is not detected by the T. equi competitive ELISA test [8]. Other divergent equine-infective Theileria species could cause similar problems.
Equid apicomplexan 18S rRNA clades C and D, which are genetically relatively similar to one another based on clustering of nodes, as compared to clade A, contain the recently described, novel Theileria species, Theileria haneyi, which was discovered in horses in the USA [12] and subsequently identified in horses from South Africa [42]. The presumptive timescale of speciation of the two fully sequenced equine Theileria species, T. equi and T. haneyi, is estimated at 33 million years [12]. This speciation event long precedes emergence of the only surviving equid genus, Equus, 4–4.5 million years ago [48]. Clades C and D also contain parasites present in common zebra (Equus quagga) and both subspecies of mountain zebra (Equus zebra), as well as dogs. Surprisingly, waterbuck (Kobus ellipsiprymnus defassa) [49], which is in the family Bovidae, and is evolutionarily distinct from equids, was also infected with a clade C parasite. A highly similar ribosomal DNA sequence was also identified in parasites from a Malaysian pony (Fig. 1), demonstrating that 18S rRNA phylotypes related to T. haneyi are present on three continents. Remarkably, the average predicted protein sequence divergence between the genomes of T. equi (Florida isolate) and T. haneyi (23%), is greater than that of the geographically separated, host cell-transforming T. parva and T. annulata (18%). Theileria parva and T. annulata also differ in wildlife host, mammalian cell tropism and tick vector. While the exact percentage difference between protein sequences in the two equid-infective Theileria may alter as annotation is improved through new data resources, such as transcriptomes, these differences suggest functional divergence in at least some of these proteins.
Equid apicomplexan rRNA clade B contains parasites identified in mountain zebra, common zebra, South African domestic horses, and East African waterbuck, the latter originating from the same geographical region as the isolate in clade C (Fig. 1). The complete genome sequence is not yet available for any of the ribosomal phylotypes that cluster in clade B, but it is reasonable to assume based on the extent of genome difference between T. equi and T. haneyi, that these are likely to be distinct at the whole genome level from the parasites clustered in other clades. The equid Theileria rRNA clade E so far only contains parasites from domestic horses (Equus caballus) in two widely separated countries, Korea and Jordan, together with a parasite detected in a dog from Saudi Arabia (Fig. 1). Similar to clade B, there is as yet no complete genome sequence originating from parasites within this ribosomal phylotype. However, the level of difference in the 18S rDNA sequence again suggests that there could be significant evolutionary divergence at the genome level. No wildlife mammalian reservoir has been identified for this group yet. Interestingly, parasites from three of the five 18S clades, namely A, C and E, have been identified in samples from dogs in Europe and the Middle East (Fig. 1), although nothing is yet known about the infection dynamics or biology of T. equi in canids. As was recently discussed [50], rRNA sequences are important for defining potential novel taxa and identifying new avenues of investigation of equid Theileria biology and infection dynamics. However, in future they need to be supported by more conventional investigation of transmission and biology in the mammalian host and tick vectors to ascertain the significance of rRNA phylotype diversity in equids and additional ‘hosts’. Additionally, 18S rRNA sequences, despite their pedigree as phylogenetic markers, represent a relatively small part of the overall parasite genetic complement. They should therefore be supported by complete genome sequences, which are more definitive and informative.
Parasites from members of several clades that have been classified as T. equi (sensu lato) frequently induce long-term asymptomatic infections in zebra. One hundred percent of Grevy’s zebra (Equus grevyi) sampled in Kenya were found to be positive for T. equi-type parasites using PCR with 18S rRNA primers. Similarly, 100% of Cape mountain zebra (Equus zebra zebra) in South Africa were positive using a multiplex real-time PCR assay [51, 52]. Common zebra (Equus quagga) are also infected in East Africa according to 18S rRNA PCR data (DOO and RPB, unpublished data), and a similar high prevalence of infection was observed in asymptomatic zebra in South Africa, with two distinct clades represented within the V4 hypervariable region of the 18S gene [44].
Theileria haneyi does not cause severe disease following experimental infection of normal horses [46], which may explain why it has remained ‘cryptic’ for so long, despite a global distribution. The same may also be true of the parasites in equid-infective ribosomal clades B, D and E, whose full distribution has yet to be systematically investigated. Recent studies of non-pathogenic T. mutans and T. velifera infections of cattle have highlighted the potential importance of co-infections in modulating disease outcomes for T. parva [53]. The same may be true for equid-infective apicomplexan parasites, as recent studies have revealed that T. equi and T. haneyi can co-infect horses [46]. Studies to determine whether clinical response to infection and efficacy of treatment differs in hosts that are co-infected with T. equi and T. haneyi, as compared to those infected with only a single species, are currently underway. These issues are important, given that the recent surveys of the prevalence of different T. equi rRNA gene clades in the field in South Africa and the Gambia revealed that co-infection is frequent [42, 47].
Given the complexities in the phylogeny and infection dynamics of equid Theileria spp., an improved understanding of the prevalence and diversity of all clades within this group of parasites is critical to improved global control of equine theileriosis.
Conclusions
Equid Theileria are promiscuous, with infections so far also documented in camelidae, tapiridae, bovidae and canidae. Further monitoring of prevalence, with a focus on co-infections, accompanied by generation of additional genome sequences from equid-infective parasites with distinct ribosomal phylotypes is therefore a priority for future research. Currently, there are insufficient whole genomes to allow meaningful application of comparative genomics to this group of parasite species. As global warming expands the habitat of competent tick vectors, spread of tick-borne disease is inevitable. Furthermore, with the growing development of resistance to acaricides and the growing public concern regarding chemically mediated control [54,55,56,57], a more complete understanding of the comparative genomics of equine Theileria spp. is crucial to improve global control of equine theileriosis with diagnostics and vaccination. In addition to the issue of host and vector range, a particular area of interest in Theileria is the relationship between infection and pathology. For example, among the equid-infective parasites, T. equi induces more severe hemolytic symptoms in horses than T. haneyi, but the underlying virulence factors encoded by these parasites are as yet unknown. Unlike other Theileria species, the erythrocyte infective stage of T. equi (sensu stricto) can be cultured in vitro, although this is not yet possible for T. haneyi. Thus, T. equi could serve as a model for development of parasite transfection systems that will enable functional analysis and identification of virulence factors in this fascinating group of pathogens.
Availability of data and materials
Not applicable.
Abbreviations
- RNA:
-
Ribonucleic acid
- rRNA:
-
Ribosomal RNA
- Mb:
-
Megabase
- ATP:
-
Adenosine triphosphate
- FAINT:
-
Frequently associated in Theileria
- ORF:
-
Open reading frame
- EMA protein:
-
Equine merozoite associated protein
- MPSP:
-
Major piroplasm surface protein
- PCR:
-
Polymerase chain reaction
References
Wise LN, Kappmeyer LS, Mealey RH, Knowles DP. Review of equine piroplasmosis. J Vet Intern Med. 2013;27:1334–46.
Scoles GA, Ueti MW. Vector ecology of equine piroplasmosis. Annu Rev Entomol. 2015;60:561–80.
Rothschild CM. Equine piroplasmosis. J Equine Vet Sci. 2013;33:497–508.
Levine N, Phylum II. Apicomplexa Levine, 1970. In: Lee JJ, Hutner SH, Bovie EC, editors. An illustrated guide to the protozoa. Lawrence: Society for Protozoologists; 1985. p. 322–74.
Escalante AA, Ayala FJ. Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes. Proc Natl Acad Sci USA. 1995;92:5793–7.
Sam-Yellowe TY. Rhoptry organelles of the Apicomplexa: their role in host cell invasion and intracellular survival. Parasitol Today. 1996;12:308–16.
Shaw MK. Cell invasion by Theileria sporozoites. Trends Parasitol. 2003;19:2–6.
Kemp LE, Yamamoto M, Soldati-Favre D. Subversion of host cellular functions by the apicomplexan parasites. FEMS Microbiol Rev. 2013;37:607–31.
Carruthers VB, Sibley L. Sequential protein secretion from three distinct organelles of Toxoplasma gondii accompanies invasion of human fibroblasts. Eur J Cell Biol. 1997;73:114–23.
Fry LM, Schneider DA, Frevert CW, Nelson DD, Morrison WI, Knowles DP. East coast fever caused by Theileria parva is characterized by macrophage activation associated with vasculitis and respiratory failure. PLoS ONE. 2016;11:e0156004.
Tretina K, Gotia HT, Mann DJ, Silva JC. Theileria-transformed bovine leukocytes have cancer hallmarks. Trends Parasitol. 2015;31:306–14.
Knowles DP, Kappmeyer LS, Haney D, Herndon DR, Fry LM, Munro JB, et al. Discovery of a novel species, Theileria haneyi n. sp., infective to equids, highlights exceptional genomic diversity within the genus Theileria: implications for apicomplexan parasite surveillance. Int J Parasitol. 2018;48:679–90.
Sivakumar T, Hayashida K, Sugimoto C, Yokoyama N. Evolution and genetic diversity of Theileria. Infect Genet Evol. 2014;27:250–63.
Schein E, Rehbein G, Voigt WP, Zweygarth E. Babesia equi (Laveran, 1901) 1. Development in horses and in lymphocyte culture. Tropenmed Parasitol. 1981;32:223–7.
Moltmann UG, Mehlhorn H, Schein E, Rehbein G, Voigt WP, Zweygarth E. Fine structure of Babesia equi (Laveran, 1901) within lymphocytes and erythrocytes of horses: an in vivo and in vitro study. J Parasitol. 1983;69:111–20.
Mekata H, Minamino T, Mikurino Y, Yamamoto M, Yoshida A, Nonaka N, et al. Evaluation of the natural vertical transmission of Theileria orientalis. Vet Parasitol. 2018;263:1–4.
Sant C, d’Abadie R, Pargass I, Basu AK, Asgarali Z, Charles RA, et al. Prospective study investigating transplacental transmission of equine piroplasmosis in thoroughbred foals in Trinidad. Vet Parasitol. 2016;226:132–7.
Hammer JF, Jenkins C, Bogema D, Emery D. Mechanical transfer of Theileria orientalis: possible roles of biting arthropods, colostrum and husbandry practices in disease transmission. Parasit Vectors. 2016;9:34.
Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91.
Gerstenberg C, Allen W, Phipps L. Mechanical transmission of Babesia equi infection in a British herd of horses. In: Proceedings of the 8th International Conference of Equine Infectious Diseases, 23–26 March, Dubai, UAE; 1998.
Kappmeyer LS, Thiagarajan M, Herndon DR, Ramsay JD, Caler E, Djikeng A, et al. Comparative genomic analysis and phylogenetic position of Theileria equi. BMC Genomics. 2012;13:603.
Ramsay JD, Ueti MW, Johnson WC, Scoles GA, Knowles DP, Mealey RH. Lymphocytes and macrophages are infected by Theileria equi, but T cells and B cells are not required to establish infection in vivo. PLoS ONE. 2013;8:e76996.
Mehlhorn H, Schein E. Redescription of Babesia equi Laveran, 1901 as Theileria equi Mehlhorn, Schein 1998. Parasitol Res. 1998;84:467–75.
McKeever DJ, Taracha EL, Innes EL, MacHugh ND, Awino E, Goddeeris BM, et al. Adoptive transfer of immunity to Theileria parva in the CD8+ fraction of responding efferent lymph. Proc Natl Acad Sci USA. 1994;91:1959–63.
Musoke AJ, Nantulya VM, Buscher G, Masake RA, Otim B. Bovine immune response to Theileria parva: neutralizing antibodies to sporozoites. Immunology. 1982;45:663–8.
Beck R, Vojta L, Mrljak V, Marinculic A, Beck A, Zivicnjak T, et al. Diversity of Babesia and Theileria species in symptomatic and asymptomatic dogs in Croatia. Int J Parasitol. 2009;39:843–8.
Rosa CT, Pazzi P, Nagel S, McClure V, Christie J, Troskie M, et al. Theileriosis in six dogs in South Africa and its potential clinical significance. J S Afr Vet Assoc. 2014;85:1114.
Qablan MA, Sloboda M, Jirků M, Oborník M, Dwairi S, Amr ZS, et al. Quest for the piroplasms in camels: identification of Theileria equi and Babesia caballi in Jordanian dromedaries by PCR. Vet Parasitol. 2012;186:456–60.
Da Silveira AW, De Oliveira GG, Menezes Santos L, da Silva Azuaga LB, Macedo Coutinho CR, Echeverria JT, et al. Natural infection of the South American tapir (Tapirus terrestris) by Theileria equi. J Wildl Dis. 2017;53:411–3.
Zhang J, Kelly P, Li J, Xu C, Wang C. Molecular detection of Theileria spp. in livestock on five Caribbean islands. Biomed Res Int. 2015;2015:624728.
Azmi K, Al-Jawabreh A, Abdeen Z. Molecular detection of Theileria ovis and Theleiria equi in livestock from Palestine. Sci Rep. 2019;9:11557.
Bishop R, Musoke A, Morzaria S, Gardner M, Nene V. Theileria: intracellular protozoan parasites of wild and domestic ruminants transmitted by ixodid ticks. Parasitology. 2004;129:S271–83.
Wise LN, Kappmeyer LS, Knowles DP, White SN. Evolution and diversity of the EMA families of the divergent equid parasites, Theileria equi and T. haneyi. Infect Genet Evol. 2019;68:153–60.
Pain A, Renauld H, Berriman M, Murphy L, Yeats CA, Weir W, et al. Genome of the host-cell transforming parasite Theileria annulata compared with T. parva. Science. 2005;309:131–3.
El-Sayed SAE, Rizk MA, Terkawi MA, Mousa A, Elsayed G, Fouda M, et al. Cocktail of Theileria equi antigens for detecting infection in equines. Asian Pac J Trop Biomed. 2015;5:977–81.
Silva MG, Graça T, Suarez CE, Knowles DP. Repertoire of Theileria equi immunodominant antigens bound by equine antibody. Mol Biochem Parasitol. 2013;188:109–15.
Zhuang W, Sugimoto C, Matsuba T, Niinuma S, Murata M, Onuma M. Analyses of antigenic and genetic diversities of Theileria sergenti piroplasm surface proteins. J Vet Med Sci. 1994;56:469–73.
Onuma M, Kakuda T, Sugimoto C. Theileria parasite infection in East Asia and control of the disease. Comp Immunol Microbiol Infect Dis. 1998;21:165–77.
Jenkins C, Bogema DR. Factors associated with seroconversion to the major piroplasm surface protein of the bovine haemoparasite Theileria orientalis. Parasit Vectors. 2016;9:106.
Schein E. Equine babesiosis. In: Ristic M, editor. Babesiosis of domestic animals and man. Boca Raton: CRC Press; 1988. p. 197–208.
Friedhoff KT, Tenter AM, Muller I. Haemoparasites of equines: impact on international trade of horses. Rev Sci Tech. 1990;9:1187–94.
Bhoora RV, Collins NE, Schnittger L, Troskie C, Marumo R, Labuschagne K, et al. Molecular genotyping and epidemiology of equine piroplasmids in South Africa. Ticks Tick Borne Dis. 2019;11:101358.
Peckle M, Pires MS, Silva CBD, Costa RLD, Vitari GLV, Senra MVX, et al. Molecular characterization of Theileria equi in horses from the state of Rio de Janeiro, Brazil. Ticks Tick Borne Dis. 2018;9:349–53.
Bhoora R, Buss P, Guthrie AJ, Penzhorn BL, Collins NE. Genetic diversity of piroplasms in plains zebra (Equus quagga burchellii) and Cape mountain zebra (Equus zebra zebra) in South Africa. Vet Parasitol. 2010;174:145–9.
Salim B, Bakheit MA, Kamau J, Nakamura I, Sugimoto C. Nucleotide sequence heterogeneity in the small subunit ribosomal RNA gene within Theileria equi from horses in Sudan. Parasitol Res. 2010;106:493–8.
Sears KP, Kappmeyer LS, Wise LN, Silva M, Ueti MW, White S, et al. Infection dynamics of Theileria equi and Theileria haneyi, a newly discovered apicomplexan of the horse. Vet Parasitol. 2019;271:68–75.
Coultous RM, Phipps P, Dalley C, Lewis J, Hammond TA, Shiels BR, et al. Equine piroplasmosis status in the UK: an assessment of laboratory diagnostic submissions and techniques. Vet Rec. 2019;184:95.
Orlando L, Ginolhac A, Zhang G, Froese D, Albrechtsen A, Stiller M, et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature. 2013;499:74–8.
Githaka N, Konnai S, Bishop R, Odongo D, Lekolool I, Kariuki E, et al. Identification and sequence characterization of novel Theileria genotypes from the waterbuck (Kobus defassa) in a Theileria parva-endemic area in Kenya. Vet Parasitol. 2014;202:180–93.
Uilenberg G, Gray J, Kahl O. Research on Piroplasmorida and other tick-borne agents: are we going the right way? Ticks Tick Borne Dis. 2018;9:860–3.
Smith RM, Bhoora RV, Kotzé A, Grobler JP, Lee Dalton D. Translocation a potential corridor for equine piroplasms in Cape mountain zebra (Equus zebra zebra). Int J Parasitol Parasites Wildl. 2019;9:130–3.
Hawkins E, Kock R, McKeever D, Gakuya F, Musyoki C, Chege SM, et al. Prevalence of Theileria equi and Babesia caballi as well as the identification of associated ticks in sympatric Grevy’s zebras (Equus grevyi) and donkeys (Equus africanus asinus) in northern Kenya. J Wildl Dis. 2015;51:137–47.
Woolhouse ME, Thumbi SM, Jennings A, Chase-Topping M, Callaby R, Kiara H, et al. Co-infections determine patterns of mortality in a population exposed to parasite infection. Sci Adv. 2015;1:e1400026.
Stone NE, Olafson PU, Davey RB, Buckmeier G, Bodine D, Sidak-Loftis LC, et al. Multiple mutations in the para-sodium channel gene are associated with pyrethroid resistance in Rhipicephalus microplus from the United States and Mexico. Parasit Vectors. 2014;7:456.
Eiden AL, Kaufman PE, Oi FM, Allan SA, Miller RJ. Detection of permethrin resistance and fipronil tolerance in Rhipicephalus sanguineus (Acari: Ixodidae) in the United States. J Med Entomol. 2015;52:429–36.
Aenishaenslin C, Michel P, Ravel A, Gern L, Waaub JP, Milord F, et al. Acceptability of tick control interventions to prevent Lyme disease in Switzerland and Canada: a mixed-method study. BMC Public Health. 2016;16:12.
Adalja A, Sell ST, McGinty M, Boddie C. Genetically modified mosquito use to reduce mosquito-transmitted disease in the US: a community opinion survey. PLoS Curr. 2016. https://doi.org/10.1371/currents.outbreaks.1c39ec05a743d41ee39391ed0f2ed8d3.
Acknowledgements
The authors wish to acknowledge the excellent technical support and animal care provided by Shelby Beckner, David Herndon, Emma Karel, Cody Evans, Ralph Horn, and Megan Jacks. Their work has enabled our group to make many of the discoveries referenced in this article.
Funding
This work was supported by the Agricultural Research Service (ARS), USA, CRIS#2090-320000-034-00D.
Author information
Authors and Affiliations
Contributions
RPB, DPK and LMF conceived and outlined the manuscript. RPB, LSK, KPS, CKO, DOO, NG and LMF wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Bishop, R.P., Kappmeyer, L.S., Onzere, C.K. et al. Equid infective Theileria cluster in distinct 18S rRNA gene clades comprising multiple taxa with unusually broad mammalian host ranges. Parasites Vectors 13, 261 (2020). https://doi.org/10.1186/s13071-020-04131-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04131-0