Abstract
Background
Mammography screening has been proven to detect breast cancer at an early stage and reduce mortality; however, it has low accuracy in young women or women with dense breasts. Blood-based diagnostic tools may overcome the limitations of mammography. This study assessed the diagnostic performance of a three-protein signature in patients with suspicious breast lesions.
Findings
This trial (MAST; KCT0004847) was a prospective multicenter observational trial. Three-protein signature values were obtained using serum and plasma from women with suspicious lesions for breast malignancy before tumor biopsy. Additionally, blood samples from women who underwent clear or benign mammography were collected for the assays. Among 642 participants, the sensitivity, specificity, and overall accuracy values of the three-protein signature were 74.4%, 66.9%, and 70.6%, respectively, and the concordance index was 0.698 (95% CI 0.656, 0.739). The diagnostic performance was not affected by the demographic features, clinicopathologic characteristics, and co-morbidities of the participants.
Conclusions
The present trial showed an accuracy of 70.6% for the three-protein signature. Considering the value of blood-based biomarkers for the early detection of breast malignancies, further evaluation of this proteomic assay is warranted in larger, population-level trials.
This Multi-protein Assessment using Serum to deTermine breast lesion malignancy (MAST) was registered at the Clinical Research Information Service of Korea with the identification number of KCT0004847 (https://cris.nih.go.kr).
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Introduction
Breast cancer incidence remains the highest in women worldwide and has substantially increased in Korea over the past several decades [1, 2]. Early detection is crucial to reducing mortality associated with breast cancer and morbidity during treatment [3]. Current breast cancer screening includes physical examination and mammography, although mammography has shown low accuracy in young women or women with dense breasts [4]. Mammography may also cause pain or discomfort during testing, and harm from irradiation may outweigh its benefits in young women [5, 6].
Blood-based markers can address the drawbacks of the current breast screening tools [7]. We recently reported a new proteomic assay that quantifies blood levels of three proteins (Mastocheck®) using multiple reaction monitoring mass spectrometry (MRM-MS) [8]. The three proteins used in the assay are apolipoprotein C-I (APOC1), carbonic anhydrase I (CA1), and neural cell adhesion molecule L1-like protein (CHL1). APOC1 was identified as a downregulated serum protein marker in an independent study [9]. Studies have suggested that the CA1 protein contributes to microcalcification in breast cancer and atherosclerosis [10, 11], and high levels of blood CA1 are detected in patients with breast cancer [10]. CHL1, a cell adhesion molecule, has been proposed as a tumor suppressor in breast cancer; however, its value as a diagnostic marker is unclear [12].
Three proteins were selected among the 124 proteins discovered in our previous proteomic experiments [13,14,15,16] based on the diagnostic performance of plasma levels of each protein in patients with breast cancer [8]. The three-protein signature was subsequently validated in a large-scale study using samples stored in biorepositories [17]. Additionally, this signature has been shown to increase the diagnostic accuracy of breast ultrasonography when used in combination [18]. Herein, we report the results of a prospective multicenter trial on the diagnostic accuracy of the three-protein signature in women with suspicious breast lesions.
Methods
Study design and participants
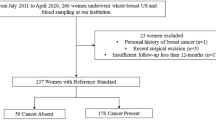
This multi-protein assessment using serum to determine breast lesion malignancy (MAST) (KCT0004847) was a prospective multicenter trial conducted in 13 teaching hospitals in Korea. From August 2019 to September 2020, plasma and serum samples were collected from 471 participants with moderate to highly suspicious breast lesions identified by breast mammography or ultrasound, for which a breast biopsy was planned (Fig. 1). Moderate to highly suspicious breast lesions were defined as category 4B, 4C, or 5 of the Breast Imaging Reporting and Data System (BI-RADS) [19]. Women younger than 18 years of age, who had a personal history of breast cancer, or other malignancy within the five years were ineligible. Up to 20 ml of whole blood was drawn before the breast biopsy and was used for the three-protein signature. Demographic and pathological information was collected after the biopsy results were reported. A total of 451 patients were included in the final analysis after excluding patients for various reasons (Fig. 1). Additionally, the blood samples of 191 participants who showed no suspicious breast lesions (BI-RADS 1 or 2) were collected for the three-protein signature assessment. The three-protein signature (Mastocheck®, a regression model based on the relative expression of CHL1, APOC1, and CA1) was approved as an in vitro diagnostic tool using plasma samples by the Korean Ministry of Food and Drug Safety in Jan 2019. In this trial, serum samples were collected to determine the correlation between plasma and serum.
This trial was registered at the Clinical Research Information Service of Korea, a member of the World Health Organization (WHO) International Clinical Trials Registry Platform (ICTRP), with the identification number KCT0004847. The original version of the study protocol is provided in Additional file 1. The detail of sample processing and statistical analysis is described in Additional file 2: Additional Methods.
Results
In this trial, 451 women with suspicious breast lesions and 191 with no suspicious lesions were included in the final analysis (Fig. 1). Among the 451 women with suspicious breast lesions, 313 were diagnosed with breast cancer through subsequent breast biopsies. Therefore, the analysis included plasma and serum samples from 313 patients with breast cancer and 329 women with biopsy-proven benign disease or no suspicious lesions in the breast. The clinical and demographic information of all participants is described in Additional file 3: Tables 1 and 2.
The sensitivity, specificity, and overall accuracy values of the three-protein signature for detecting breast malignancies in 642 women were 74.4%, 66.9%, and 70.6%, respectively (Table 1). The concordance index was 0.698 (95% CI 0.656, 0.739). The performance of the three-protein signature was not affected by the various clinical or demographic features of the participants and was not biased by site (p = 0.383) (Table 2). Notably, accuracy was not influenced by the mammographic density of the participants (p = 0.878). The three-protein signature assay was repeated using serum samples to determine the concordance rate with the results from the plasma samples. The results were concordant in 605 (94.2%) patients (Additional file 3: Table 1).
Although the three-protein signature showed trends for higher sensitivity for in situ tumors and the American Joint Committee on Cancer (AJCC) stage I cancers, the sensitivity of the three-protein signature did not show statistically significant differences across AJCC stages (p = 0.859, Fig. 2a). Additionally, breast cancer subtypes, as defined by hormone receptor status and human epidermal growth factor receptor 2 (HER2) expression, did not show statistically significant association with the sensitivity of the three-protein signature (p = 0.902, Fig. 2b). Figure 2c shows the levels of the three protein markers among all participants. The levels of APOC1 and CA1 showed statistically significant differences between 313 patients with breast cancer and 329 participants with BI-RADS C1 or 2 breast imaging or biopsy-proven benign breast lesions. However, the CHL1 did not show such differences in the present study. The levels of APOC1 showed statistically significant differences between participants who had no suspicious lesion, patients who had biopsy-proven benign breast lesions, and patients with malignant breast lesions (p < 0.001, Fig. 2d). While the CA1 levels of patients with breast malignancy were significantly higher when compared to those of participants with biopsy-proven benign lesions or participants with no suspicious lesions (p = 0.019 and p < 0.001, respectively), the levels of CA1 did not show significant difference between the latter two groups (p = 0.482) (Fig. 2d).
Results of the three-protein signature analysis. a, b depict the sensitivity of the three-protein signature in different cancer stages and subtypes, respectively. The sensitivity of the three-protein signature showed no statistical differences across AJCC stages and molecular subtypes (p = 0.859 and p = 0.902, respectively). c The concentration of each protein in all participants. APOC1 and CA1 levels were statistically different between 313 patients with breast cancer (Cancer group) and 329 participants (Non-cancer group) with no suspicious breast lesions (Normal group) or biopsy-proven benign breast lesions (Benign group), whereas CHL1 did not show such a difference. Wilcoxon rank sum test was used to compare differences between two groups because the concentration of each protein was not normally distributed. d Depict the concentrations of the three markers in three participants’ group. APOC1 were significantly different between Normal group, Benign group, and Cancer group. While CA1 levels of Cancer group were higher than those of Normal group and Benign group, CA1 did not show significant difference between Normal group and Benign group. p-values calculated using Dunn’s nonparametric comparison for post hoc Kruskal–Wallis testing. *p < 0.05, **p < 0.01, ***p < 0.001 and ns non-significant
Discussion
To our knowledge, this is the first report of a prospective multicenter trial that addressed the accuracy of blood-based proteomic diagnostic markers for breast cancer. Although many studies have focused on discovering blood-based protein biomarkers for the early detection of breast cancer, the clinical value of these biomarkers has not yet been tested in a prospective clinical trial setting [20]. Our results demonstrated that the sensitivity of the three-protein signature was 74.4%, which is comparable with the results of our previous reports using archived blood samples [17]. Interestingly, the sensitivity of the three-protein signature was not affected by the mammographic density of participants or other clinicopathological factors. As low sensitivity has been a major hurdle for using traditional serum markers of breast cancer in the clinic [21], the present findings suggest that this proteomic-based assay may be a useful tool for breast cancer diagnosis in women with dense breasts. Indeed, our previous study using large-scale biorepository samples suggested that combining the three-protein signature with mammography can improve diagnostic accuracy in women with dense breasts [22].
Our study carries several limitations. First, no follow-up data were available for the 191 participants who showed no suspicious lesions in breast imaging. Second, detailed pathologic information on the 61 (19.5%) participants with breast cancer was not available. While the stage or subtype did not influence the accuracy of the three-protein signature in the remaining 252 patients, it would be important to prospectively address this issue in a larger prospective cohort since the current trial was not designed for determination of the stage- or subtype-specific accuracy of the assay. Third, unlike APOC1 and CA1, which showed significant differences between benign and cancer patients, CHL1 levels failed to show such differences in the current trial. Further efforts to discover additional biomarker candidates are warranted to improve the diagnostic accuracy of the assay. Finally, the demographic features and co-morbidities showed differences between patients with breast malignancy and participants without suspicious lesions or with benign breast lesions (Additional file 3: Table 2). This finding also suggest that it require further data form a large cohort to verify the value of three-protein signature.
Screening tests are subject to high specificity to ensure a low rate of false positives, minimizing unnecessary diagnostic workups. The results of this trial revealed that the three-protein signature showed a moderate degree of specificity (60.9 (52.7–69.0)%) for 451 women with suspicious lesions and all participants (66.9 (61.8–72.0)%). A larger clinical trial with asymptomatic women undergoing breast cancer screening is required to determine the recall rates of the three-protein signature. Considering the benefits of blood-based biomarkers in breast cancer screening [7], our data warrant further efforts to validate the value of this proteomic assay. This prospective study showed that a three-protein signature based on MRM-MS is a sensitive tool for breast cancer diagnosis. A future large clinical trial is warranted to determine its value in breast cancer screening.
Availability of data and materials
All clinical data generated or analyzed during this study are summarized in this published article and its Additional files. The proteomic data that support the findings of this study are available from the corresponding author on reasonable request.
Abbreviations
- MAST:
-
Multi-protein assessment using serum to determine breast lesion malignancy
- MRM-MS:
-
Multiple reaction monitoring mass spectrometry
- APOC1:
-
Apolipoprotein C-I
- CA1:
-
Carbonic anhydrase I
- CHL1:
-
Neural cell adhesion molecule L1-like protein
- BI-RADS:
-
Breast imaging reporting and data system
- WHO:
-
World Health Organization
- ICTRP:
-
International clinical trials registry platform
- AJCC:
-
American Joint Committee on Cancer
- HER2:
-
Human epidermal growth factor receptor 2
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Kang SY, Kim YS, Kim Z, Kim HY, Kim HJ, Park S, et al. Breast cancer statistics in Korea in 2017: data from a breast cancer registry. J Breast Cancer. 2020;23(2):115–28.
Duffy SW, Vulkan D, Cuckle H, Parmar D, Sheikh S, Smith RA, et al. Effect of mammographic screening from age 40 years on breast cancer mortality (UK Age trial): final results of a randomised, controlled trial. Lancet Oncol. 2020;21(9):1165–72.
Lauby-Secretan B, Scoccianti C, Loomis D, Benbrahim-Tallaa L, Bouvard V, Bianchini F, et al. Breast-cancer screening-viewpoint of the IARC Working Group. N Engl J Med. 2015;372(24):2353–8.
Sharp PC, Michielutte R, Freimanis R, Cunningham L, Spangler J, Burnette V. Reported pain following mammography screening. Arch Intern Med. 2003;163(7):833–6.
Myers ER, Moorman P, Gierisch JM, Havrilesky LJ, Grimm LJ, Ghate S, et al. Benefits and harms of breast cancer screening: a systematic review. JAMA. 2015;314(15):1615–34.
Li J, Guan X, Fan Z, Ching LM, Li Y, Wang X, et al. Non-invasive biomarkers for early detection of breast cancer. Cancers (Basel). 2020;12(10):2767.
Lee HB, Kang UB, Moon HG, Lee J, Lee KM, Yi M, et al. Development and validation of a novel plasma protein signature for breast cancer diagnosis by using multiple reaction monitoring-based mass spectrometry. Anticancer Res. 2015;35(11):6271–9.
Sun Y, Zhang J, Guo F, Zhao W, Zhan Y, Liu C, et al. Identification of apolipoprotein C-I peptides as a potential biomarker and its biological roles in breast cancer. Med Sci Monit. 2016;22:1152–60.
Zheng Y, Xu B, Zhao Y, Gu H, Li C, Wang Y, et al. CA1 contributes to microcalcification and tumourigenesis in breast cancer. BMC Cancer. 2015;15:679.
Yuan L, Wang M, Liu T, Lei Y, Miao Q, Li Q, et al. Carbonic anhydrase 1-mediated calcification is associated with atherosclerosis, and methazolamide alleviates its pathogenesis. Front Pharmacol. 2019;10:766.
Martin-Sanchez E, Mendaza S, Ulazia-Garmendia A, Monreal-Santesteban I, Blanco-Luquin I, Cordoba A, et al. CHL1 hypermethylation as a potential biomarker of poor prognosis in breast cancer. Oncotarget. 2017;8(9):15789–801.
Kang UB, Ahn Y, Lee JW, Kim YH, Kim J, Yu MH, et al. Differential profiling of breast cancer plasma proteome by isotope-coded affinity tagging method reveals biotinidase as a breast cancer biomarker. BMC Cancer. 2010;10:114.
Kim DH, Bae J, Lee JW, Kim SY, Kim YH, Bae JY, et al. Proteomic analysis of breast cancer tissue reveals upregulation of actin-remodeling proteins and its relevance to cancer invasiveness. Proteomics Clin Appl. 2009;3(1):30–40.
Suh EJ, Kabir MH, Kang UB, Lee JW, Yu J, Noh DY, et al. Comparative profiling of plasma proteome from breast cancer patients reveals thrombospondin-1 and BRWD3 as serological biomarkers. Exp Mol Med. 2012;44(1):36–44.
Chang JW, Kang UB, Kim DH, Yi JK, Lee JW, Noh DY, et al. Identification of circulating endorepellin LG3 fragment: potential use as a serological biomarker for breast cancer. Proteomics Clin Appl. 2008;2(1):23–32.
Kim Y, Kang UB, Kim S, Lee HB, Moon HG, Han W, et al. A validation study of a multiple reaction monitoring-based proteomic assay to diagnose breast cancer. J Breast Cancer. 2019;22(4):579–86.
Ha SM, Kim HK, Kim Y, Noh DY, Han W, Chang JM. Diagnostic performance improvement with combined use of proteomics biomarker assay and breast ultrasound. Breast Cancer Res Treat. 2022;192:541–52.
Mercado CL. BI-RADS update. Radiol Clin North Am. 2014;52(3):481–7.
Nunez C. Blood-based protein biomarkers in breast cancer. Clin Chim Acta. 2019;490:113–27.
Duffy M. Serum tumor markers in breast cancer: are they of clinical value? Clin Chem. 2006;52:345–51.
Kim Y, Moon H-G, Lee H-B, Moon WK, Cho N, Chang J-M, et al. Efficacy of mastocheck for screening of early breast cancer: comparison with screening mammography. J Breast Dis. 2019;7(2):59–64.
Acknowledgements
We thank everyone who contributed to this study, including all patients who participated in this study. DN and HM are the members of inventors of three protein markers for breast cancer diagnosis (APOC1, CA1, and CHL1) and Korean patent numbers are 10-1431063 (2014), 10-1431064 (2014) and 10-1431065 (2014), respectively.
Funding
This study was supported by the Bertis Inc. The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
EL, YK, DN and HM contributed to the conception and design of this study. HS, KH, JM, MK, SA, SJ, HS, MC, TY, SJ, SW, and JK contributed to the patient sample and data collection. EL and HM contributed to the acquisition, analysis, or interpretation of data. ES, DN and HM drafted and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Institutional review board or independent ethics committee approval was obtained at each participating site (as follows) and the study was conducted in accordance with the principles of the Declaration of Helsinki. Written informed consent has been obtained from the participants involved. This trial was registered at the Clinical Research Information Service of Korea, a member of the WHO International Clinical Trials Registry Platform (ICTRP) with the identification number of KCT0004847.
List of approval number at each site
Seoul National University Hospital (1905-175-1036), Seoul National University Bundang Hospital (E-1909-564-406), Seoul Metropolitan Government Seoul National University Boramae Medical Center (20-2019-61), Dankook University Hospital (2019-09-001), Chung-Ang University Hospital (1982-004-384), Hallym University Kangnam Sacred Heart Hospital (2019-08-009), National Cancer Center (NCC2019-0298), Myongji Hospital (2019-09-010), Hanyang University Hospital (2019-09-011), Catholic University of Korea Seoul St. Mary's Hospital, College of Medicine (KC19TDDI0796), Korea University Anam Hospital (2020AN0088), Korea University Guro Hospital (2020GR0119) and Gyeongsang National University Hospital (2020-01-030).
Consent for publication
All informed consent was obtained. This manuscript does not contain any individual person’s information.
Competing interests
This study was supported by the Bertis Inc. The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. HM and HS reported being Outside Director; receiving consulting fees as a member of Bertis Inc.;holding stock options of the Bertis Inc. YK reported receiving consulting fees from Bertis Inc. during the conduct of the study; holding stock options of the Bertis Inc. HS reported his family members hold stocks of the Bertis Inc. DN reported being a member of Bertis Inc.;holding equity in Bertis Inc. EL reported receiving honoraria for consultation or advisory board participation from Bertis Inc. No other disclosures were reported.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. MAST trial protocol.
Additional file 2
. Additional Methods.
Additional file 3
. Additional Table 1 and Additional Table 2.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lee, ES., Kim, Y., Shin, HC. et al. Diagnostic accuracy of a three-protein signature in women with suspicious breast lesions: a multicenter prospective trial. Breast Cancer Res 25, 20 (2023). https://doi.org/10.1186/s13058-023-01616-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13058-023-01616-5