Abstract
Background
Ductal carcinoma in situ (DCIS) of the breast is a precursor of invasive breast carcinoma. DNA methylation alterations are thought to be an early event in progression of cancer, and may prove valuable as a tool in clinical decision making and for understanding neoplastic development.
Results
We generate genome-wide DNA methylation profiles of 285 breast tissue samples representing progression of cancer, and validate methylation changes between normal and DCIS in an independent dataset of 15 normal and 40 DCIS samples. We also validate a prognostic signature on 583 breast cancer samples from The Cancer Genome Atlas. Our analysis reveals that DNA methylation profiles of DCIS are radically altered compared to normal breast tissue, involving more than 5,000 genes. Changes between DCIS and invasive breast carcinoma involve around 1,000 genes. In tumors, DNA methylation is associated with gene expression of almost 3,000 genes, including both negative and positive correlations. A prognostic signature based on methylation level of 18 CpGs is associated with survival of breast cancer patients with invasive tumors, as well as with survival of patients with DCIS and mixed lesions of DCIS and invasive breast carcinoma.
Conclusions
This work demonstrates that changes in the epigenome occur early in the neoplastic progression, provides evidence for the possible utilization of DNA methylation-based markers of progression in the clinic, and highlights the importance of epigenetic changes in carcinogenesis.
Similar content being viewed by others
Background
Epigenetic marks (and DNA methylation in particular) are known to be deregulated in cancer. Cancer-specific changes include hypermethylation of CpGs in gene promoters [1], hypomethylation of non-CpG island CpGs [2], and overall increase in variation of methylation [3],[4]. DNA methylation patterns are also associated with histopathological parameters such as hormone receptor status, TP53 mutation status, histologic grade, stage, and survival time [5]-[9].
Ductal carcinoma in situ (DCIS) of the breast is a neoplasm where the cells are confined by the basement membrane of breast ducts. DCIS is a precursor of an invasive breast carcinoma (IBC). Treatment of DCIS consists of surgical excision in the form of either breast-conserving surgery (that is, wide local resection or sector resection) or removal of the entire breast parenchyma (mastectomy). Treatment by mastectomy results in very few recurrences but is considered over-treatment in a majority of cases. Approximately 30% of patients treated by breast-conserving surgery alone are reported to develop a local recurrence after 15 years follow-up and the risk of local recurrence is reduced by half if postoperative radiotherapy is given [10],[11]. To avoid overtreatment of patients with DCIS it would be of great value to be able to predict which patients have potentially malignant and invasive tumors and are likely to experience recurrence of disease.
Epigenetic studies of breast tissue report aberrant methylation levels already present in benign neoplastic breast lesions such as columnar cell lesions and ductal hyperplasia [12],[13]. More studies have reported aberrant methylation levels in DCIS (summarized in [11]). Most of these studies reported methylation levels of only one or a few genes, while two studies have used genome-wide approaches. These studies reported 108 and 214 CpG islands (CGIs), respectively, to be hypermethylated in DCIS compared with normal tissue and that these CGIs were enriched for homeobox genes [14],[15].
Studies of benign or premalignant tumors from a variety of organs have revealed that some of these lesions have epigenetic characteristics that separate them both from normal tissue and from the malignant tumors. These studies include results from meningiomas of the brain [16],[17], gastric lesions [18],[19], cystadenomas of the ovary [20], colorectal lesions [21],[22] and uterine leiomyoma [23],[24]. Taken together, these data suggest that epigenetic changes occur early in cancer development and as such have great potential as biomarkers in addition to increase our biological understanding of progression of cancer.
In this study, we investigate methylation patterns in a total of 285 fresh frozen tissue samples, including 46 normal breast tissue samples from healthy women, 22 pure DCIS, 31 mixed DCIS-IBC and 186 IBC of stage I and II. Validation was performed using 583 breast cancer samples from The Cancer Genome Atlas (TCGA), as well as in an independent set of DCIS and adjacent normal tissues.
The aim of this study was to investigate DNA methylation patterns during progression of breast cancer. Genome-wide profiling allows identification of molecular changes that occur during neoplasia as well as changes that are required for a tumor to acquire invasive capabilities. Additionally, the association between methylation and survival of patients was studied, and a prognostic signature was identified. The correlation between methylation and expression was incorporated into the analyses. These findings could improve our understanding of the biological mechanisms that occur during progression of breast cancer, and contribute to identification of biomarkers for risk-related classification of patients with in situ and invasive breast cancer.
Results
Tumor classification based on DNA methylation
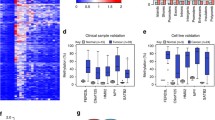
Genome-wide DNA methylation analysis was performed on a total of 239 breast tumor samples and 46 normal samples. The tumor samples included 22 pure DCIS, 31 mixed DCIS-IBC and 186 pure IBC. A gene region collapsed data set was constructed to reduce the dimensions of the data and to study the methylation in the functional regions of the genes. Each gene is represented by up to six methylation values representing the respective functional regions as described in Materials and methods). Hierarchical clustering was performed to explore the structure of the DNA methylation data, using methylation levels of the 500 most variable gene regions (Figure 1). The samples were divided into two main clusters, where one contained all normal samples as well as tumor samples, and the other contained only tumor samples. Basal-like tumors were enriched in the cluster containing the normal samples while luminal A and luminal B tumors were mostly found in the other cluster. DCIS and mixed DCIS-IBC tumors were found in both clusters.
Hierarchical clustering of the methylation level of the 500 most variable gene regions. Tissue types (green, healthy breast; blue, DCIS; purple, mixed DCIS-IBC; red, IBC) and PAM50 subtype (dark blue, luminal A; light blue, luminal B; pink, HER2-enriched; red, basal-like; green, normal-like) are indicated.
Differentially methylated gene regions were identified between the five intrinsic subtypes of breast cancer. For a locus to be considered differentially methylated, the minimum difference between the median methylation levels in the groups was 0.1 (10%) and the false discovery rate (FDR) q-value was smaller than 0.01 (1%); 16,723 gene regions were differentially methylated between the intrinsic subtypes of breast cancer (listed in Additional file 1). Hierarchical clustering of the invasive tumors using these regions confirmed that the basal-like and normal-like tumors showed clearly distinct profiles compared with the luminal-like tumors (Additional file 2).
Correlation between DNA methylation and gene expression
Correlation between DNA methylation level and gene expression was investigated to assess to what degree gene expression may be influenced by DNA methylation in breast cancer. Gene expression level was tested for correlation to both methylation level of single CpGs within 100 kb of a transcription start site (TSS) and methylation level of gene regions.
Pearson correlation was calculated between gene expression and methylation level of CpGs within 100 kb of a TSS, and an association was considered significant if the Bonferroni corrected P-value was smaller than 0.05. By this definition, the methylation level of 9,800 CpGs was significantly correlated with the expression of 2,960 genes (Additional file 3). The expression level of 2,558 genes was negatively correlated with the methylation level of at least one CpG, while the expression level of 852 genes was positively correlated with the methylation level of at least one CpG. The positive correlations were quite evenly distributed relative to TSSs (±100 kb; Figure 2A). The negative correlations were also found at all distances relative to TSSs (±100 kb; Figure 2A), but they were found to be enriched close to TSSs (1,000 bp upstream to 5,000 bp downstream). The CpGs that correlated with expression were distributed across the whole genome, and were enriched on chromosomes 1, 17 and 19 (Figure 2B).
CpGs whose methylation level significantly correlated with gene expression (Bonferroni corrected P -value <0.05). (A) Significance level of correlation between methylation level and gene expression plotted against distance between the CpG and transcription start site (TSS). Red dots represent negative correlation and blue dots represent positive correlation. (B) Significance level and genome-wide distribution of correlation between methylation level and gene expression.
Pearson correlation was also calculated between gene expression and the methylation level of each respective gene region. The expression of 1,719 genes significantly correlated with methylation level in at least one gene region (Additional file 4). The expression of 1,445 genes negatively correlated with the methylation level of at least one gene region, and the expression of 355 genes positively correlated with the methylation level of at least one gene region (Figure 3). Of the negative correlations between methylation and expression, almost 40% were found upstream of TSSs (TSS1500 and TSS200 subregions), while only about 15% of the positive correlations were found upstream of a TSS. The rest of the negative correlations were distributed in the 5’ UTR, first exon and gene body, while less than 10% of the negative correlations were found in the 3' UTR. Of the positive correlations, 40% were found in the gene body and 30% were found in the 3' UTR, meaning that more than 70% of positive correlations were found outside of promoter regions.
Significant correlation (Bonferroni corrected P -value <0.05) between gene expression and methylation level of gene regions. Bar plot showing the distribution of negative and positive correlations relative to the functional regions of genes. The distribution is notably different for the negative versus positive correlations.
Methylation changes during progression of breast cancer
To identify differentially methylated CpGs during progression of breast cancer, Significance Analysis of Microarrays (SAM) was applied to the complete methylation data set with all CpGs. For a locus to be considered differentially methylated, the difference between the median methylation levels in the two groups had to be at least 0.1 (10%) and the FDR q-value had to be smaller than 0.01 (1%). The differences in methylation levels between normal tissue and DCIS were substantial. A high degree of CpG methylation deregulation during neoplastic transformation may have important implications for a better understanding of breast cancer progression. Thus, to identify the most biologically relevant alterations we examined differences between normal tissue and DCIS in two independent patient cohorts. Only the significant CpGs or regions that were differentially methylated in both datasets are reported. Comparing normal tissue and DCIS revealed that 16,949 CpGs were differentially methylated, representing 5,659 genes. Comparing DCIS and IBC revealed that 2,000 CpGs were differentially methylated. These CpGs represented 1,076 genes, and 1,745 of the CpGs were hypermethylated while 255 of the CpGs were hypomethylated (Table 1). All differentially methylated CpGs are shown in Additional files 5 and 6.
SAM was also applied to the gene region collapsed data, and comparing normal tissue and DCIS in the two independent patient cohorts revealed 1,249 differentially methylated gene regions representing 1,011 genes. In comparison, 166 gene regions representing 154 genes were differentially methylated between DCIS and IBC (Table 1). All differentially methylated gene regions are shown in Additional files 7 and 8.
To identify pathways for which the differentially methylated genes between normal tissue and DCIS were enriched Ingenuity Pathways Analysis was performed. This analysis was performed on the genes represented by differentially methylated gene regions rather than single CpGs. The differentially methylated genes between normal tissue and DCIS approached a significant threshold for an enrichment in the agranulocyte adhesion and diapedesis pathway (P = 0.053) and the granulocyte adhesion and diapedesis pathway (P = 0.084) (Table 2).
The methylation level of four genes (CPA1, CUL7, LRRTM2 and POU2AF1) increased both from normal to DCIS and from DCIS to IBC, while 10 genes (ARSJ, CES8, FAIM2, GPRC5B, ICAM2, P4HA3, PGLYRP2, PLOD1, PNMAL2, STAP2) showed a decrease in methylation between normal and DCIS, but an increase in methylation from DCIS to IBC.
Survival analysis
To identify CpGs for which methylation level may predict survival, Lasso regularization was performed. The analysis was performed using methylation level of single CpGs, preselected to be correlated with gene expression. A survival signature that consisted of 18 CpGs was identified (Table 3). The methylation level of these CpGs correlated with the expression level of 26 genes, including IRF6, TBX5, CSNK1G2, MACF1, KCTD21 and EPN3 (Table 4). Of the genes associated with the signature, 15 negatively correlated with methylation level and 11 positively correlated with methylation level. No canonical pathways were found significantly enriched among the 26 genes. Of the genes in the signature, 17 were differentially methylated between normal and DCIS, and 4 were differentially methylated between DCIS and IBC. The survival signature was applied to patients with invasive tumors (n = 176) as explained in Materials and methods. The patients segregated exceptionally well into high- and low-risk groups according to breast cancer-specific survival (hazards ratio (HR) = 13.7, P < 2.2e-16; Figure 4A).
Application of the DNA methylation-based prognostic signature for patients. (A) In the original data (n = 176); (B) in the TCGA validation (n = 583); (C) with either DCIS or mixed DCIS-IBC (n = 52). (D) Classification with the DNA methylation-based prognostic signature was complementary to classification by lymph node status.
To validate the prognostic value of the discovered signature, the signature was applied to a validation set of breast cancer patients collected by TCGA (n = 583). The patients were divided into two groups with significantly different prognosis (HR = 2.31, P = 6.23e-4; Figure 4B).
The prognostic signature derived from patients with IBC was then applied to patients with DCIS and mixed DCIS-IBC tumors (n = 52). The patients were separated into groups with significantly different prognosis (P = 3.69e-2; Figure 4C). The good prognosis group included 14 pure DCIS and 15 mixed DCIS-IBC, while the bad prognosis group included 7 pure DCIS and 16 mixed DCIS-IBC. Comparing prognosis in DCIS versus mixed DCIS-IBC showed that the patients with mixed lesions had significantly adverse prognosis. In fact, only one breast cancer-specific death was observed among the patients with pure DCIS.
Multivariable Cox proportional hazard models were calculated for the patients in the training set (n = 176) as well as patients in the TCGA validation (n = 583) adjusting for estrogen receptor (ER) status, TP53 mutation status (only training set), T status, and lymph node status. Classification by the prognostic signature was significantly associated with survival in both data sets (P < 0.001 and P = 0.008, respectively; Table 5). In addition to the prognostic signature, lymph node status was significantly associated with survival in the TCGA validation set. Importantly, combining lymph node status and classification by the prognostic signature provided an even better segregation of patients (P = 8.26e-5; Figure 4D). Patients that were both lymph node-negative and had a low index had the best prognosis, while patients that were lymph node-positive and had a high index had the worst prognosis. PAM50 classification was not significant in either of the patient cohorts.
Discussion
Here we report the DNA methylation profiles of a breast cancer progression series, including normal breast tissue, DCIS, IBC and mixed lesions. Interestingly, most of the aberrations in the epigenetic profile were observed already in the pre-invasive DCIS stage. The affected pathways suggested that many of the changes may not occur in the tumor, but in infiltrating cells or at least in genes that enable cross-talk to such cells. Also of interest was that DNA methylation profiles of the basal type of breast cancer were more similar to normal tissue than were the luminal-like tumors. These data suggest that the methylation profiles may be a function of the cell of origin as much as a marker of progression. We also report a signature comprising DNA methylation levels of 18 CpGs that was prognostic for breast cancer patients with invasive tumors as well as for patients with DCIS and mixed lesions of DCIS and IBC. The signature was discovered in a training data set of 176 patients and validated in 583 patients from the TCGA. In the validation patient group the prognostic signature and lymph node status were complementary, potentially providing valuable information for clinical decision-making. The patients that were classified with good prognosis by DNA methylation and additionally were lymph node-negative might benefit from reduced or no adjuvant treatment, while patients that were classified with adverse prognosis by DNA methylation and were lymph node-positive could potentially benefit from more aggressive treatment.
A great advantage of DNA methylation is that it is relatively easy to design an assay that may be used in a clinical setting. DNA methylation can be measured on an absolute scale (from 0 to 100%), is stable in the cell over time, and is relatively insensitive to handling in the laboratory. This work clearly shows the potential of DNA methylation-based signatures for clinical utilization.
With data from two independent cohorts of normal tissue and DCIS, we report that the DNA methylation profiles of DCIS were radically changed compared with normal breast tissue, involving more than 5,000 genes. One cohort consisted of fresh frozen tissue and normal controls from healthy women, while the other cohort consisted of formalin-fixed paraffin-embedded (FFPE) DCIS samples and matched adjacent normal breast tissue. Thus, the reported changes in methylation levels across these genes appear to be independent of tissue preparation and the normal tissue's proximity to tumor tissue. Comparably, the changes between DCIS and IBC were more modest, involving around 1,000 genes. These findings suggest that the epigenome is severely altered in the early neoplastic setting in the breast. Previous studies of breast cancer progression have also reported early aberrant DNA methylation in DCIS, but they characterized fewer genes (summarized in [11]). The current study has the advantage of a high coverage methylation assay (Illumina HumanMethylation450) and leverages true normal controls from healthy women. Our observation that extensive epigenetic alterations occur early in cancer progression has been reported for other cancer types, including colorectal cancer. For example, studies using the HumanMethylation450 assay reported that precancerous adenomas demonstrate heterogeneity similar to invasive tumors, and that aberrant DNA methylation occurs early in colorectal cancer formation [22].
Classification of breast cancer by hierarchical clustering showed that basal-like tumors clustered with the normal samples in one cluster, and luminal A and luminal B tumor clustered together in the second cluster (Figure 1). This observation largely recapitulates and extends the results from a previous study [8]. Since DNA methylation aberrations occur early in carcinogenesis, it is possible that DNA methylation changes may play a role in the development of molecular subtypes of breast cancer, although it is also possible that the correlation with methylation is a consequence of subtype. Future studies are needed to define the mechanistic effects that DNA methylation and other epigenetic marks may have on early development of cancer.
DCIS lesions tend to grow slower and show less inter-tumor heterogeneity than IBC lesions. Consequently, it would be pertinent to perform subtype-specific analyses of differences between DCIS and IBC. In the present study, however, the number of DCIS samples was too few to perform subtype-specific analyses. Future studies should aim to collect enough DCIS samples to divide both DCIS and IBC samples into intrinsic subtypes of breast cancer while including enough samples for statistical analyses. The inter-sample heterogeneity in the normal samples (mammoplastic reductions and needle biopsies from healthy women) was low compared with the neoplastic lesions (Figure 1).
Correlation between DNA methylation and gene expression was found throughout the genome and involved almost 3,000 genes. CpGs whose methylation level correlated with expression were enriched close to TSSs, but also found at distances up to 100 kb from them. Interestingly, about a quarter of the genes whose expression level correlated with methylation level showed a positive correlation, meaning that a higher methylation level was associated with higher expression. Viewed in relation to functional regions in genes, 70% of the positive correlations between methylation level and expression were found in the 3' UTR or the gene body. Similar findings have been reported in chronic lymphocytic leukemia [25] and support that promoter hypermethylation is an important mechanism for gene silencing, while DNA methylation elsewhere may have more complex functions that are yet to be fully understood. Possible mechanisms for regulation of gene expression by non-promoter methylation include interplay between nucleosome positioning and chromatin structure, regulation of enhancer region availability, and/or gene body regulation of alternative promoters [25],[26]. Statistical significance of correlation between DNA methylation and gene expression was corrected for multiple testing by Bonferroni correction. This method is very strict, and may underestimate the association between DNA methylation and gene expression.
The survival signature segregated patients with DCIS and mixed DCIS-IBC into two groups with significantly different prognosis. The signature classified most of the patients with mixed DCIS-IBC that experienced breast cancer-specific death into the bad prognosis group. Additionally, the single patient with pure DCIS that experienced breast cancer-specific death was also classified into the bad prognosis group. Since only one of the patients with pure DCIS died of breast cancer, it was not possible to perform the analysis on only patients with pure DCIS. Taken together, the signature may have great potential to classify patients with DCIS or mixed lesions according to prognosis, but more patients must be studied to further validate the clinical value.
Several of the genes in the survival signature have roles in tumor suppressive functions. The protein product of IRF6 has been shown to function synergistically with the tumor suppressor maspin to regulate mammary epithelial differentiation [27], and has also been shown to have tumor suppressor activity in squamous cell carcinoma [28]. TBX5 is a transcription factor that has been implicated as a tumor suppressor in colon cancer and has been found silenced by DNA methylation [29]. A SNP (rs1265507) located between TBX5 and TBX3 was also associated with mammographic density in a genome-wide association study [30]. In the present study, high methylation levels of CpGs in TBX5 were associated with lower expression levels of TBX5 and adverse prognosis. DIEXF is thought to be involved in the turnover of p53 [31], and CEND1 has been shown to affect cyclin D1 levels [32]. ZNF259 has been shown to be involved in regulation of the cell cycle through interactions with the epidermal growth factor receptor [33], and KCTD21 is thought to act as a tumor suppressor in medulloblastoma by modulating Hedgehog signaling through degradation of histone deacetylase 1 [34].
Some genes in the survival signature have also been associated with functions related to motility and invasion: EPN3 over-expression has been shown to promote cancer cell invasion [35], MACF1 has been shown to be involved in cell mobility and steering by interactions with HER2 [36], and CSNK1G2 is thought to modulate the activity of metastasis-associated MTA1 while itself a target of ER [37]. Taken together, many of the genes associated with the survival signature have tumor suppressive functions or are involved in regulation of motility and ability to invade.
A strong immune component in breast tumors observed by measuring DNA methylation has previously been reported [38]. The genes that were differentially methylated between DCIS and IBC were borderline significantly enriched in the agranulocyte and granulocyte adhesion and diapedesis pathways, suggesting that many of the observed changes may occur in infiltrating cells or in genes that enable cross-talk to such cells.
CUL7 (cullin 7) methylation levels increased from both normal to DCIS and DCIS to IBC. CUL7 encodes a ubiquitin ligase that forms complexes with p53 and Parc. It was shown to regulate apoptosis independently of p53 [39]. In another report [40], CUL7 was shown to function as an antiapoptotic oncogene through cooperation with Myc in a p53-dependent manner. Also, CUL7 has been shown to be involved in liver carcinogenesis [41]. Importantly, CUL7 has not previously been reported in breast cancer. ICAM2 (Intercellular adhesion molecule 2) methylation levels decreased between normal and DCIS, and increased between DCIS and IBC. ICAM2 is involved in cell adhesion and thought to play a role in immune response. In pancreatic cancer, ICAM2 has been reported to have tumor suppressor function through immune surveillance [42].
Conclusion
DNA methylation profiles of DCIS were radically changed compared with normal breast tissue while the changes between DCIS and IBC were comparably modest. A DNA methylation-based prognostic signature was reported that has potential in patients both with invasive breast cancer and with in situ carcinoma. Correlation between DNA methylation and gene expression was observed in a substantial part of the genome, and both positive and negative correlations were observed.
Materials and methods
Patient material
Material for this study was obtained from 285 fresh frozen tissue samples representing different progression stages of breast cancer; 46 normal samples, 22 pure DCIS, 31 mixed DCIS-IBC and 186 pure IBC were included. Of the 46 normal samples, 17 were tissue from mammoplastic reductions of healthy women collected at Akershus University Hospital (institutional review board (IRB) approval number 429-04148). Twenty-nine needle biopsies from healthy women and 49 IBC samples were collected at the Norwegian Radium Hospital (IRB approval number S-02036) [43]. DCIS samples, mixed DCIS-IBC samples, and 15 of the IBC samples were collected at Uppsala Academic Hospital (IRB approval number Dnr 2005:118) [44],[45]. Of the pure DCIS samples, 18 of 22 had a tumor component of >75% [44]. The 123 IBC samples were collected from hospitals in the Oslo region (IRB approval number S-97103) [46]. The IBC samples were predominantly stage I and II. All studies are in compliance with the Helsinki Declaration and were approved by local ethical committees and local authorities.
DNA methylation analysis
The DNA methylation status of more than 450,000 CpG sites was interrogated using Illumina Infinium HumanMethylation450 microarray. The returned value of each CpG probe is called β and is calculated as the methylated signal divided by the sum of the methylated and the unmethylated signal. β thus represents the fraction of methylated DNA molecules at a specific locus.
Preprocessing of DNA methylation data
Preprocessing and normalization involved steps of probe filtering, color bias correction, background subtraction and subset quantile normalization as previously described [47]. After preprocessing of the data, 468,424 CpG probes were included. The normalized data as well as the raw data are available in Gene Expression Omnibus (GEO) with accession number GSE60185.
Gene expression analysis
mRNA expression data were available for a subset of 104 of the IBC samples studied here. An Agilent whole genome 4x44K oligo array was used for the mRNA analysis as previously described [48]. The mRNA expression data are available in GEO with accession number GSE19783. Molecular subtypes of breast cancer (luminal A, luminal B, HER2-enriched, basal-like and normal-like) were determined using the PAM50 gene list.
Methylation data processing
Statistical and bioinformatical analyses of the methylation level of the 285 samples were performed on two individual datasets, one including methylation levels of all 468,424 CpGs, and one including only 'gene region collapsed' data. The 'gene region collapsed' methylation data were constructed to reduce the dimensions of the methylation data and to focus the analysis on regions that are most relevant for gene function. A CpG that is mapped to a gene is located in one of six subregions: TSS1500, TSS200, 5’ UTR, first exon, body and 3' UTR. These subregions represent: 1) CpGs that are between 1,500 and 200 bp upstream of the TSS; 2) CpGs that are between 200 bp upstream of the TSS and the TSS itself; 3) CpGs in the 5’ UTR; 4) CpGs in the first exon; 5) CpGs in other exons or in introns (body); and 6) CpGs in the 3' UTR. Methylation levels for each subregion were summarized using the median. In this approach intergenic CpGs will not be included. The resulting gene region collapsed dataset had 88,909 targets.
Methylation changes during progression of breast cancer
SAM was used to identify differentially methylated loci between normal and DCIS, and between DCIS and IBC. The analysis was performed using the SAM function of the R package samr[49] with 100 permutations. For a locus to be considered differentially methylated, the difference between the median methylation levels in the two groups had to be at least 0.1 (10%) and the FDR q-value had to be smaller than 0.01 (1%).
Hierarchical clustering
Hierarchical clustering was performed using the methylation level of the 500 most variable gene regions. The distance matrix was calculated using Pearson correlation and average linkage was applied.
Correlation between DNA methylation and gene expression
Correlation between DNA methylation level and gene expression was investigated by two approaches. First, if a CpG was within 100 kb of the TSS of a gene, the methylation level of the CpG and expression of the gene were tested for non-zero correlation using Pearson correlation (eMap1 function in the R package eMap) [50]. For both analyses an association was considered significant if the Bonferroni corrected P-value was smaller than 0.05. Genome-wide correlation between methylation and expression was visualized using the R package quantsmooth[51]. Second, the median methylation level of CpGs in the 'gene region collapsed' data and gene expression of the corresponding gene was tested for non-zero correlation using Pearson correlation (R function corr.test).
Survival analysis
Lasso regularization [52],[53] was applied to identify CpGs for which methylation level predicted survival (cv.glmnet function in the R package glmnet) [54]. This approach gives a signature of targets that together capture the variation that is associated with survival of patients. Pre-selection was performed before regression in order to reduce the number of possible CpG sites and to focus on the ones correlated with expression. Univariate absolute correlation between methylation level and expression with nominal P-value lower than 0.05 were used to preselect data, and 182,653 CpGs were included in the analysis. The analysis was run 100 times and the probes that were present in 80% of the resulting lists were used in the final signature. The coefficients were calculated as the mean of the coefficients in all lists where the probe was present. Patients were divided into high- and low-risk groups according to the following index for patient i:
where g is the target (CpG or gene), n is the number of targets, βg is the Lasso coefficient for target g and Xgi is the methylation value for target g in patient i. Kaplan-Meier estimator and log-rank tests were performed using the functions Surv, survfit and survdiff (R package survival) [55]. Breast cancer-specific survival was used in all analyses.
Multivariate Cox proportional hazard survival analysis was performed using the function coxph (R package survival) to adjust for ER status, TP53 mutation status, T status and lymph node status. Each parameter in the multivariate model was investigated for violations of the assumption of proportional hazards using the function cox.zph (R package survival).
Validation cohort of adjacent normal tissue and DCIS
To validate the methylation changes between normal tissue and DCIS, an independent set of DCIS and adjacent normal tissues was profiled using the Illumina Infinium HumanMethylation450 array. FFPE pure DCIS (n = 40) and adjacent normal tissue (n = 15) underwent pathology review and 2 mm core punches were taken for processing as described in the Illumina Infinium FFPE Restoration solution protocol. The methylation data were preprocessed using the R package ChAMP [56] and 397,600 probes out of 485,577 remained after quality control. A gene region collapsed data set was also constructed for this data set as described above.
Validation cohort from The Cancer Genome Atlas
To validate the prognostic signature, DNA methylation data and clinical information were downloaded from TCGA data portal [9]. Only breast cancer patients for whom there were both overall survival data and tumor DNA methylation analysis had been performed by Illumina HumanMethylation450 were used for validation (n = 583). Probes with more than 50% missing values were removed, and further missing values were imputed using the function pamr.knnimpute (R package pamr) [57] with k = 10.
Data analysis
All analyses were performed using the R computing framework [58]. Gene lists were analyzed with Ingenuity Pathways Analysis (Ingenuity® Systems, Redwood, California, USA).
Authors' contributions
TF conceived and designed the approach, analyzed the data, interpreted the results, and wrote and revised the manuscript. AF conceived and designed the approach, analyzed the data, interpreted the results, and revised the manuscript. KCJ performed laboratory experiments, data analysis, and revised the manuscript. HE conceived and designed the approach, analyzed the data, interpreted the results, and revised the manuscript. NT performed data analysis, and revised the manuscript. JK performed laboratory experiments, data analysis, and revised the manuscript. MLHR responsible for the patient cohort and clinical database, and revised the manuscript. VDH responsible for the patient cohort and clinical database, and revised the manuscript. FW responsible for the patient cohort and clinical database, and revised the manuscript. BN responsible for the patient cohort and clinical database, and revised the manuscript. VDH responsible for the patient cohort and clinical database, and revised the manuscript. A-LB-D conceived and designed the approach, interpreted the results, and revised the manuscript. JT conceived and designed the approach, interpreted the results, and revised the manuscript. BCC conceived and designed the approach, interpreted the results, and revised the manuscript. VNK conceived and designed the approach, analyzed the data, interpreted the results, and wrote and revised the manuscript. All authors read and approved the final manuscript.
Additional files
Abbreviations
- bp:
-
base pair
- CGI:
-
CpG island
- DCIS:
-
ductal carcinoma in situ
- ER:
-
estrogen receptor
- FDR:
-
false discovery rate
- FFPE:
-
formalin-fixed paraffin-embedded
- GEO:
-
Gene Expression Omnibus
- HR:
-
hazards ratio
- IBC:
-
invasive breast carcinoma
- IRB:
-
institutional review board
- SAM:
-
Significance Analysis of Microarrays
- TCGA:
-
The Cancer Genome Atlas
- TSS:
-
transcription start site
- UTR:
-
untranslated region
References
Herman JG, Baylin SB: Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003, 349: 2042-2054. 10.1056/NEJMra023075.
Esteller M: Epigenetics in cancer. N Engl J Med. 2008, 358: 1148-1159. 10.1056/NEJMra072067.
Feinberg AP, Irizarry RA: Evolution in health and medicine Sackler colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci U S A. 2010, 107: 1757-1764. 10.1073/pnas.0906183107.
Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, Briem E, Zhang K, Irizarry RA, Feinberg AP: Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011, 43: 768-775. 10.1038/ng.865.
Bediaga NG, Acha-Sagredo A, Guerra I, Viguri A, Albaina C, Ruiz DI, Rezola R, Alberdi MJ, Dopazo J, Montaner D, Renobales M, Fernandez AF, Field JK, Fraga MF, Liloglou T, de Pancorbo MM: DNA methylation epigenotypes in breast cancer molecular subtypes. Breast Cancer Res. 2010, 12: R77-10.1186/bcr2721.
Holm K, Hegardt C, Staaf J, Vallon-Christersson J, Jonsson G, Olsson H, Borg A, Ringner M: Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns. Breast Cancer Res. 2010, 12: R36-10.1186/bcr2590.
Kamalakaran S, Varadan V, Giercksky Russnes HE, Levy D, Kendall J, Janevski A, Riggs M, Banerjee N, Synnestvedt M, Schlichting E, Karesen R, Shama PK, Rotti H, Rao R, Rao L, Eric Tang MH, Satyamoorthy K, Lucito R, Wigler M, Dimitrova N, Naume B, Borresen-Dale AL, Hicks JB: DNA methylation patterns in luminal breast cancers differ from non-luminal subtypes and can identify relapse risk independent of other clinical variables. Mol Oncol. 2011, 5: 77-92. 10.1016/j.molonc.2010.11.002.
Ronneberg JA, Fleischer T, Solvang HK, Nordgard SH, Edvardsen H, Potapenko I, Nebdal D, Daviaud C, Gut I, Bukholm I, Naume B, Borresen-Dale AL, Tost J, Kristensen V: Methylation profiling with a panel of cancer related genes: association with estrogen receptor, TP53 mutation status and expression subtypes in sporadic breast cancer. Mol Oncol. 2011, 5: 61-76. 10.1016/j.molonc.2010.11.004.
Comprehensive molecular portraits of human breast tumours. Nature. 2012, 490: 61-70. 10.1038/nature11412.
Bijker N, Meijnen P, Peterse JL, Bogaerts J, Van HI, Julien JP, Gennaro M, Rouanet P, Avril A, Fentiman IS, Bartelink H, Rutgers EJ: Breast-conserving treatment with or without radiotherapy in ductal carcinoma-in-situ: ten-year results of European Organisation for Research and Treatment of Cancer randomized phase III trial 10853'a study by the EORTC Breast Cancer Cooperative Group and EORTC Radiotherapy Grou. J Clin Oncol. 2006, 24: 3381-3387. 10.1200/JCO.2006.06.1366.
Pang JM, Dobrovic A, Fox SB: DNA methylation in ductal carcinoma in situ of the breast. Breast Cancer Res. 2013, 15: 206-10.1186/bcr3420.
Verschuur-Maes AH, de Bruin PC, van Diest PJ: Epigenetic progression of columnar cell lesions of the breast to invasive breast cancer. Breast Cancer Res Treat. 2012, 136: 705-715. 10.1007/s10549-012-2301-4.
van Hoesel AQ, Sato Y, Elashoff DA, Turner RR, Giuliano AE, Shamonki JM, Kuppen PJ, van de Velde CJ, Hoon DS: Assessment of DNA methylation status in early stages of breast cancer development. Br J Cancer. 2013, 108: 2033-2038. 10.1038/bjc.2013.136.
Tommasi S, Karm DL, Wu X, Yen Y, Pfeifer GP: Methylation of homeobox genes is a frequent and early epigenetic event in breast cancer. Breast Cancer Res. 2009, 11: R14-10.1186/bcr2233.
Faryna M, Konermann C, Aulmann S, Bermejo JL, Brugger M, Diederichs S, Rom J, Weichenhan D, Claus R, Rehli M, Schirmacher P, Sinn HP, Plass C, Gerhauser C: Genome-wide methylation screen in low-grade breast cancer identifies novel epigenetically altered genes as potential biomarkers for tumor diagnosis. FASEB J. 2012, 26: 4937-4950. 10.1096/fj.12-209502.
Kishida Y, Natsume A, Kondo Y, Takeuchi I, An B, Okamoto Y, Shinjo K, Saito K, Ando H, Ohka F, Sekido Y, Wakabayashi T: Epigenetic subclassification of meningiomas based on genome-wide DNA methylation analyses. Carcinogenesis. 2012, 33: 436-441. 10.1093/carcin/bgr260.
Gao F, Shi L, Russin J, Zeng L, Chang X, He S, Chen TC, Giannotta SL, Weisenberger DJ, Zada G, Mack WJ, Wang K: DNA methylation in the malignant transformation of meningiomas. PLoS One. 2013, 8: e54114-10.1371/journal.pone.0054114.
Kang GH, Shim YH, Jung HY, Kim WH, Ro JY, Rhyu MG: CpG island methylation in premalignant stages of gastric carcinoma. Cancer Res. 2001, 61: 2847-2851.
Kang GH, Lee S, Kim JS, Jung HY: Profile of aberrant CpG island methylation along multistep gastric carcinogenesis. Lab Invest. 2003, 83: 519-526. 10.1097/01.LAB.0000064704.53132.65.
Bhagat R, Chadaga S, Premalata CS, Ramesh G, Ramesh C, Pallavi VR, Krishnamoorthy L: Aberrant promoter methylation of the RASSF1A and APC genes in epithelial ovarian carcinoma development. Cell Oncol (Dordr). 2012, 35: 473-479. 10.1007/s13402-012-0106-4.
Yamamoto E, Suzuki H, Yamano HO, Maruyama R, Nojima M, Kamimae S, Sawada T, Ashida M, Yoshikawa K, Kimura T, Takagi R, Harada T, Suzuki R, Sato A, Kai M, Sasaki Y, Tokino T, Sugai T, Imai K, Shinomura Y, Toyota M: Molecular dissection of premalignant colorectal lesions reveals early onset of the CpG island methylator phenotype. Am J Pathol. 2012, 181: 1847-1861. 10.1016/j.ajpath.2012.08.007.
Luo Y, Wong CJ, Kaz AM, Dzieciatkowski S, Carter KT, Morris SM, Wang J, Willis JE, Makar KW, Ulrich CM, Lutterbaugh JD, Shrubsole MJ, Zheng W, Markowitz SD, Grady WM: Differences in DNA methylation signatures reveal multiple pathways of progression from adenoma to colorectal cancer. Gastroenterology. 2014, 147: 418-429. 10.1053/j.gastro.2014.04.039.
Navarro A, Yin P, Monsivais D, Lin SM, Du P, Wei JJ, Bulun SE: Genome-wide DNA methylation indicates silencing of tumor suppressor genes in uterine leiomyoma. PLoS One. 2012, 7: e33284-10.1371/journal.pone.0033284.
Maekawa R, Sato S, Yamagata Y, Asada H, Tamura I, Lee L, Okada M, Tamura H, Takaki E, Nakai A, Sugino N: Genome-wide DNA methylation analysis reveals a potential mechanism for the pathogenesis and development of uterine leiomyomas. PLoS One. 2013, 8: e66632-10.1371/journal.pone.0066632.
Kulis M, Heath S, Bibikova M, Queiros AC, Navarro A, Clot G, Martinez-Trillos A, Castellano G, Brun-Heath I, Pinyol M, Barberan-Soler S, Papasaikas P, Jares P, Bea S, Rico D, Ecker S, Rubio M, Royo R, Ho V, Klotzle B, Hernandez L, Conde L, Lopez-Guerra M, Colomer D, Villamor N, Aymerich M, Rozman M, Bayes M, Gut M, Gelpi JL, et al: Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012, 44: 1236-1242. 10.1038/ng.2443.
Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF: Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010, 466: 253-257. 10.1038/nature09165.
Bailey CM, Hendrix MJ: IRF6 in development and disease: a mediator of quiescence and differentiation. Cell Cycle. 2008, 7: 1925-1930. 10.4161/cc.7.13.6221.
Botti E, Spallone G, Moretti F, Marinari B, Pinetti V, Galanti S, De Meo PD, De NF, Ganci F, Castrignano T, Pesole G, Chimenti S, Guerrini L, Fanciulli M, Blandino G, Karin M, Costanzo A: Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. Proc Natl Acad Sci U S A. 2011, 108: 13710-13715. 10.1073/pnas.1110931108.
Yu J, Ma X, Cheung KF, Li X, Tian L, Wang S, Wu CW, Wu WK, He M, Wang M, Ng SS, Sung JJ: Epigenetic inactivation of T-box transcription factor 5, a novel tumor suppressor gene, is associated with colon cancer. Oncogene. 2010, 29: 6464-6474. 10.1038/onc.2010.370.
Stevens KN, Lindstrom S, Scott CG, Thompson D, Sellers TA, Wang X, Wang A, Atkinson E, Rider DN, Eckel-Passow JE, Varghese JS, Audley T, Brown J, Leyland J, Luben RN, Warren RM, Loos RJ, Wareham NJ, Li J, Hall P, Liu J, Eriksson L, Czene K, Olson JE, Pankratz VS, Fredericksen Z, Diasio RB, Lee AM, Heit JA, DeAndrade M, et al: Identification of a novel percent mammographic density locus at 12q24. Hum Mol Genet. 2012, 21: 3299-3305. 10.1093/hmg/dds158.
Tao T, Shi H, Guan Y, Huang D, Chen Y, Lane DP, Chen J, Peng J: Def defines a conserved nucleolar pathway that leads p53 to proteasome-independent degradation. Cell Res. 2013, 23: 620-634. 10.1038/cr.2013.16.
Tsioras K, Papastefanaki F, Politis PK, Matsas R, Gaitanou M: Functional interactions between BM88/Cend1, Ran-binding protein M and Dyrk1B kinase affect Cyclin D1 levels and cell cycle progression/exit in mouse neuroblastoma cells. PLoS One. 2013, 8: e82172-10.1371/journal.pone.0082172.
Galcheva-Gargova Z, Konstantinov KN, Wu IH, Klier FG, Barrett T, Davis RJ: Binding of zinc finger protein ZPR1 to the epidermal growth factor receptor. Science. 1996, 272: 1797-1802. 10.1126/science.272.5269.1797.
De Smaele E, Di ML, Moretti M, Pelloni M, Occhione MA, Infante P, Cucchi D, Greco A, Pietrosanti L, Todorovic J, Coni S, Canettieri G, Ferretti E, Bei R, Maroder M, Screpanti I, Gulino A: Identification and characterization of KCASH2 and KCASH3, 2 novel Cullin3 adaptors suppressing histone deacetylase and Hedgehog activity in medulloblastoma. Neoplasia. 2011, 13: 374-385.
Coon BG, Direnzo DM, Konieczny SF, Aguilar RC: Epsins' novel role in cancer cell invasion. Commun Integr Biol. 2011, 4: 95-97.
Zaoui K, Benseddik K, Daou P, Salaun D, Badache A: ErbB2 receptor controls microtubule capture by recruiting ACF7 to the plasma membrane of migrating cells. Proc Natl Acad Sci U S A. 2010, 107: 18517-18522. 10.1073/pnas.1000975107.
Mishra SK, Yang Z, Mazumdar A, Talukder AH, Larose L, Kumar R: Metastatic tumor antigen 1 short form (MTA1s) associates with casein kinase I-gamma2, an estrogen-responsive kinase. Oncogene. 2004, 23: 4422-4429. 10.1038/sj.onc.1207569.
Dedeurwaerder S, Desmedt C, Calonne E, Singhal SK, Haibe-Kains B, Defrance M, Michiels S, Volkmar M, Deplus R, Luciani J, Lallemand F, Larsimont D, Toussaint J, Haussy S, Rothe F, Rouas G, Metzger O, Majjaj S, Saini K, Putmans P: Hames G, van BN, Coulie PG, Piccart M, Sotiriou C, Fuks F: DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol Med. 2011, 3: 726-741. 10.1002/emmm.201100801.
Dowell JD, Tsai SC, Dias-Santagata DC, Nakajima H, Wang Z, Zhu W, Field LJ: Expression of a mutant p193/CUL7 molecule confers resistance to M. Biochim Biophys Acta. 2007, 1773: 358-366. 10.1016/j.bbamcr.2006.11.020.
Kim SS, Shago M, Kaustov L, Boutros PC, Clendening JW, Sheng Y, Trentin GA, Barsyte-Lovejoy D, Mao DY, Kay R, Jurisica I, Arrowsmith CH, Penn LZ: CUL7 is a novel antiapoptotic oncogene. Cancer Res. 2007, 67: 9616-9622. 10.1158/0008-5472.CAN-07-0644.
Paradis V, Albuquerque M, Mebarki M, Hernandez L, Zalinski S, Quentin S, Belghiti J, Soulier J, Bedossa P: Cullin7: a new gene involved in liver carcinogenesis related to metabolic syndrome. Gut. 2013, 62: 911-919. 10.1136/gutjnl-2012-302091.
Hiraoka N, Yamazaki-Itoh R, Ino Y, Mizuguchi Y, Yamada T, Hirohashi S, Kanai Y: CXCL17 and ICAM2 are associated with a potential anti-tumor immune response in early intraepithelial stages of human pancreatic carcinogenesis. Gastroenterology. 2011, 140: 310-321. 10.1053/j.gastro.2010.10.009.
Haakensen VD, Biong M, Lingjaerde OC, Holmen MM, Frantzen JO, Chen Y, Navjord D, Romundstad L, Luders T, Bukholm IK, Solvang HK, Kristensen VN, Ursin G, Borresen-Dale AL, Helland A: Expression levels of uridine 5’-diphospho-glucuronosyltransferase genes in breast tissue from healthy women are associated with mammographic density. Breast Cancer Res. 2010, 12: R65-10.1186/bcr2632.
Muggerud AA, Hallett M, Johnsen H, Kleivi K, Zhou W, Tahmasebpoor S, Amini RM, Botling J, Borresen-Dale AL, Sorlie T, Warnberg F: Molecular diversity in ductal carcinoma in situ (DCIS) and early invasive breast cancer. Mol Oncol. 2010, 4: 357-368. 10.1016/j.molonc.2010.06.007.
Muggerud AA, Ronneberg JA, Warnberg F, Botling J, Busato F, Jovanovic J, Solvang H, Bukholm I, Borresen-Dale AL, Kristensen VN, Sorlie T, Tost J: Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer. Breast Cancer Res. 2010, 12: R3-10.1186/bcr2466.
Naume B, Zhao X, Synnestvedt M, Borgen E, Russnes HG, Lingjaerde OC, Stromberg M, Wiedswang G, Kvalheim G, Karesen R, Nesland JM, Borresen-Dale AL, Sorlie T: Presence of bone marrow micrometastasis is associated with different recurrence risk within molecular subtypes of breast cancer. Mol Oncol. 2007, 1: 160-171. 10.1016/j.molonc.2007.03.004.
Touleimat N, Tost J: Complete pipeline for Infinium((R)) Human Methylation 450 K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012, 4: 325-341. 10.2217/epi.12.21.
Enerly E, Steinfeld I, Kleivi K, Leivonen SK, Aure MR, Russnes HG, Ronneberg JA, Johnsen H, Navon R, Rodland E, Makela R, Naume B, Perala M, Kallioniemi O, Kristensen VN, Yakhini Z, Borresen-Dale AL: miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One. 2011, 6: e16915-10.1371/journal.pone.0016915.
Tibshirani R, Chu G, Balasubramanian N, Jun L: samr: SAM: Significance Analysis of Microarrays. R package version 2.0. 2011, [], [http://CRAN.R-project.org/package=samr]
Sun W: eMap: map gene expression qtl. R package version 1.2. 2010, [], [http://www.bios.unc.edu/~weisun/software/eMap_1.2.tar.gz]
Oosting J, Eilers P, Menezes R: quantsmooth: Quantile smoothing and genomic visualization of array data. R package version 1.26.0. 2012, [], [http://bioconductor.org/packages/release/bioc/html/quantsmooth.html]
Tibshirani R: Regression shrinkage and selection via the Lasso. J Roy Stat Soc B, Series B. 1996, 58: 267-288.
Bovelstad HM, Nygard S, Storvold HL, Aldrin M, Borgan O, Frigessi A, Lingjaerde OC: Predicting survival from microarray data-a comparative study. Bioinformatics. 2007, 23: 2080-2087. 10.1093/bioinformatics/btm305.
Friedman J, Hastie T, Tibshirani R: Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010, 33: 1-22.
Therneau T: A Package for Survival Analysis in S. R package version 2.37-4. 2013, [], [http://CRAN.R-project.org/package=survival]
Morris T, Butcher L, Feber A, Teschendorff A, Chakravarthty A, Beck S: ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics. 2014, 30: 428-430. 10.1093/bioinformatics/btt684.
Hastie T, Tibshirani R, Narasimhan BC: pamr: Pam: prediction analysis for microarrays. R package version 1.55. 2014, [], [http://CRAN.R-project.org/package=pamr]
R Development Core Team: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. 2014, [], [http://www.R-project.org/]
Acknowledgements
TF was a PhD fellow at the Faculty of Medicine, University of Oslo. This research was supported by NFR- Kreft grant no. 193387/H10, K.G. Jebsen Center for breast cancer research, DNK: The genetic makeup of breast cancer patients grant no. 368039-6260-33220, HSØ: OSBRAC 39346 (OUS no.), HSØ: The participation of Ahus on the K.G. Jebsen Center grant no. 2639032, and NIH grant P20GM104416 (BCC).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Electronic supplementary material
13059_2014_435_MOESM1_ESM.txt
Additional file 1: Significance Analysis of Microarrays (SAM) analysis of methylation level of gene regions between the five PAM50 derived subtypes. Gene region, false discovery rate (FDR) q-value and median methylation for each PAM50 subtype. (TXT 766 KB)
13059_2014_435_MOESM2_ESM.png
Additional file 2: Hierarchical clustering and heatmap of invasive tumors using gene regions differentially methylated between the five gene expression-derived subtypes.(PNG 302 KB)
13059_2014_435_MOESM3_ESM.txt
Additional file 3: Correlation between DNA methylation and gene expression; individual CpGs. Illumina probe ID, gene, transcription start site (TSS), chromosome, position of CpG, CpG position relative to CpG islands (CGIs) and gene region, correlation coefficient and uncorrected P-value. (TXT 716 KB)
13059_2014_435_MOESM4_ESM.txt
Additional file 4: Correlation between DNA methylation and gene expression; functional gene regions. Gene, gene region (whose methylation level correlates with expression), correlation coefficient and uncorrected P-value. (TXT 87 KB)
13059_2014_435_MOESM5_ESM.txt
Additional file 5: SAM analysis of methylation level of individual CpGs between healthy breast tissue and DCIS. Illumina probe ID, gene, CpG position relative to CGIs and gene region. Analyses performed in two independent datasets. Only concordant results in both datasets are reported. (TXT 517 KB)
13059_2014_435_MOESM6_ESM.txt
Additional file 6: SAM analysis of methylation level of individual CpGs between DCIS and IBC. Illumina probe ID, gene, CpG position relative to CGIs and gene region. (TXT 59 KB)
13059_2014_435_MOESM7_ESM.txt
Additional file 7: SAM analysis of methylation level of gene regions between healthy breast tissue and DCIS. Analyses performed in two independent datasets. Only concordant results in both datasets are reported. (TXT 31 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Fleischer, T., Frigessi, A., Johnson, K.C. et al. Genome-wide DNA methylation profiles in progression to in situand invasive carcinoma of the breast with impact on gene transcription and prognosis. Genome Biol 15, 435 (2014). https://doi.org/10.1186/s13059-014-0435-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13059-014-0435-x