Abstract
Background
We previously showed that BRCA-like profiles can be used to preselect individuals with the highest risk of carrying BRCA mutations but could also indicate which patients would benefit from double-strand break inducing chemotherapy. A simple, robust, and reliable assay for clinical use that utilizes limited amounts of formalin-fixed, paraffin-embedded tumor tissue to assess BRCAness status in both ER-positive and ER-negative breast cancer (BC) is currently lacking.
Methods
A digital multiplex ligation-dependent probe amplification (digitalMLPA) assay was designed to detect copy number alterations required for the classification of BRCA1-like and BRCA2-like BC. The BRCA1-like classifier was trained on 71 tumors, enriched for triple-negative BC; the BRCA2-like classifier was trained on 55 tumors, enriched for luminal-type BC. A shrunken centroid-based classifier was developed and applied on an independent validation cohort. A total of 114 cases of a randomized controlled trial were analyzed, and the association of the classifier result with intensified platinum-based chemotherapy response was assessed.
Results
The digitalMLPA BRCA1-like classifier correctly classified 91% of the BRCA1-like samples and 82% of the BRCA2-like samples. Patients with a BRCA-like tumor derived significant benefit of high-dose chemotherapy (adjusted hazard ratio (HR) 0.12, 95% CI 0.04–0.44) which was not observed in non-BRCA-like patients (HR 0.9, 95% CI 0.37–2.18) (p = 0.01). Analysis stratified for ER status showed borderline significance.
Conclusions
The digitalMLPA is a reliable method to detect a BRCA1- and BRCA2-like pattern on clinical samples and predicts platinum-based chemotherapy benefit in both triple-negative and luminal-type BC.
Similar content being viewed by others
Background
Breast cancers arising in women with a BRCA1 or BRCA2 germline mutation are characterized by genomic instability. As both BRCA1 and BRCA2 play a role in the process of homologous recombination, non-functioning BRCA genes result in incorrect DNA repair, leading to gross genomic instability. Previously, our group has shown that BRCA1- and BRCA2-mutated tumors have a specific pattern of chromosomal gains and losses [1,2,3]. These specific genomic regions were used to develop a BRCA1-like and a BRCA2-like classifier. In follow-up studies, we showed that these classifiers could be used to identify germline BRCA1- or BRCA2-mutated cases, but also tumors with other mechanisms of BRCA1 or BRCA2 inactivation such as BRCA1 promoter methylation [1, 4, 5]. In addition, the classifiers have been used in the classification of BRCA1 variants of unknown significance (VUS) [2].
Interestingly, we showed that the classifiers could not only be used to identify mechanisms of BRCA inactivation, but could also be used to predict treatment benefit. We hypothesized that a BRCA1-like or BRCA2-like profile is a read-out for a homologous recombination deficiency (HRD) phenotype and could therefore indicate tumors which would be highly sensitive to DNA-damaging agents. In several studies, we showed a remarkable benefit of high-dose alkylating chemotherapy in patients with a BRCA1-like or BRCA2-like profile [6,7,8,9]. In addition to high-dose chemotherapy, a gene expression-based BRCA1ness test, derived from the DNA-based BRCA1-like classifier, could predict benefit from the addition of veliparib/carboplatin to paclitaxel in the ISPY2 trial [10]. The currently presented BRCA-like test could function as a companion diagnostic test.
For a clinical test, it is essential that the method is highly reliable, has a fast turnaround time, and works on small quantities of paraffin-embedded tissue material. Multiplex ligation-dependent probe amplification (MLPA) is an established method and a standardized assay applied in routine diagnostics worldwide [11,12,13,14,15]. MLPA is a method based on the amplification and relative quantification of the ligated adjacent probes, which can target up to 50 different genomic regions that show diagnostically or clinically significant copy number changes in patient samples [11]. There are several advantages of MLPA over other copy number profiling methods: its fast turnaround time, suitable for degraded paraffin-embedded material, and use of standard (diagnostic) molecular laboratory equipment. In the past, we developed a multiplex ligation-dependent probe amplification (MLPA) assay to perform a BRCA1-like classification based on 34 MLPA probes [16]. The BRCA2-like copy number profile contains threefold as many genomic regions as the BRCA1-like profile, making it impossible to design a MLPA. Recently, digitalMLPA was introduced as a novel technology to measure copy number aberrations on 700 genomic locations [17], making it a perfect technique for combined BRCA1-like and BRCA2-like profiling.
In the current study, we developed a digitalMLPA assay to assess both the BRCA1-like and BRCA2-like patterns. We validated the digitalMLPA assay on an independent sample cohort. In addition, we profiled samples from a randomized controlled trial [18], to determine if the digitalMLPA has treatment predictive value. We showed that the assay could identify both triple-negative and luminal-type tumors with a remarkable good survival after high-dose double-strand break (DSB)-inducing chemotherapy.
Methods
Sample selection
Three series of breast cancer specimens were used for this study. (1) For the development and testing of the BRCA1-like classifier, a set of samples enriched for triple-negative (TN) breast cancer was used, as was done in the original BRCA1-like classifier development study [2]. (2) For the BRCA2-like classification, we used a mixed set of different subtypes, similar to the original BRCA2 classifier study [1]. For both the training and the test sets, samples were equally divided and stratification was based on ER status, material type (paraffin-embedded or fresh frozen), and array-based BRCA-like classification. Samples were scored as positive for ER and/or PR by immunohistochemistry (IHC), when at least 10% of the tumor cell nuclei showed staining of the ER or PR, respectively. A sample was scored as being HER2 positive when either a strong membrane staining (3+) could be observed by IHC or if CISH revealed amplification of HER2 in samples with moderate (2+) membrane staining at IHC. Mutation/methylation status was available for a minority of the patients that had undergone clinical genetic testing. The other patients are indicated as “unknown.” Supplemental figure 1 shows the sample flow through the training and validation series for the BRCA1-like and BRCA2-like digitalMLPA development. (3) To assess whether the digitalMLPA BRCA1-like and BRCA2-like classifiers had treatment predictive value, we analyzed samples from the Dutch high dose trial [18]. In this trial, patients were randomized between standard dose 5-fluorouracil-epirubicin-cyclophosphamide (FEC) chemotherapy and high-dose cyclophosphamide-thiotepa-carboplatin (HD-CTC) chemotherapy. We were able to include samples from 122 women for the digitalMLPA analysis; as for quite some trial patients, no tissue blocks or DNA was left for further analysis. Table 1 gives an overview of the sample series. The institutional review board of The Netherlands Cancer Institute approved the study. All trial participants provided written informed consent. The issue of non-trial participants was obtained under an opt-out regime in this study conform to Dutch regulations and the Code of Conduct of Federa-COREON (https://www.federa.org/codes-conduct).
Development of digitalMLPA probe mix for BRCAness
Array comparative genomic hybridization (Array-CGH)-based classifiers recognizing genomic alterations specific for BRCA1-mutated and BRCA2-mutated tumors were developed and previously published [1, 2]. Array CGH was performed on a microarray containing 3.5 bacterial artificial chromosomes (BACs). Using the shrunken centroids algorithm, BAC clones located in the most important regions contributing to the respective classifiers were identified for both a BRCA1-like and a BRCA2-like pattern. For each BAC clone selected based on high centroid value, at least one digitalMLPA probe was designed to be included in the BRCAness digitalMLPA assay. This resulted in a set of 84 digitalMLPA probes for BRCA1ness and 206 digitalMLPA probes for BRCA2ness classification (Supplemental Table 1).
In addition to the BRCAness-specific probes, a set of 206 “digital karyotyping” probes was included that covers all chromosome arms. This set was used for determining gross chromosomal aberrations and for data normalization. A set of 128 internal quality control probes was also included to determine reaction quality, amount of input DNA, and sample contamination and for troubleshooting purposes [17]. Quality tests were performed at MRC Holland to test the performance of each probe included in the BRCAness digitalMLPA assay (product details available at request). Probes showing high variability in DNA samples with normal copy numbers (standard deviation > 0.1) or probes sensitive to pipetting mistakes or technical errors during digitalMLPA experiments were replaced by other probes that performed better in the quality tests.
digitalMLPA experiments
digitalMLPA experiments were performed as described previously [17]. Briefly, 40 ng of each DNA test sample was mixed with 2 μL of a unique barcode solution (MRC Holland), followed by DNA denaturation. Reactions were incubated with the digitalMLPA D004-X2 BRCAness probemix overnight at 60 °C to ensure hybridization of the probes to the sample DNA. The next day, the probes were ligated and amplified by PCR. The PCR-amplified products were then loaded onto an Illumina MiSeq sequencer (Illumina, San Diego, CA) for quantification using the MiSeq Reagent Kit version 3 (150 cycles; Illumina). Reference samples included were triplicates of Promega Human Genomic male DNA (Promega Benelux, Leiden, the Netherlands).
Data analysis
Analysis of MiSeq data was done as described previously [17]. First, reads were mapped to the corresponding digitalMLPA probes present in the probemix. After correct identification, read counts for each probe were first compared with the median value of all reference probes in each sample. In the second step, the normalized values of each individual probe were compared with the median value of the corresponding probe in the reference samples. These final ratios were used as input for class prediction.
Class prediction
Data values from all the 290 probes of interest (84 digitalMLPA probes for BRCA1-like and 206 digitalMLPA probes for BRCA2-like classification) were used for prediction analysis for microarrays (PAM) in R. In brief, the nearest shrunken centroid method [19] was applied to the training set with maximum distinct clinical courses (first set), and the classification performance was evaluated by a 10-times-repeated 10-fold cross-validation. A cutoff was chosen to make the best discrimination (highest accuracy) between a BRCA1-like and a non-BRCA1-like profile for the BRCA1-like classifier and between a BRCA2-like and a non-BRCA2-like profile for the BRCA2-like classifier (supplementary figure 2).
Subsequently, the obtained classifiers were tested on the respective test sets using the predefined cutoffs. The BRCA1 classifier and BRCA2-like results obtained by array CGH [1, 2] were considered as the gold standard.
Statistics
In the Dutch high dose trial, we performed the Kaplan-Meier method and compared survival by log-rank tests (using R version 3.6 and package Survminer). Cox regression analysis was used to calculate hazard ratios in 114 trial patients where clinical information was complete; for 8 patients, histological grade was unknown. Multivariate Cox regression models were stratified for the number of lymph nodes and TN status and adjusted for pT-stage, histological grade, and BRCA-like status. The interaction with treatment was calculated between BRCA-like and non-BRCA-like patients adjusted as mentioned above. These analyses were repeated for the patients with a triple-negative tumor as well as for the ER/PR-positive Her2-negative (luminal-type) tumors.
This study was designed according to the REporting recommendations for tumor MARKer prognostic studies (REMARK) guidelines (Supl table 3) [20].
Results
Training and validation of digitalMLPA-based BRCAness classifiers
Table 1 gives an overview of the sample series used to develop (training set) and test (test set) the classifier. We tested how the digitalMLPA BRCA1-like classifier performed against the array-based test result, which was considered the gold standard. A threshold for the digitalMLPA BRCA1-like was set at 0.14 using the array-based test result (supplemental figure 2A). Using this threshold, the BRCA1-like classification based on the digitalMLPA data resulted in an accuracy of 99% in the training set (Table 2). All BRCA1-like were correctly classified, and only one non-BRCA1-like sample was scored as a BRCA1-like sample with digitalMLPA. Performance in the test set was excellent with an accuracy of 91%. Six samples were not correctly classified by digitalMLPA using the specified cutoff values: three BRCA1-like and three non-BRCA1-like samples. Two of the misclassified samples had a classifier result just below the threshold. Two other samples had a copy number pattern with many chromosomal aberrations and some noise, due to a lower quality DNA sample. For the other two misclassified samples, we could not find an explanation. The digitalMLPA assay performed equally well on paraffin-embedded or fresh frozen samples (data not shown).
Results of the classifier in the prediction of BRCA2-like status
For BRCA2-like classification, the cutoff was set at 0.21, using the already established BRCA2-like array-based classification (Supplemental figure 2B). The BRCA2-like classifier had an accuracy of 89% in the training set; four BRCA2-like and two non-BRCA2-like samples were not correctly classified (Table 3). In the test set, the accuracy was 82%; seven BRCA2-like and three non-BRCA2-like samples were misclassified.
Classifier performance in BRCA-mutated and BRCA-methylated samples
As a next step, we assessed if the digitalMLPA classifiers correctly classified samples with a BRCA1 mutation, BRCA2 mutation, and BRCA1 promoter methylation (Table 4). As previously shown by us, the BRCA1-like and BRCA2-like classifiers detected 88–90% of BRCA1-mutated, BRCA1-methylated, or BRCA2-mutated tumors [2, 4, 5]. The digitalMLPA classified 88% of the BRCA1-mutated tumors and 96% of BRCA1-methylated tumors as BRCA1-like. Ninety-three percent of the BRCA2-mutated tumors were classified as BRCA2-like.
Survival analysis Dutch high dose study
Most importantly, we tested if the digitalMLPA assay had treatment predictive value. In the Dutch high dose trial, patients were randomized between conventional FEC-based chemotherapy and HD-CTC. We previously showed that patients with BRCA1-like and BRCA2-like tumors had remarkably better responses on HD-CTC than on FEC [8, 21]. To test if the digitalMLPA BRCAness assay could be used for treatment response prediction, we analyzed 122 samples of the same trial using digitalMLPA (Table 1 (C, D)). Median follow-up for these 122 patients was 8 years. After 5 years, 88 out of 122 patients were alive (72%) and 76 out of 122 did not have a recurrence within 5 years (data not shown). Supplemental Table 2 shows the association of BRCA1- and BRCA2-like tumors with known clinical risk factors in trial patients. The digitalMLPA BRCA1-like pattern was associated with younger age, ER-negative tumors, and high grade, as previously shown by us using array-based BRCAness classification methods [5, 21, 22]. The BRCA2-like profile was associated with ER- and PR-positive tumors.
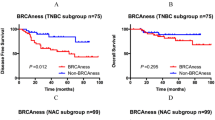
Figure 1 shows Kaplan-Meier plots for overall survival for all patients, non-BRCA-like, BRCA1-like, BRCA2-like, and BRCA-like (either a BRCA1-like or a BRCA2-like profile) tumors, stratified for treatment. HR in this subset of the original trial population was 0.38 (95% CI 0.20–0.72; p = 0.003) for HD-CTC compared to FEC chemotherapy. Patients with a BRCA-like profile had a significantly better overall survival after HD-CTC treatment compared with FEC chemotherapy (adjusted HR 0.12, 95% CI 0.04–0.44, p = 0.001) (Table 5). This difference was not observed in non-BRCA-like patients (adjusted HR 0.90, 95% CI 0.37–2.18; p = 0.818). The effect of HD-CTC over FEC treatment was significantly different between BRCA-like and non-BRCA-like patients (p interaction 0.011). The different effect of treatment for BRCA-like and non-BRCA-like tumors was borderline significant when the analysis was stratified for triple-negative and luminal breast cancer (p interaction was 0.072 and 0.07, respectively) (Table 5 and Fig. 1). Subgroup analysis should be interpreted with caution, due to small numbers.
Kaplan-Meier survival plots for overall survival according to digitalMLPA BRCAness status and stratified for treatment in the Dutch high-dose study. Survival is subsequently shown all patients (a), for non-BRCA-like patients (b), BRCA1-like patients (c), BRCA2-like patients (d), BRCA-like patients (e), ER+ non-BRCA-like patients (f), and ER+ BRCA-like patients (g). p values have been obtained by log-rank test
Discussion
In this study, we developed a novel digitalMLPA assay to assess BRCA1-like and BRCA2-like copy number profiles within one assay. We showed an accuracy of 91% for the BRCA1-like classification and 82% for the BRCA2-like classification using the array-based BRCA-like classification as the gold standard. The digitalMLPA correctly classified most BRCA1-mutated, BRCA1-methylated, and BRCA2-mutated tumors, with respectively 88%, 96%, and 93% correct classification. The digitalMLPA had treatment predictive power: the test could identify both TN and luminal-type tumors with remarkable good responses on high-dose chemotherapy.
The BRCA1-like and BRCA2-like classifiers were originally developed to identify BRCA1 and BRCA2 deficiency and could support decision-making in genetic counseling. We previously showed that the classifiers could identify tumors with other mechanisms of BRCA1 or BRCA2 inactivation such as promoter methylation [1, 4, 5]. In addition to the role of the test in clinical genetics decision-making, the BRCA1-like and BRCA2-like classifiers have shown to have treatment predictive value [6,7,8, 21]. As BRCA1 and BRCA2 play a role in the process of DNA double-strand break repair, we believe that a copy number profile resembling BRCA1- or BRCA2-mutated tumors could indicate the specific chromosomal instability associated with defective DNA repair, i.e., HRD. A BRCA1-like or BRCA2-like profile could therefore be considered as a readout for HRD. In addition to our BRCA1/2-like tests, other (commercially) available HRD tests are on the market for breast and ovarian cancer. These are either based on genomics scars [23], mutational signatures [24], or point mutations in homologous recombination repair genes [25]. In ovarian cancer, the highest benefit of niraparib was seen in patients with HRD tumors [26, 27]. In breast cancer, trials in the (neo) adjuvant setting showed higher responses on platinum salts or alkylating chemotherapy for patients with HRD tumors vs. patients with non-HRD tumors [28, 29]. However, the lack of a control arm was a flaw in these studies [30, 31]. Of note, a randomized trial in the neoadjuvant setting comparing paclitaxel single agent with cisplatin single agent showed higher responses for HRD tumors in both arms, indicating that this test is not predictive for selective benefit of DNA-damaging agents [32]. Furthermore, in the advanced breast cancer TNT trial, no association was observed between HRD status and response rate [33]. In this latter trial, an additional explanation for genomic scars having less predictive power is that most patients were already pre-treated by other chemotherapy agents. Resistance might already have been occurred in these patients [34]. Although patients with BRCAness tumors might have benefit from conventional types of DNA-damaging therapy, like FEC, we believe that BRCAness patients will have additional benefit of high-dose DNA-damaging therapies, like the regimen applied in the Dutch high dose trial, as these are in particular highly vulnerable for this kind of tumors [6, 7]. Results of in vitro and in vivo studies point in the same direction [35].
Our BRCA1-like and BRCA2-like classifiers are breast cancer subtype specific. The BRCA1-like profile is mainly observed in TN tumors, while the BRCA2-like profile is mainly observed in luminal-type cancer. As TN and luminal-type breast tumors have distinct molecular profiles, developing two distinct classifiers, based on each subtype-specific genomic profiles, resulted in highly specific classifiers. The BRCA1-like classifier has been validated in three different studies as a predictor of high-dose chemotherapy benefit [6,7,8]. Currently, a phase III trial is running in stage 3 triple-negative breast cancer, applying the BRCA1-like MLPA test (NCT02810743). For luminal-type breast cancer, the digitalMLPA is an easy-to-use, robust assay, to assess the BRCA2-like phenotype. With such a test, we could easily test retrospective series and generate data on lumina-type breast cancer, which is currently lacking in the literature due to the focus on TN tumors. There is evidence from the literature that a small group of luminal tumors may also have benefit from HRD-inducing chemotherapy. Manie et al. showed that HRD testing using genomic instability assays in luminal-type tumors resulted in a threefold increase in the identification of HRD tumors than by HRD testing solely based on BRCA mutation profiling [36]. Our BRCA2-like test has the potential to identify a subpopulation of luminal-type tumors that might benefit from high-dose DSB-inducing chemotherapy. The fact that the BRCA1-like and BRCA2-like tests are combined in one assay facilitates routine testing in a diagnostic lab.
Advantages of digitalMLPA, as compared to array- or NGS-based BRCA-like testing, include a high dynamic range for copy number detection, a robust assay, the good performance on degraded DNA, and the low input amount (≥ 20 ng) of DNA [17]. Especially, in patient samples with limited tissue material, such as core biopsies, a low input amount is a prerequisite. With digitalMLPA, a large number of genomic loci of interest can be analyzed for copy number alterations in a single reaction with low hands-on time, and results are available within 36 h. Because of the targeted approach, data analysis and result interpretation are easier than with array-based analysis and can be performed in any routine diagnostic setting.
A limitation of the current study is that we could not profile the whole randomized trial population, due to missing tissue blocks or a limited amount of DNA available. The series profiled was enriched for triple-negative and BRCA1-like tumors, as these were the samples with enough DNA left for successful digital MLPA analysis (Table 1 (D)). As we know that BRCA1-like tumors have a very low HR (ranging from 0.05 to 0.19 [6, 7]) for high-dose chemotherapy, the overall HR (0.38) in our study population is lower than in the total HER2-negative trial population (HR 0.68) [37]. However, even in this selected subgroup, we showed a significant interaction between treatment benefit and digitalMLPA BRCAness status, indicating the strong predictive power of our digitalMLPA assay. Independent validation of a different cohort will be necessary; we are currently exploring possibilities. Another suggestion for improvement would be to train a specific classifier to predict treatment benefit. However, the BRCA-like classifiers are based on the biology of BRCA tumors and thus are not merely “data-driven,” which could be seen as a strength. The third limitation is the slightly worse performance of the BRCA2-like classifier. We noticed before that BRCA2-like tumors are more alike sporadic luminal-type tumors. It appeared more difficult to find specific discriminatory regions [1, 38].
Conclusions
DigitalMLPA BRCAness analysis shows results comparable to previously published array-based BRCA-like testing. The digitalMLPA is a fast and reliable method to detect copy number changes in relevant regions for BRCA1- and BRCA2-like classification in one experiment in breast cancer samples. We showed that the digitalMLPA assay had treatment predictive power, in both TN and luminal-type tumors. Independent validation of patient cohorts treated with DNA DSB-inducing chemotherapy is warranted.
Availability of data and materials
The dataset supporting the conclusions of this article is available upon reasonable request. Requests should be made to Dr. E.H. Lips (e.lips@nki.nl).
Abbreviations
- BC:
-
Breast cancer
- BAC:
-
Bacterial artificial chromosome
- CGH:
-
Comparative genomic hybridization
- FEC:
-
5-Fluorouracil-epirubicin-cyclophosphamide
- HD-CTC:
-
High-dose cylclophosphamide-thiotepa-carboplatin
- HR:
-
Hazard ratio
- HRD:
-
Homologous recombination deficiency
- MLPA:
-
Multiplex ligation-dependent probe amplification
- TN:
-
Triple-negative
- VUS:
-
Variants of unknown significance
References
Joosse SA, Brandwijk KI, Devilee P, Wesseling J, Hogervorst FB, Verhoef S, et al. Prediction of BRCA2-association in hereditary breast carcinomas using array-CGH. Breast Cancer Res Treat. 2012;132(2):379–89.
Joosse SA, van Beers EH, Tielen IH, Horlings H, Peterse JL, Hoogerbrugge N, et al. Prediction of BRCA1-association in hereditary non-BRCA1/2 breast carcinomas with array-CGH. Breast Cancer Res Treat. 2009;116(3):479–89.
Wessels LF, van Welsem T, Hart AA, Van’t Veer LJ, Reinders MJ, Nederlof PM. Molecular classification of breast carcinomas by comparative genomic hybridization: a specific somatic genetic profile for BRCA1 tumors. Cancer Res. 2002;62(23):7110–7.
Joosse SA, Brandwijk KI, Mulder L, Wesseling J, Hannemann J, Nederlof PM. Genomic signature of BRCA1 deficiency in sporadic basal-like breast tumors. Genes Chromosomes Cancer. 2011;50(2):71–81.
Lips EH, Mulder L, Oonk A, van der Kolk LE, Hogervorst FB, Imholz AL, et al. Triple-negative breast cancer: BRCAness and concordance of clinical features with BRCA1-mutation carriers. Br J Cancer. 2013;108(10):2172–7.
Schouten PC, Gluz O, Harbeck N, Mohrmann S, Diallo-Danebrock R, Pelz E, et al. BRCA1-like profile predicts benefit of tandem high dose epirubicin-cyclophospamide-thiotepa in high risk breast cancer patients randomized in the WSG-AM01 trial. Int J Cancer. 2016;139(4):882–9.
Schouten PC, Marme F, Aulmann S, Sinn HP, van Essen HF, Ylstra B, et al. Breast cancers with a BRCA1-like DNA copy number profile recur less often than expected after high-dose alkylating chemotherapy. Clin Cancer Res. 2015;21(4):763–70.
Vollebergh MA, Lips EH, Nederlof PM, Wessels LF, Schmidt MK, van Beers EH, et al. An aCGH classifier derived from BRCA1-mutated breast cancer and benefit of high-dose platinum-based chemotherapy in HER2-negative breast cancer patients. Ann Oncol. 2011;22(7):1561–70.
Vollebergh MA, Lips EH, Nederlof PM, Wessels LF, Wesseling J, Vd Vijver MJ, et al. Genomic patterns resembling BRCA1- and BRCA2-mutated breast cancers predict benefit of intensified carboplatin-based chemotherapy. Breast Cancer Res. 2014;16(3):R47.
Severson TM, Wolf DM, Yau C, Peeters J, Wehkam D, Schouten PC, et al. The BRCA1ness signature is associated significantly with response to PARP inhibitor treatment versus control in the I-SPY 2 randomized neoadjuvant setting. Breast Cancer Res. 2017;19(1):99.
Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30(12):e57.
Apostolou P, Fostira F, Kouroussis C, Kalfakakou D, Delimitsou A, Agelaki S, et al. BRCA1 and BRCA2 germline testing in Cretan isolates reveals novel and strong founder effects. Int J Cancer. 2020;147(5):1334–42.
Hogervorst FB, Nederlof PM, Gille JJ, McElgunn CJ, Grippeling M, Pruntel R, et al. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res. 2003;63(7):1449–53.
Homig-Holzel C, Savola S. Multiplex ligation-dependent probe amplification (MLPA) in tumor diagnostics and prognostics. Diagn Mol Pathol. 2012;21(4):189–206.
Schwab CJ, Jones LR, Morrison H, Ryan SL, Yigittop H, Schouten JP, et al. Evaluation of multiplex ligation-dependent probe amplification as a method for the detection of copy number abnormalities in B-cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2010;49(12):1104–13.
Lips EH, Laddach N, Savola SP, Vollebergh MA, Oonk AM, Imholz AL, et al. Quantitative copy number analysis by multiplex ligation-dependent probe amplification (MLPA) of BRCA1-associated breast cancer regions identifies BRCAness. Breast Cancer Res. 2011;13(5):R107.
Benard-Slagter A, Zondervan I, de Groot K, Ghazavi F, Sarhadi V, Van Vlierberghe P, et al. Digital multiplex ligation-dependent probe amplification for detection of key copy number alterations in T- and B-cell lymphoblastic leukemia. J Mol Diagn. 2017;19(5):659–72.
Rodenhuis S, Bontenbal M, Beex LV, Wagstaff J, Richel DJ, Nooij MA, et al. High-dose chemotherapy with hematopoietic stem-cell rescue for high-risk breast cancer. N Engl J Med. 2003;349(1):7–16.
Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99(10):6567–72.
Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS Med. 2012;9(5):e1001216.
Vollebergh MA, Lips EH, Nederlof PM, Wessels LF, Wesseling J, Vd Vijver MJ, et al. Genomic patterns resembling BRCA2-mutated breast cancers predict benefit of intensified carboplatin-based chemotherapy. Breast Cancer Res. 2014;16(3):R47.
Lips EH, Mulder L, Hannemann J, Laddach N, Vrancken Peeters MT, van de Vijver MJ, et al. Indicators of homologous recombination deficiency in breast cancer and association with response to neoadjuvant chemotherapy. Ann Oncol. 2011;22(4):870–6.
Timms KM, Abkevich V, Hughes E, Neff C, Reid J, Morris B, et al. Association of BRCA1/2 defects with genomic scores predictive of DNA damage repair deficiency among breast cancer subtypes. Breast Cancer Res. 2014;16(6):475.
Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, Zou X, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23(4):517–25.
Hodgson DR, Dougherty BA, Lai Z, Fielding A, Grinsted L, Spencer S, et al. Candidate biomarkers of PARP inhibitor sensitivity in ovarian cancer beyond the BRCA genes. Br J Cancer. 2018;119(11):1401–9.
Mirza MR, Monk BJ, Herrstedt J, Oza AM, Mahner S, Redondo A, et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med. 2016;375(22):2154–64.
Swisher EM, Lin KK, Oza AM, Scott CL, Giordano H, Sun J, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18(1):75–87.
Sharma P, Barlow WE, Godwin AK, Pathak H, Isakova K, Williams D, et al. Impact of homologous recombination deficiency biomarkers on outcomes in patients with triple-negative breast cancer treated with adjuvant doxorubicin and cyclophosphamide (SWOG S9313). Ann Oncol. 2018;29(3):654–60.
Telli ML, Hellyer J, Audeh W, Jensen KC, Bose S, Timms KM, et al. Homologous recombination deficiency (HRD) status predicts response to standard neoadjuvant chemotherapy in patients with triple-negative or BRCA1/2 mutation-associated breast cancer. Breast Cancer Res Treat. 2018;168(3):625–30.
Schouten PC, Linn SC. Challenges in the use of DNA repair deficiency as a biomarker in breast cancer. J Clin Oncol. 2015;33(17):1867–9.
Miquel-Cases A, Schouten PC, Steuten LM, Retel VP, Linn SC, van Harten WH. (Very) early technology assessment and translation of predictive biomarkers in breast cancer. Cancer Treat Rev. 2017;52:117–27.
Mayer EL, Abramson VG, Jankowitz RC, Falkson CI, Marcom PK, Traina TA, et al. TBCRC 030: a randomized phase II study of preoperative cisplatin versus paclitaxel in TNBC—evaluating the homologous recombination deficiency (HRD) biomarker. J Clin Oncol. 2019;37(15_suppl):507.
Tutt A, Tovey H, Cheang MCU, Kernaghan S, Kilburn L, Gazinska P, et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat Med. 2018;24(5):628–37.
Pellegrino B, Mateo J, Serra V, Balmana J. Controversies in oncology: are genomic tests quantifying homologous recombination repair deficiency (HRD) useful for treatment decision making? ESMO Open. 2019;4(2):e000480.
Pajic M, Blatter S, Guyader C, Gonggrijp M, Kersbergen A, Kucukosmanoglu A, et al. Selected alkylating agents can overcome drug tolerance of G0-like tumor cells and eradicate BRCA1-deficient mammary tumors in mice. Clin Cancer Res. 2017;23(22):7020–33.
Manie E, Popova T, Battistella A, Tarabeux J, Caux-Moncoutier V, Golmard L, et al. Genomic hallmarks of homologous recombination deficiency in invasive breast carcinomas. Int J Cancer. 2016;138(4):891–900.
Rodenhuis S, Bontenbal M, van Hoesel QG, Smit WM, Nooij MA, Voest EE, et al. Efficacy of high-dose alkylating chemotherapy in HER2/neu-negative breast cancer. Ann Oncol. 2006;17(4):588–96.
Schouten PC, van Dyk E, Braaf LM, Mulder L, Lips EH, de Ronde JJ, et al. Platform comparisons for identification of breast cancers with a BRCA-like copy number profile. Breast Cancer Res Treat. 2013;139(2):317–27.
Acknowledgements
We would like to acknowledge the NKI-AVL Core Facility Molecular Pathology & Biobanking (CFMPB) for supplying NKI-AVL Biobank material and laboratory support.
Funding
This research was supported by the Center for Translational Molecular Medicine (project Breast CARE (030–104)) and co-funded by the Cancer Research UK and the Dutch Cancer Society (grant number C38317/A24043).
Author information
Authors and Affiliations
Contributions
EHL, SS, and PMN designed and supervised the study. ABS, MO, and CES generated the data. ABS, MO, and EL analyzed the data. EHL wrote the manuscript with contributions from JW, FBLH, SCL, SS, and PMN. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Institutional Review Boards of the NKI-AVL. All trial participants provided written informed consent prior to study enrollment.
Consent for publication
Not applicable
Competing interests
Anne Benard-Slagter and Suvi Savola are employees of MRC Holland, which supplies the digital MLPA probe mixes. All other authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure 1.
Consort diagram for training and validation the BRCA1 –like (A) and BRCA2-like (B) digitalMLPA classifiers. The Number of correctly classified samples by digitalMLPA is indicated as well as the number of BRCA1 mutated, BRCA1 methylated and BRCA2 mutated samples. Figure 2. Determination of cut-off for BRCA1-like (A) and BRCA2-like (B) classification in training set. Cut-off value for BRCA1-like was set 0.14 and cut-off value for BRCA1-like was set at 0.21. Table 2. Association of BRCA-like test result and clinical characteristics.
Additional file 2:
Proble list digitalMLPA BRCAness probemix.
Additional file 3:.
Description of the study according to REMARK criteria.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lips, E.H., Benard-Slagter, A., Opdam, M. et al. BRCAness digitalMLPA profiling predicts benefit of intensified platinum-based chemotherapy in triple-negative and luminal-type breast cancer. Breast Cancer Res 22, 79 (2020). https://doi.org/10.1186/s13058-020-01313-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13058-020-01313-7