Abstract
Background
Melophagus ovinus (Diptera: Hippoboscidae), a hematophagous ectoparasite, is mainly found in Europe, Northwestern Africa, and Asia. This wingless fly infests sheep, rabbits, and red foxes, and causes inflammation, wool loss and skin damage. Furthermore, this parasite has been shown to transmit diseases, and plays a role as a vector. Herein, we investigated the presence of various Rickettsia species in M. ovinus.
Methods
In this study, a total of 95 sheep keds were collected in Kuqa County and Alaer City southern region of Xinjiang Uygur Autonomous Region, northwestern China. First, collected sheep keds were identified on the species level using morphological keys and molecular methods based on a fragment of the 18S ribosomal DNA gene (18S rDNA). Thereafter, to assess the presence of rickettsial DNA in sheep keds, the DNA of individual samples was screened by PCR based on six Rickettsia-specific gene fragments originating from six genes: the 17-kilodalton antigen gene (17-kDa), 16S rRNA gene (rrs), surface cell antigen 4 gene (sca4), citrate synthase gene (gltA), and outer membrane protein A and B genes (ompA and ompB). The amplified products were confirmed by sequencing and BLAST analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome).
Results
According to its morphology and results of molecular analysis, the species was identified as Melophagus ovinus, with 100% identity to M. ovinus from St. Kilda, Australia (FN666411). DNA of Rickettsia spp. were found in 12 M. ovinus samples (12.63%, 12/95). Rickettsia raoultii and R. slovaca were confirmed based on phylogenetic analysis, although the genetic markers of these two rickettsial agents amplified in this study showed molecular diversity.
Conclusions
This is the first report of R. raoultii and R. slovaca DNA in M. ovinus. Rickettsia slovaca was found for the first time around the Taklimakan Desert located in China. This finding extends the geographical range of spotted fever group rickettsiae.
Similar content being viewed by others
Background
Melophagus ovinus, also referred to as the louse fly or sheep ked, is a wingless insect that belongs to the family Hippoboscidae (Diptera: Hippoboscoidea). Sheep keds are one of the most common and economically important blood-feeding ectoparasites [1]. This wingless arthropod is roughly 4 to 6 mm long and has a small head with strong piercing mouthparts. The abdominal area is wide, and the three pairs of legs are tipped with claws. As a Palaearctic species, M. ovinus has a wide geographical distribution [2]. For example, M. ovinus is native to most part of Europe, Northwestern Africa, Mongolia and North India, and this ectoparasite has been introduced into and established in Kenya, South Africa, Japan, Australia, New Zealand and most of North America [3]. In China, a small number of reports have recorded the presence of this arthropod in Qinghai, Shandong Province, and Xinjiang Uygur Autonomous Region (XUAR) [2, 4]. Although sheep are generally considered as the main host, M. ovinus has been observed to infest a broader range of domestic animals (goats and dogs), and wild animals (European bisons, rabbits and red foxes) as well as human beings [2, 5, 6].
Melophagus ovinus lives (as adult) on the hairs or fleece of their hosts, visiting the skin to feed on blood. The life-cycle of M. ovinus comprises the larva, pupa, nymph and adult stages [5]. The female produces a single fully developed larva every 6–8 days that firmly attaches to the wool and becomes a puparium in 6–12 h. The puparium later develops into an adult within 19–30 days [2]. As M. ovinus is regarded as a permanent ectoparasite, the transfer of keds from an infested to a non-infested sheep occurs by direct contact [1]. Some studies have indicated that infestation by M. ovinus results in pruritus, wool loss and skin damage, due to scratching, biting and rubbing, that leads to inflammation [2, 7]. Additionally, damaged areas of the skin are entry portals for bacterial infections and cutaneous myiasis, which may result in tissue necrosis, and ultimately reduces the value of the hide [5].
Melophagus ovinus is a biological vector of Trypanosoma melophagium, an apathogenic protozoan [1, 8]. Luedke et al. reported that M. ovinus is able mechanically to transmit bluetongue virus, which is responsible for a severe infectious disease of ruminants [9]. Additionally, M. ovinus may be a carrier for two organisms, e.g. Bartonella schoenbuchensis and B. chomeli, found in the USA [10], and Anaplasma ovis has been detected in M. ovinus in Hungary [11]. Similarly, Chu et al. reported that the DNA of Borrelia garinii and B. valaisiana-related group was present in M. ovinus [12]. Moreover, recent reports by Kumsa et al. revealed the presence of Acinetobacter spp. in sheep keds in Ethiopia [13].
In 2011, Hornok et al. reported the DNA of unidentified Rickettsia species in M. ovinus, based on the presence of Rickettsia citrate synthase gene (gltA) [11]. Herein, Rickettsia agents were detected in M. ovinus using six rickettsial genetic markers, the 17-kilodalton antigen gene (17-kDa), gltA, 16S rRNA gene (rrs), outer membrane protein A gene (ompA), surface cell antigen 4 gene (sca4), and outer membrane protein B gene (ompB).
Methods
Study areas and animals
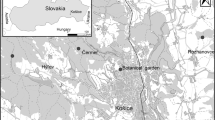
In April 2016, sheep keds were collected from sheep in two locations near the Taklimakan Desert in the southern region of XUAR: (1) in Wuzun Town, Kuqa County (n = 89) (1,070.0 m above sea level; 41°72′N, 83°06′E), and (2) in Tuokayi Town, Alaer City (n = 6) (1,016.0 m above sea level; 40°53′N, 81°12′E).
Sheep ked collection and morphological identification
Each sheep ked was manually removed using forceps or by hand to avoid any damage, and were then placed into 70% ethanol for subsequent identification. All sheep keds that were collected from the same infested animal were placed into pre-labeled vials and transported to the Laboratory of the School of Medicine, Shihezi University. Photographs of the sheep keds were taken with a Leica stereo microscope M165 C (LEICA M165 C, Solms, Germany). The sex and stage were determined according to standard morphological keys [13].
DNA extraction and molecular analyses
Prior to DNA extraction, each sheep ked specimen was rinsed twice in sterile water for 15 min and then dried on sterile filter paper. Genomic DNA was individually extracted by using the TIANamp Genomic DNA Kit (TIANGEN, Beijing, China), according to the instructions provided by the manufacturer. The DNA from each specimen was eluted in 60 μl of Tris-EDTA buffer solution and stored at -20 °C under sterile conditions to preclude contamination until the sample was used for polymerase chain reaction (PCR) analysis.
To examine the phylogenetic relationships within Hippoboscidae, all DNA samples were subjected to PCR to amplify a ~985 bp fragment of the 18S ribosomal DNA gene (18S rDNA). The primers 18S-F: 5′-GTC TCA AAG ATT AAG CCA TGC ATG-3′ and 18S-R: 5′-CTT GTT AGG TTC ACC TAC GGA AAC-3′ were used in this study (the primers were designed by Primer Premier 5.0 software). The thermocycling conditions were as follows: 95 °C for 5 min, 35 cycles at 94 °C for 40 s, 57 °C for 40 s and 72 °C for 1 min and 30 s, followed by a final extension at 72 °C for 10 min. Moreover, in parallel with each amplification reaction, a negative control (distilled water) was included. Out of all the samples, 20 PCR products were randomly purified using the TIANgel Midi Purification Kit (TIANGEN, Beijing, China) and sequenced by Sangon Biotech Co., Ltd (Shanghai, China).
Detection of rickettsial agents and sequence analysis
Identification of Rickettsia spp. was performed by PCR. Primers targeted six Rickettsia-specific gene fragments as follows: a 434 bp fragment of the 17-kDa, 834 bp of gltA, 629 bp of ompA, 1332 bp of rrs, 920 bp of sca4, and 865 bp of ompB, according to previous descriptions [14–17]. Sterile water was used as a negative control and the DNA of spotted fever group rickettsiae amplified in our laboratory was used as a positive control [18–20]. The primers and cycling conditions are shown in Additional file 1. Purification and sequencing of the PCR products were conducted as described above. Phylogenetic trees were made based on the sequence distance method using the neighbor-joining (NJ) and maximum-likelihood (ML) algorithms implemented in the Molecular Evolutionary Genetics Analysis (MEGA) 6 software [21].
Results
A total of 95 sheep keds were collected from three sheep flocks (n = 230). Morphologically, these ectoparasites were identified as Melophagus ovinus (Fig. 1a-f). The length of the 18S rDNA sequences amplified from M. ovinus was 985 bp, which was longer than the available sequences of M. ovinus in GenBank (FN666411, from St. Kilda, Australia). Interestingly, molecular analysis of 20 M. ovinus samples showed that two different lineages exist due to their diversity (99.8–100%) in 18S rDNA flanking fragments. The full length sequences from our study were deposited in GenBank (KX506727 and KX506728).
Out of 95 M. ovinus samples 12 were found to be positive for the six Rickettsia genetic markers (17-kDa, gltA, ompA, rrs, sca4 and ompB). Out of the 12 positive samples, four were confirmed as R. raoultii, and the remaining eight were identified as R. slovaca on the basis of the 17-kDa - gltA - ompA - rrs - sca4 - ompB concatenated sequence (Fig. 2).
Phylogenetic tree of the 17-kDa - gltA - ompA - rrs - sca4 - ompB concatenated sequence of Rickettsia slovaca (▲) and R. raoultii (◆) from Melophagus ovinus obtained in this study and sequences from Rickettsia species retrieved from the GenBank database. The tree was constructed on the basis of neighbor-joining (NJ; 500 bootstrap replicates) and maximum-likelihood (ML, 1,000 bootstrap replicates) analyses using MEGA6. The scale bar represents the inferred substitutions per nucleotide site. The relative support for clades in the tree was produced from the NJ and ML analyses
Concerning R. raoultii, the loci rrs and 17-kDa were identical in each sample and showed identities of 100% (1,196/1,196 bp) and 99.51% (407/409 bp), respectively, when compared with the R. raoultii strain Khabarovsk (CP010969). However, in the remaining four genetic markers the differences appeared to be more pronounced. For example, genetic analysis revealed two different sequences for gltA, three sequences for ompA, three sequences for ompB, and two sequences for sca4.
Concerning R. slovaca, sequences of the 17-kDa, gltA, rrs, and sca4 genes appeared to be conserved, and showed similarities of 100% (411/411 bp) for 17-kDa, 100% (834/834 bp) for gltA, 100% (1,196/1,196 bp) for rrs, and 99.88% (867/868 bp) for sca4 when compared with those of R. slovaca strain D-CWPP (CP003375). However, for both ompA and ompB, three different sequences were obtained, respectively. The detailed similarities and divergences of the sequences in this study are shown in the Additional file 2. All sequences from this study were deposited in the GenBank database (16S: KX506722–KX506723; 17-KDa: KX506725–KX506726; gltA: KX506730–KX506732; ompA: KX506733–KX506738; ompB: KX506739–KX506744; sca4: KX506745–KX506747).
Discussion
An overall prevalence of Rickettsia spp. in M. ovinus collected from sheep in southern XUAR was 12.63% (12/95). In these samples the presence of R. raoultii and R. slovaca DNA was confirmed by conventional PCR followed by sequencing. To the best of our knowledge, this is the first molecular evidence of R. raoultii and R. slovaca DNA in M. ovinus.
Rickettsia raoultii was first identified in 1999 [22]. In the past five years, this species has been detected (among the others) in Mongolia, Georgia, Germany, Slovakia and China [23–27]. To date, the DNA of R. raoultii was shown to be present in 14 tick species of the genera Dermacentor, Rhipicephalus, Haemaphysalis, Hyalomma, Ixodes and Amblyomma [18]. Rickettsia slovaca, a member of the spotted fever group (SFG) rickettsiae, was first isolated in 1968 from Dermacentor marginatus in Slovakia [28], and later on described as the causative agent of tick-borne lymphadenopathy [29]. This species has been detected subsequently in Georgia [24], Germany [25], Greece and Turkey [30, 31]. In 2012, R. slovaca and R. raoultii were reported for the first time in Dermacentor silvarum ticks in XUAR, northwest China [32]. However, R. raoultii has seldom been reported in other arthropods. In this study, molecular evidence is provided for the presence of R. raoultii DNA in sheep keds (M. ovinus).
In a previous study, an unidentified Rickettsia species was detected in M. ovinus collected in Hungary, with a prevalence of 1.67% (1/60) based on the gltA gene detection [11]. Recently, two reports have shown the absence of Rickettsia spp. in M. ovinus collected in Ethiopia and the Czech Republic [13, 33].
Herein, a high prevalence (12.63%, 12/95) of R. raoultii and R. slovaca DNA was demonstrated in M. ovinus from the Taklimakan Desert in China. The observations from this study have extended the spectrum of pathogens potentially present in M. ovinus. Future investigations are warranted to elucidate the genetic diversity of R. raoultii and R. slovaca in M. ovinus. Additionally, the presence of these Rickettsia spp. should be examined in a broader range of arthropods.
Conclusions
This is the first report of genetic markers of R. raoultii and R. slovaca in M. ovinus. Rickettsia slovaca was found for the first time around the Taklimakan Desert located in China. This findings extend our knowledge on the geographical distribution of spotted fever group rickettsiae.
Abbreviations
- 17-kDa :
-
17-kilodalton antigen gene
- gltA :
-
Citrate synthase gene
- MEGA:
-
Molecular evolutionary genetics analysis
- ML:
-
Maximum-likelihood
- NJ:
-
Neighbor-joining
- ompA :
-
Outer membrane protein A gene
- ompB :
-
Outer membrane protein B gene
- PCR:
-
Polymerase chain reaction
- rrs :
-
16S rRNA gene
- sca4 :
-
Surface cell antigen 4 gene
- SFG:
-
Spotted fever group
- XUAR:
-
Xinjiang Uygur Autonomous Region
References
Gibson W, Pilkington JG, Pemberton JM. Trypanosoma melophagium from the sheep ked Melophagus ovinus on the island of St Kilda. Parasitology. 2010;137(12):1799–804.
Small RW. A review of Melophagus ovinus, (L.), the sheep ked. Vet Parasitol. 2005;130(1–2):141–55.
ZipcodeZoo. Melophagus ovinus. http://zipcodezoo.com/index.php/Melophagus_ovinus. php/Melophagus ovinus#pane1. Accessed 30 Jul 2016.
Wang Y, Sun BJ, Yu CY. Melophagus ovinus Linnaeus intercepted and captured from Canadian pickled goatskins at Linyi port. Chin J Vector Bio Control. 2010;21(1):35.
Kumsa B, Parola P, Raoult D, Socolovschi C. Bartonella melophagi in Melophagus ovinus (sheep ked) collected from sheep in northern Oromia, Ethiopia. Comp Immunol Microbiol Infect Dis. 2014;37(1):69–76.
Bequaert J. A monograph of the Melophaginae, or ked-flies, of sheep, goats, deer and antelopes (Diptera, Hippoboscidae). Entomol Americana. 1942;22:1–220.
Soulsby EJL. Helminths, arthropods and protozoa of domesticated animals (sixth edition of MSnnig’s Veterinary helminthology and entomology). Can Vet J. 1969;10(8):223.
Martinkovic F, Matanovic K, Rodrigues AC, Garcia HA, Teixeira MM. Trypanosoma (Megatrypanum) melophagium in the sheep ked Melophagus ovinus from organic farms in Croatia: phylogenetic inferences support restriction to sheep and sheep keds and close relationship with Trypanosomes from other ruminant species. J Eukaryot Microbiol. 2012;59(2):134–44.
Luedke AJ, Jochim MM, Bowne JG. Preliminary bluetongue transmission with the sheep ked Melophagus ovinus (L.). Can J Comp Med Vet Sci. 1965;29(9):229–31.
Halos L, Jamal T, Maillard R, Girard B, Guillot J, Chomel B, et al. Role of Hippoboscidae flies as potential vectors of Bartonella spp. infecting wild and domestic ruminants. Appl Environ Microbiol. 2004;70(10):6302–5.
Hornok S, de la Fuente J, Biró N, de Fernández Mera IG, Meli ML, Elek V, et al. First molecular evidence of Anaplasma ovis and Rickettsia spp. in keds (Diptera: Hippoboscidae) of sheep and wild ruminants. Vector Borne Zoonotic Dis. 2011;11(10):1319–21.
Chu CY, Jiang BG, Qiu EC, Zhang F, Zuo SQ, Yang H, et al. Borrelia burgdorferi sensu lato in sheep keds (Melophagus ovinus), Tibet, China. Vet Microbiol. 2011;149(3–4):526–9.
Kumsa B, Socolovschi C, Parola P, Rolain JM, Raoult D. Molecular detection of Acinetobacter species in lice and keds of domestic animals in Oromia Regional State, Ethiopia. PLoS One. 2012;7(12), e52377.
Anstead CA, Chilton NB. A novel Rickettsia species detected in vole ticks (Ixodes angustus) from Western Canada. Appl Environ Microbiol. 2013;9(24):7583–9.
Mcintosh D, Bezerra RA, Luz HR, Faccini JL, Gaiotto FA, Gine GA, et al. Detection of Rickettsia bellii and Rickettsia amblyommii in Amblyomma longirostre (Acari: Ixodidae) from Bahia state, Northeast Brazil. Braz J Microbiol. 2015;46(3):879–83.
Zhang L, Jin J, Fu X, Raoult D, Fournier PE. Genetic differentiation of Chinese isolates of Rickettsia sibirica by partial ompA gene sequencing and multispacer typing. J Clin Microbiol. 2006;44(7):2465–7.
Zhao SS, Li HY, Yin XP, Liu ZQ, Chen CF, Wang YZ. First detection of Candidatus Rickettsia barbariae in the flea Vermipsylla alakurt from north-western China. Parasit Vectors. 2016;9(1):1–5.
Guo LP, Mu LM, Xu J, Jiang SH, Wang AD, Chen CF, et al. Rickettsia raoultii in Haemaphysalis erinacei from marbled polecats, China-Kazakhstan border. Parasit Vectors. 2015;8(1):1–3.
Guo LP, Jiang SH, Liu D, Wang SW, Chen CF, Wang YZ. Emerging spotted fever group rickettsiae in ticks, northwestern China. Ticks Tick Borne Dis. 2016;7(6):1146–50.
Wei QQ, Guo LP, Wang AD, Mu LM, Zhang K, Chen CF, et al. The first detection of Rickettsia aeschlimannii and Rickettsia massiliae in Rhipicephalus turanicus ticks, in northwest China. Parasit Vectors. 2015;8(1):1–4.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30(12):2725–9.
Rydkina E, Roux V, Rudakov N, Gafarova M, Tarasevich I, Raoult D. New Rickettsiae in ticks collected in territories of the former Soviet Union. Emerg Infect Dis. 1999;5(6):811–4.
Speck S, Derschum H, Damdindorj T, Dashdavaa O, Jiang J, Kaysser P, et al. Rickettsia raoultii, the predominant Rickettsia, found in Mongolian Dermacentor nuttalli. Ticks Tick Borne Dis. 2012;3(4):227–31.
Jiang J, You BJ, Liu E, Apte A, Yarina TR, Myers TE, et al. Development of three quantitative real-time PCR assays for the detection of Rickettsia raoultii, Rickettsia slovaca, and Rickettsia aeschlimannii, and their validation with ticks from the country of Georgia and the Republic of Azerbaijan. Ticks Tick Borne Dis. 2012;3(5–6):327–31.
Wächter M, Wölfel S, Pfeffer M, Dobler G, Kohn B, Moritz A, et al. Serological differentiation of antibodies against Rickettsia helvetica, R. raoultii, R. slovaca, R. monacensis and R. felis in dogs from Germany by a micro-immunofluorescent antibody test. Parasit Vectors. 2015;8(1):585–5.
Špitalská E, Štefanidesová K, Kocianová E, Boldiš V. Rickettsia slovaca and Rickettsia raoultii in Dermacentor marginatus and Dermacentor reticulatus ticks from Slovak Republic. Exp Appl Acarol. 2012;57(2):189–97.
Wen J, Jiao D, Wang JH, Yao DH, Liu ZX, Zhao G, et al. Rickettsia raoultii, the predominant Rickettsia found in Dermacentor silvarum ticks in China-Russia border areas. Exp Appl Acarol. 2014;63(4):579–85.
Rehácěk J, Řeháček J. Rickettsia slovaca, the organism and its ecology. Acta SC Nat Brno. 1984;18:1–50.
Killmaster LF, Zemtsova GE, Montgomery M, Schumacher L, Burrows M, Levin ML. Isolation of a Rickettsia slovaca-like agent from Dermacentor variabilis ticks in vero cell culture. Vector Borne Zoonotic Dis. 2016;16(1):61–2.
Kostopoulou V, Chochlakis D, Kanta C, Katsanou A, Rossiou K, Rammos A, et al. A case of human infection by Rickettsia slovaca in Greece. Jpn J Infect Dis. 2016;69(4):335–7.
Orkun Ö, Karaer Z, Çakmak A, Nalbantoğlu S. Spotted fever group rickettsiae in ticks in Turkey. Ticks Tick Borne Dis. 2014;5(2):213–8.
Tian ZC, Liu GY, Shen H, Xie JR, Luo J, Tian MY. First report on the occurrence of Rickettsia slovaca and Rickettsia raoultii in Dermacentor silvarum in China. Parasit Vectors. 2012;5(1):1–4.
Rudolf I, Betášová L, Bischof V, Venclíková K, Blažejová H, Mendel J, et al. Molecular survey of arthropod-borne pathogens in sheep keds (Melophagus ovinus), Central Europe. Parasitol Res. 2016;115(10):3679–82.
Acknowledgements
We would like to thank Dr Sándor Hornok (Department of Parasitology and Zoology, University of Veterinary Medicine, Budapest, Hungary) who revised the manuscript.
Funding
This research was supported in part by grants from the National Natural Science Foundation of China (Grant U1503283 and No. 81560338), China Agriculture Research System (CARS-40-11), Special Fund for Agro-scientific Research in the Public Interest (No. 201303037), the International Cooperation Projects of Xinjiang Uygur Autonomous Region (No.20156016), and the Co-innovation Center for the High Incidence of Zoonotic Disease Prevention and Control in Western China (2013–179).
Availability of data and materials
The newly-generated sequences were deposited in the GenBank database (16S: KX506722–KX506723; 17-KDa: KX506725–KX506726; gltA: KX506730–KX506732; ompA: KX506733–KX506738; ompB: KX506739–KX506744; sca4: KX506745–KX506747).
Authors’ contributions
YZW and CFC conceived and designed the study and critically revised the manuscript. ZQL, SWW, HZW and CCT performed the sheep ked collection. DL and HZ performed the laboratory studies. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
The contents of this manuscript do not warrant publication consent.
Ethical approval and consent to participate
This study was approved by the Animal Ethics Committee of Shihezi University (Approval No. AECSU2015-11).
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1:
PCR protocol for the detection of Rickettsia spp., Xinjiang, China. (DOCX 22 kb)
Additional file 2:
Closest relative sequences of the Rickettsia raoultii and Rickettsia slovaca detected in the sheep keds (Melophagus ovinus), Northwest of China. (DOC 109 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liu, D., Wang, YZ., Zhang, H. et al. First report of Rickettsia raoultii and R. slovaca in Melophagus ovinus, the sheep ked. Parasites Vectors 9, 600 (2016). https://doi.org/10.1186/s13071-016-1885-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-016-1885-7