Abstract
Background
Bovine babesiosis and theileriosis is an important hemoprotozoal disease in cattles and yaks in tropical and subtropical regions leading to significant economic losses. In the field, the risk of co-infection between the bovine Babesia and Theileria species is very high. Thus, it is necessary to develop a simple, accurate, rapid and cost-effective method for large-scale epidemic investigation, in particular for the detection of co-infection in field.
Methods
In this study, DNA sequences of a ribosomal protein S8 (RPS8) gene from eight species of cattle piroplasms in China were used to develop a species-specific PCR-RFLP diagnostic tool. The eight Theileria and Babesia species could be differentiated by digesting the RPS8 PCR product with Mbo I.
Results
The sensitivity of the PCR assays was 0.1 pg DNA for Babesia species but 1 pg DNA for Theileria species. The clearly different size of the PCR-RFLP products allowed for a direct discrimination between eight bovine Theileria and Babesia species (T. annulata, T. sinensis, T. sergenti, B. ovata, B. bovis, B. bigemina, B. major and Babesia species Kashi isolate).
Conclusion
Our results indicated that the established method based on the RPS8 gene was a reliable molecular diagnostic tool for the simultaneous detection and identification of bovine Babesia and Theileria species in China, which could be applicable for the survey of parasite dynamics, epidemiological studies as well as prevention and control of the disease.
Similar content being viewed by others
Background
Piroplasms, comprising mainly the genera Babesia and Theileria, are tick-transmitted protozoa that are pathogenic to ruminants, horses, pigs, dogs, cats and cattle, and in some cases, even to humans. In the vertebrate hosts, the infection usually causes fever, anemia and haemoglubinuria, and in severe cases, death [1, 2]. Animals that recover from acute or primary infections remain chronically infected, and act as reservoirs for ticks [3].
Eight species of bovine Babesia and Theileria (B. bigemina, B. bovis, B. major, B. ovata, an unidentified Babesia sp., T. annulata, T. sergenti and T. sinensis) have been identified in China [4]. The eight species of bovine Babesia and Theileria species can cause a significant loss in meat and milk production from cattles in some parts of China. Boophilus microplus has been demonstrated to be the vector of B. bigemina and B. bovis [5], while Haemaphysalis longicornis and H. punctata are potential vectors of B. major [6, 7]. H. longicornis is also considered to be the vector of B. ovata and T. sergenti [4, 8]. Nymphal progeny derived from female Hyalomma anatolicum anatolicum collected from the field were shown to be capable of transmitting an unidentified Babesia sp. (Designated Babesia Usp.) to calves [9]. Hyalomma spp., including Hyalomma detritum, Hy. a. anatolicum and Hy. rufipes, are distributed mainly in semi-dry and desert-land in Northern China, and have been reported to be vectors of T. annulata [10]. T. sinensis is transmitted by H. qinghaiensis [11]. In the field, the risk of co-infection between the eight bovine Babesia and Theileria species is very high. The species are morphylogically indistinguishable, and molecular techniques have become the key to species identification. So it is necessary to develop a simple, reliable and cost-effective method that is suitable for large-scale epidemic investigation, particularly for the detection of co-infection in field [12].
In the work described here, an informative molecular target has been identified in the ribosomal protein S8 (RPS8) gene from bovine Babesia and Theileria species endemic in China. The amplified gene fragment containing non-coding regions varied extensively both in length and in sequence, and allowed the development of an assay for species differentiation based solely on fragment size when combined with a simple PCR-restriction fragment length polymorphism (RFLP) protocol.
Methods
Ethics statement
All animal experiments were performed according to the protocols approved by the Animal Care and Use Committee of the Lanzhou Veterinary Research Institute (permit number 2009–26).
Parasite species
The isolates used in this study were listed in Table 1. Babesia bovis (Shanxian and Lushi) [13], B. bigemina (Kunming and Lushi) [14], B. major (Yili) [7], B. ovata (Wenchuan and Lushi and Zhangjiachuan) [15], Babesia sp. Kashi2 (Kashi) [16],Theileria annulata (Xingjiang and Ningxia and Sanmenxia) [17], T. sergenti (Lushi) [4], T. sinensis (Weiyuan and Lintan and Lintao) [18].
DNA extraction
The calves, aged between 12 and 24 months old, were infected by inoculating 5 ml of cryopreserved infected blood stock of these Babesia and Theileria isolates into the jugular vein. When the parasitemia reached 5 %, blood was collected into heparin vacutainer tubes via jugular venipuncture. The infected blood was resuspended in PSG buffer in the presence of SDS (final concentration was 2 %) and proteinase K (final concentration was 1 mg/ml). The solution was incubated at 42 °C for 14 h. Parasite DNA was extracted by conventional phenol/chloroform for deproteinization of the aqueous solution containing the desired nucleic acid. The purified DNAs were precipitated by the addition of two volumes of cold absolute ethanol. The pellet was dried, dissolved in sterile distilled water and kept at −20 °C until use. Control DNA was isolated from blood of uninfected cattle and blood of Trypanosoma brucei evansi infected mouse [19].
PCR-RFLP analysis
To develop a PCR-RFLP technique for species- and strain-specific diagnosis of bovine Babesia and Theileria parasites, sequences conserved in all Babesia and Theileria species were identified from sequence alignment and used as primers in a single PCR protocol. For the PCR step, a PCR product that was about 707–855 bp long was amplified using primers 5′- ATGGGTATTTCACGTGACAG -3′ and 5′- GCGTTTCTTCTTATCCATACG -3′. Each PCR mixture (total volume, 50 μl) contained 5 μl of 10 × PCR buffer, 6 mM MgCl2, deoxynucleoside triphosphate at a concentration of 200 μM each, primer at a concentration of 200 nM each, 2.5 U of Taq polymerase, and 20 ng of DNA template. A total of 35 cycles, each consisting of 94 °C for 45 s, 54 °C for 45 s, and 72 °C for 1 min, were performed; an initial hot start at 94 °C for 3 min and a final extension step at 72 °C for 7 min were also included.
For restriction fragment analysis, 20 μl of the PCR products was digested in a 50-μl reaction mixture containing 20 U of MboI (Takara) and 5 μl of the appropriate restriction buffer at 37 °C for 1 h, under conditions recommended by the supplier. The digested products were fractionated on a 3.0 % agarose gel and visualized by ethidium bromide staining. In additional, predicted restriction fragment length polymorphism (RFLP) patterns were produced in silico using the web-based software In Silico [20]. Image analysis of the electrophoretic gels was performed with 1-day Manager Software (TDI, Madrid, Spain).
Results and discussion
PCR-RFLP analysis
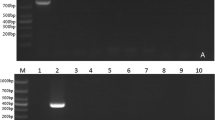
PCR amplification of RPS8 gene from the DNA yielded a product of 709 bp for T. annulata isolates, 713 bp for T. sergenti Lushi isolate, 707 bp for T. sinensis isolates. Similarly, Babesia species yielded products that were similar or identical in size. PCR products of B. bigemina isolates, B. major, B. ovata isolates, B. bovis isolates, and Babesia sp. Kashi2 were 849, 847, 849, 820, 855 bp, respectively (Table 2, Fig. 1). Specificity for Babesia and Theileria was confirmed by the absence of products from samples of Trypanosoma brucei evansi and cattle genomic DNA (Fig. 1). The single PCR was quite sensitive (0.1 pg genomic DNA of Babesia species and 1 pg genomic DNA of Theileria species), as demonstrated by the amplification of serial diluted DNA samples (data not shown). Amplicon size alone could not distinguish the species. However, on digestion with Mbo I, fragment polymorphism was visible post gel electrophoresis of the digested DNA (Table 2, Fig. 2). Thus RFLP will clearly distinguish among Babesia- and Theileria- infected cattles. However, this is based on a limited sample size and we need to confirm that there is no intra-specific restriction polymorphism, particularly for the complex Theileria buffeli/orientalis group [21, 22].
PCR products of a fragment of the RPS8 gene. Lane 1, 2000 bp size markers; lane 2: T. sergenti; lane 3, T. annulata; lane 4, B. bovis; lane 5, B. major; lane 6, B. bigemina; lane 7, T. sinensis; lane 8, B. ovata; lane 9, Babesia sp. Kashi 2; lane 10, Trypanosoma brucei evansi;lane 11, Negative control, cattle genomic DNA
Fragments of the RPS8 (coding and non-coding regions) gene digested with MboI. Lane 1, 2000 bp size marker; lane 2, B. bovis (Shanxian and Lushi isolates); lane 3, T. sergenti (Lushi isolate); lane 4, B. major (Yili isolate); lane 5, T. annulata (Sanmenxia and Xinjiang and Ningxia isolates); lane 6, Babesia sp. Kashi (Kashi isolate) 2; lane 7, B. bigemina (Kunming and Lushi isolates); lane 8, T. sinensis (Weiyuan and Lintan and Lintao isolates); lane 9, B. ovata (Wenchuan and Lushi and Zhangjiachuan isolates)
A more practical assay is required to classify piroplasms such as Theileria and Babesia isolates since current serological and morphological tests cannot discriminate between closely related species [12]. Although reverse line blot (RLB) assay for the simultaneous identification of bovine Babesia and Theileria species has been developed, its use for routine diagnosis is restricted by various factors. These include the availability of reagents, complexity of operating procedures, special equipment needs and high susceptibility in the subjective interpretation of the hybridisation signal [23, 24]. Although nucleic acid-based tests such as real-time PCR and Loop-mediated isothermal amplification (LAMP) demonstrate significant sensitivity and specificity, they are only suitable for single species differentiation [25–29]. It would be desirable to have a ‘universal’ PCR-based test for the simultaneous detection and identification of these parasites. This requires the analysis of a molecular target conserved among piroplasms, yet variable enough to design a reliable species identification protocol.
In our previous study, RPS8 rDNA was confirmed to be a useful and novel genetic marker for defining species boundaries and for detecting closely related species, similar to 18S rDNA, because it tends to have little intra-species variation but considerable inter-species difference. It is relatively simple to amplify RPS8 rDNA by polymerase chain reaction (PCR) based on the highly conserved rDNA flanking both RPS8 regions [30, 31]. In this study, our results indicated that the RPS8-based PCR-restriction fragment length polymorphism was a simple and reliable molecular diagnostic tool for the simultaneous detection and identification of bovine Babesia and Theileria species in China, which could be applicable for the survey of parasite dynamics, epidemiological studies as well as prevention and control of the disease.
Conclusions
In this work, we found that utilizing PCR with restriction fragment length polymorphism (RFLP) on the RPS8 gene can be useful for the differentiation of the most common pathogenic Babesia and Theileria species infecting cattles in China. However, more samples are needed to verify the usefulness of the RPS8 (coding and non-coding regions) gene as a marker for the detection of the most Babesia and Theileria species, particularly for some closely related species.
References
Li Y, Chen Z, Liu Z, Liu J, Yang J, Li Q, et al. First report of Theileria and Anaplasma in the Mongolian gazelle, Procapra gutturosa. Parasit Vectors. 2014;7:614.
Zanet S, Trisciuoglio A, Bottero E, de Mera IG, Gortazar C, Carpignano MG, et al. Piroplasmosis in wildlife: Babesia and Theileria affecting free-ranging ungulates and carnivores in the Italian Alps. Parasit Vectors. 2014;7:70.
Yin H, Schnittger L, Luo J, Seitzer U, Ahmed JS. Ovine theileriosis in China: a new look at an old story. Parasitol Res. 2007;101:191–5.
Luo JX, Yin H, Liu GY, Guan GQ, Liu ZJ, Liu AH, et al. Collection and identification of piroplasma infected to cattle and sheep in China. Chin J Parasitol Parasit Dis. 2006;24:S48–53 (in Chinese).
Lu WS, Yin H, Lu WX, Yu F, Zhang QC, Dou HF. Studies on the transmission ability of Babesia bovis and B. bigemina by Boophilus microplus. Chin J Vet Sci Technol. 1989;7:10–1 (in Chinese).
Lu WS, Yin H, Lu WX, Yu F, Zhang QC, Dou HF. Experimental studies on the transovarial transmission of Babesia major from bovine by tick Haemaphysalis longicornis. Chin J Vet Sci Technol. 1990;6:5–6 (in Chinese).
Yin H, Lu WS, Luo JX, Zhang QC, Lu WX, Dou HF. Experiments on the transmission of Babesia major and Babesia bigemina by Haemaphysalis punctata. Vet Parasitol. 1996;67:89–98.
Bai Q, Liu GY, Han GF, Hui Y. Isolation and complementary transmission of Babesia ovata in Zhangjiachuan of Gansu Province. Chin J Vet Sci Technol. 1994;24:9–10 (in Chinese).
Luo JX, Yin H, Guan GQ, Zhang QC, Lu WS. Description of a new Babesia sp. Infective for cattle in China. Parasitol Res. 2002;88:S13–5.
Luo JX, Lu WS. Cattle theileriosis in China. Trop Anim Health Prod. 1997;29:4–7.
Yin H, Luo J, Guan G, Gao Y, Lu B, Zhang Q, et al. Transmission of an unidentified Theileria species to small ruminants by Haemaphysalis qinghaiensis ticks collected in the field. Parasitol Res. 2002;88:S25–7.
Figueroa JV, Buening GM. Nucleis acid probes as a diagnostic method for tick-borne hemoparasites of veterinary importance. Vet Parasitol. 1995;57:75–92.
Bai Q, Liu GY, Zhou JY. Isolation of single species of Babesia bovis. Chin J Vet Sci Technol. 1991;1:20–2 (in Chinese).
Bai Q, Yin SX, Chen ZH. Isolation of single species of Babesia bigemina. Chin J Vet Sci Technol. 1987;9:25–7 (in Chinese).
Bai Q, Liu GY, Zhang L. Discovey and isolation of Babesia ovata in China. Chin J Vet Sci Technol. 1990;16:2–4 (in Chinese).
Luo J, Chen F, Lu W, Guan G, Ma M, Yin H. Experimental transmission of an unnamed bovine Babesia by Hyalomma spp., Haemaphysalis longicornis and Boophilus microplus. Vet Parasitol. 2003;116:115–24.
Yin H, Lu W, Luo JX. Babesiosis in China. Trop Anim Health Prod. 1997;29:11–5.
Bai Q, Liu GY, Yin H, Zhao QZ, Liu DK. Theileria sinensis sp nov: a New species of bovine Theileria - classical taxonomic studies. Acta Veterinar ia et Zootechnica Sinica. 2002;33:73–7 (in Chinese).
Tian Z, Liu G, Xie J, Shen H, Zhang L, Zhang P, et al. The internal transcribed spacer 1 (ITS-1), a controversial marker for the genetic diversity of Trypanosoma evansi. Exp Parasitol. 2011;129:303–6.
Bikandi J, San MR, Rementeria A, Garaizar J. In silico analysis of complete bacterial genomes: PCR, AFLP–PCR, and endonuclease restriction. Bioinformatics. 2004;20:798–9.
Gubbels MJ, Hong Y, van der Weide M, Qi B, Nijman IJ, Guangyuan L, et al. Molecular characterisation of the Theileria buffeli/orientalis group. Int J Parasitol. 2000;30:943–52.
Kamau J, de Vos AJ, Playford M, Salim B, Kinyanjui P, Suqimoto C. Emergence of new types of Theileria orientalis in Australian cattle and possible cause of theileriosis outbreaks. Parasit Vectors. 2011;4:22.
Gubbels JM, de Vos AP, van der Weide M, Viseras J, Schouls LM, de Vries E, et al. Simultaneous detection of bovine Theileria and Babesia species by reverse line blot hybridization. J Clin Microbiol. 1999;37:1782–9.
Ros-García A, Barandika JF, García-Pérez AL, Juste RA, Hurtado A. Assessment of exposure to piroplasms in sheep grazing in communal mountain pastures by using a multiplex DNA bead-based suspension array. Parasit Vectors. 2013;6:277.
Ros-García A, Nicolás A, García-Pérez AL, Juste RA, Hurtado A. Development and evaluation of a real-time PCR assay for the quantitative detection of Theileria annulata in cattle. Parasit Vectors. 2012;5:171.
Liu A, Guan G, Du P, Gou H, Zhang J, Liu Z, et al. Rapid identification and differentiation of Theileria sergenti and Theileria sinensis using a loop-mediated isothermal amplification (LAMP) assay. Vet Parasitol. 2013;191:15–22.
Criado-Fornelio A, Buling A, Asenzo G, Benitez D, Florin-Christensen M, Gonzalez-Oliva A, et al. Development of fluorogenic probe-based PCR assays for the detection and quantification of bovine piroplasmids. Vet Parasitol. 2009;162:200–6.
Iseki H, Alhassan A, Ohta N, Thekisoe OM, Yokoyama N, Inoue N, et al. Development of a multiplex loop-mediated isothermal amplification (mLAMP) method for the simultaneous detection of bovine Babesia parasites. J Microbiol Methods. 2007;71:281–7.
Kim C, Iseki H, Herbas MS, Yokoyama N, Suzuki H, Xuan X, et al. Development of TaqMan-based real-time PCR assays for diagnostic detection of Babesia bovis and Babesia bigemina. Am J Trop Med Hyg. 2007;77:837–41.
Tian ZC, Liu GY, Yin H, Luo JX, Guan GQ, Luo J, et al. RPS8—a New informative DNA marker for phylogeny of Babesia and Theileria parasites in China. PLoS One. 2013;8, e79860.
Tian Z, Liu G, Yin H, Luo J, Guan G, Luo J, et al. Discrimination between ovine Babesia and Theileria species in China based on the ribosomal protein S8 (RPS8) gene. Vet Parasitol. 2013;197:354–9.
Acknowledgements
The authors thank Michelle Li for improving the manuscript. This work was supported by the National Natural Sciences Foundation of China (No. 31201899), the Natural Science Foundation of Gansu Province (No. 096RJZA128), ASTIP, FRIP (2014ZL010), CAAS; NBCIS CARS-38.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and disgned the study: TZ, YH; performed experiments: TZ, LA, YJ; analyzed data: DJ, LX, LG; wrote the paper:TZ, YH; all authors read and approved the final version of the manuscript.
Authors’ information
1State Key Laboratory of Veterinary Etiological Biology, Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, 730046, P. R. China. tianzhancheng@caas.cn.
2State Key Laboratory of Veterinary Etiological Biology, Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, 730046, P. R. China. dujunzheng@caas.cn.
3State Key Laboratory of Veterinary Etiological Biology, Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, 730046, P. R. China. yangjifei@caas.cn.
4State Key Laboratory of Veterinary Etiological Biology, Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, 730046, P. R. China. liuaihong@caas.cn.
5State Key Laboratory of Veterinary Etiological Biology, Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, 730046, P. R. China. 814329371@qq.com.
6State Key Laboratory of Veterinary Etiological Biology, Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, 730046, P. R. China. liuguangyuan@caas.cn.
7State Key Laboratory of Veterinary Etiological Biology, Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, 730046, P. R. China. yinhong@caas.cn.
8Jiangsu Co-innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou, 225009, P. R. China. yinhong@caas.cn.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Tian, Z., Du, J., Yang, J. et al. A PCR-RFLP Assay targeting RPS8 gene for the discrimination between bovine Babesia and Theileria species in China. Parasites Vectors 8, 475 (2015). https://doi.org/10.1186/s13071-015-1085-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-015-1085-x