Abstract
Objective
Our aim is to analyzed the expression pattern of sirtuin(SIRT) superfamily and evaluated their prognostic values in serous ovarian cancer patients.
Methods
We first analyzed the differential expression of SIRT members among fallopian tube epithelium (FTE), primary serous ovarian cancers/tubal cancers (PSOCs/PSTCs), and omental metastases using GSE10971 and GSE30587 datasets. The prognostic values of SIRT members were evaluated using TCGA and GSE9891 dataset.
Results
SIRT3 and SIRT5 expression were significantly decreased and increased in PSOCs/PSTCs compared with that in normal counterparts, respectively. SIRT6 and SIRT7 were overexpressed in ometal metastases compared with corresponding primary counterparts. With respect to recurrence free survival, however, SIRT7 overexpression was correlated with better prognosis. A similar trend was observed by multivariable analysis. Regarding overall survival (OS), increased expression of SIRT3, SIRT5, and SIRT7 were associated with better survival by univariable analysis. Subsequent multivariable analysis showed that SIRT3 remained an independent favorable prognostic factor for OS. The SIRT3-related nomogram illustrated age at initial diagnosis as sharing the largest contribution to OS, followed by SIRT3 expression and FIGO stage. The C-index for OS prediction was 0.65 (95%CI, 0.61–0.69) in training cohort (TCGA dataset) and 0.65 (95%CI, 0.59–0.71) in validation cohort (GSE9891 dataset), respectively. The calibration plots showed optimal agreement between the prediction by SIRT3-related nomogram and actual observation for 1-, 3-, and 5-year OS probability.
Conclusion
In conclusion, SIRT3 was an independent favorable prognostic factor for OS in serous ovarian cancer, and added prognostic value to the traditional clinicopathological factors used to evaluate patients’ prognosis.
Similar content being viewed by others
Introduction
Ovarian cancer, characterized by late-stage presentation and metastatic bulky disease burden, is the leading cause of gynecologic cancer death [1]. After initial cytoreductive surgery combined with platinum based chemotherapy, most of the patients who achieving complete remission still face a high risk of recurrence and death, and their 5 year survival rate is ranged from 20 to 30%. Thus, it is important to identify new biomarkers to predict ovarian cancer prognosis, which will facilitate timely inclusion into clinical trials or personalized treatment strategies.
Sirtuins (SIRTs), initially identified as adenosine diphosphate (ADP) ribosyltransferases in bacterium Salmonella typhimurium, are highly conserved histone deacetylases (HDAC) dependent on nicotine adenine dinucleotide (NAD) and/or mono ADP-ribosyltransferases [2, 3]. Currently, a superfamily of seven SIRTs (SIRT1–7) has been identified in mammalian, and each has its own unique characteristics and functions. SIRT1, SIRT6 and SIRT7 are predominantly localized in the nucleus; SIRT3, SIRT4 and SIRT5 are mitochondrial proteins and primarily reside within the mitochondria; and SIRT2 is localized mainly in the cytoplasm [3]. In certain conditions, SIRT1 and SIRT2 are able to shuttle between the nucleus and cytoplasm.
SIRTs received increasing attention due to their divergent role in cancer biology. The role of SIRTs is of extreme complexity, with both tumor promoter and tumor suppressor functions dependent on cell contexts. In ovarian cancer, SIRTs has been implicated in the regulation of various cellular processes, including but not limited to cell growth, apoptosis, invasion, and metastasis [4,5,6,7,8,9,10]. However, the role of SIRTs in predicting progression and prognosis of ovarian cancer patients are still largely elusive.
In this study, we analyzed the expression pattern of the SIRT superfamily (SIRT1-SIRT7) and evaluated their prognostic values in serous ovarian cancer patients by using the high-throughput expression data deposited in Gene Expression Omnibus(GEO) database and The Cancer Genome Altas (TCGA) database.
Materials and methods
Databsets used in present study
The publicly available gene expression data used in our study is described in TCGA and GEO database(GSE10971, GSE30587 and GSE9891 datasets). GSE10971 dataset was used to determine the differential expression of SIRT members between fallopian tube epithelium (FTE) (n = 24) and primary serous ovarian cancer/tubal cancers (PSOCs/PSTCs) (n = 13). GSE30587 dataset was used to determine the differential expression of SIRT members between paired PSOC tissues(n = 9) and their omental metastasis counterparts(n = 9). TCGA dataset was used to explore the association of SIRT members’ expression with clinicopathological factors and to develop SIRT-related nomograms to predict recurrence free survival (RFS) and overall survival (OS). GSE9891 dataset was used to validate the nomograms developed from TCGA dataset. For both TCGA and GSE9891 dataset, only serous ovarian cancer patients with adequate information (including age at initial diagnosis, histological grade, FIGO stage, debulking status, days to recurrence, recurrence status, days to death, and vital status) were included in our analysis. Collectively, there were 462 patients in TCGA dataset and 226 patients in GSE9891 dataset included in our final analysis. TCGA dataset and GSE9891 dataset were obtained from the “curatedOvarianData” Bioconductor package (version 2.12 for R 3.0.3).
Statistical analysis
Continuous data between two groups were compared using two independent samples t test. Categorical data were compared using chi-square or Fisher’s exact test where appropriate. Both univariable and multivariable analyses were performed in survival analysis. The statistical analyses mentioned above were analyzed using IBM SPSS Statistics (version 22.0). P values < 0.05 were considered significant (P < 0.05).
We developed the SIRT-related nomogram using rms package. Harrell’s concordance index (c-index) was calculated through a bootstrap method with 1000 resamples and used to evaluate the predictive efficiency of nomogram. Calibration curves were used to compare the prediction by nomogram and actual observation graphically. The statistical analyses mentioned above were analyzed using R (version 3.5.2). P values < 0.05 were considered significant (P < 0.05).
Results
The expression pattern of SIRT family members in serous ovarian cancer
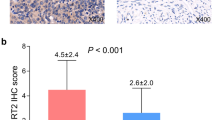
We first determined the differential expression of SIRT family members between FTE and PSOCs/ PSTCs. We found that SIRT3 expression was significantly decreased in PSOCs/PSTCs compared with that in FTE (Fig. 1C), whereas SIRT5 was obviously increased in PSOCs/PSTCs compared with that in FTE (Fig. 1E). The expression of the rest members of SIRT family was comparable between FTE and PSOCs/PSTCs (Fig. 1A, B, D, F, G). Then, GSE30587 dataset was used to determine the differential expression of SIRT members between paired PSOC tissues and their omental metastasis counterparts. It was found that SIRT6 and SIRT7 were overexpressed in ometal metastases compared with corresponding primary counterparts (Fig. 2F, G). The expression of the rest members of SIRT family was comparable between PSOCs and correponding omental metastases (Fig. 2A-E).
Differential Expression of SIRT members between FTE and PSOCs/PSTCs. (a-g) There was no significant difference in the expression of SIRT1 (a), SIRT2 (b), SIRT4 (d), SIRT6 (f), and SIRT7 (g) between FTE and PSOCs/PSTCs. SIRT3 (c) and SIRT5 (e) expression were significantly decreased and increased in PSOCs/PSTCs compared with that in normal counterparts, respectively
Differential Expression of SIRT members between PSOCs and paired omental metastases. (a-g) There was no significant difference in the expression of SIRT1 (a), SIRT(b), SIRT3 (c), SIRT4 (d), and SIRT5 (e) between PSOCs and paired omental metastases. SIRT6 (f) and SIRT7 (g) were significantly overexpressed in ometal metastases compared with corresponding primary counterparts
The prognostic value of SIRT members in serous ovarian cancer patients
Next, we evaluated the prognostic value of each SIRT member in predicting RFS and OS in serous ovarian cancer patients using the TCGA dataset. The results of the univariable and multivariable analysis are listed in Table 1. With respect to RFS, univariable analysis showed that increased FIGO stage decreased SIRT7 expression were correlated with worse prognosis. A similar trend was observed by multivariable analysis (Table 1). Regarding OS, younger age at initial diagnosis was associated with better prognosis. Increased FIGO stage, high grade, and suboptimal debulking had a negative impact on survival. With respect to SIRT members, increased expression of SIRT3, SIRT5, and SIRT7 were all correlated with better survival. All factors reaching statistical significance in the univariable analysis were entered into the multivariable Cox regression analysis. Age at initial diagnosis, FIGO stage, and SIRT3 expression remained independent prognostic factors for OS (Table 1).
Development and validation of SIRT3-related nomogram predictive of OS
To precisely predict the OS of serous ovarian cancer patients, we tried to develop a prognostic nomogram. Besides independent prognostic factors indicated by multivariable analysis, another two established prognostic factors, including histological grade and debulking status, were also incorporated to develop the prognostic nomogram (Fig. 3). The nomogram illustrated age at initial diagnosis as sharing the largest contribution to OS, followed by SIRT3 expression and FIGO stage. Histological grade and debulking status had a moderate impact on the OS. The C-index for OS prediction was 0.65 (95%CI, 0.61–0.69). The calibration plots showed optimal agreement between the prediction by nomogram and actual observation for 1-, 3-, and 5-year OS probability in TCGA dataset (Fig. 4A-C).
Calibration plots of the SIRT3-related nomogram to predict OS. (a-c) Calibration curves of the SIRT3-related nomogram to predict OS at 1 year (a), 3 years (b), and 5 years (c) in TCGA dataset. (d-f) Calibration curves of the SIRT3-related nomogram to predict OS at 1 year (d), 3 years (e), and 5 years (f) in GSE9891 dataset
GSE9891 dataset was used as a validation cohort. The C-index for OS prediction was 0.65 (95%CI, 0.59–0.71) in GSE9891 dataset. The calibration plots also showed optimal agreement between the prediction by nomogram and actual observation for 1-, 3-, and 5-year OS probability (Fig. 4D-F).
Discussion
The expression and prognostic value of some SIRT members have been explored in ovarian cancer. However, the small sample sizes in previous studies have yielded inconsistent results, raising doubts in generalizing their findings. In this study, we explored the expression and clinical significance of all the SIRT members in serous ovarian cancer with a larger sample size by using high throughput mRNA profile datasets from both TCGA database and GEO database.
Among the SIRT family, SIRT1 is the most extensively studied member. However, its role in ovarian cancer are still uncertain. Ray et al. indicated that increased expression and activity of SIRT1 impaired the invasion of ovarian cancer cells by targeting the EMT inducer ZEB1 [7]. Downregulation of SIRT1 also led to decreased migration and angiogenesis of ovarian cancer cells by repressing HMGB1 [5], and was associated with decreased OS in serous ovarian cancer. However, in contrast, other studies showed that SIRT1 overexpression or activation resulted in acquired chemoresistance and increased invasiveness of ovarian cancer cells [8, 11], and was associated with a poorer prognosis in serous ovarian cancer patients [10]. Our data indicated that SIRT1 expression appeared to be downregulated in omental metastasis compared with primary couterparts, and was associated with extended RFS in TCGA dataset though without reaching statistical significance.
SIRT3 is a well known and the best characterized mitochondrial sirtuin (mtSIRT) and capable of regulating multiple major aspect of mitochondrial biology [12]. The role of SIRT3 in cancer biology is still in debate [13, 14]. The expression of SIRT3 was aberrantly decreased in head and neck squamous cell carcinoma (HNSCC), gastric cancer, and mantle cell lymphoma [15,16,17]. However, in certain cancers, SIRT3 were significantly upregulated [18,19,20]. Whereas in breast cancer, contradictory results have been observed [21,22,23]. In ovarian cancer, SIRT3 expression was significantly downregulated in cancer tissues as demonstrated by both our study and a previous study [24]. Moreover, SIRT3 was also downregulated in the metastatic tissues and highly metastatic cell line of ovarian cancer [9]. Upregulation of SIRT3 impaired the viability, migration, and invasion of ovarian cancer cells [4, 9, 24]. Similar to the dual roles of SIRT3 in cancer biology, the prognostic value of SIRT3 in cancer is also dichotomous. Some studies indicated that decreased expression of SIRT3 was associated with poor prognosis in pancreatic, gastric, and hepatocellular cancer [25, 26]. In certain cancer, in contrast, increased expression of SIRT3 was correlated with a worse clinical outcome [27]. However, the prognostic value of SIRT3 in ovarian cancer has not yet been evaluated. In present study, we found that SIRT3 is an independent predictor of better OS in serous ovarian cancer. Moreover, our SIRT3-related nomogram indicated that SIRT3 shared a larger contribution to OS, compared with FIGO stage, histological grade, and debulking status.
In contrast to SIRT3, the roles of the other two mtSIRTs, SIRT4 and SIRT5, are not yet explored in ovarian cancer, though their dual puzzling functions as tumor suppressors and tumor promoters have been reported in other cancers [28,29,30]. In ovarian cancer, It seemed that increased expression of SIRT4 was associated with a better prognosis by our univariable analysis, though without reaching statistical significance. Whereas SIRT5 appeared to a risk factor for worse prognosis. Their functions in ovarian cancer remains to be explored in future studies.
The roles of SIRT 6 and SIRT7 in cancer were also cellular context dependent. Some studies indicated that SIRT6 could inhibit ovarian cancer cell growth and was a favorable prognostic factor [31, 32]. However, Bae et al. indicated SIRT6 promoted invasion and migration of ovarian cancer cells, without affecting cell proliferation [6]. Moreover, SIRT6 was associated with higher FIGO stage, higher histological grade, platinum-resistance, and shorter OS [6]. Consistently, our work revealed that SIRT6 was elevated in omental metastases compared with paired PSOCs. However, survival analysis failed to demonstrate a prognostic value of SIRT6 in serous ovarian cancer. With respect to SIRT7, it was reported that SIRT7 was unregulated in ovarian cancer cells compared with normal couterparts, and could promote cell growth and colony formation, and reduce cell apoptosis [33]. Our data suggested that SIRT7 was upregulated in omental metastases compared with paired PSOCs. However, our univariable analysis revealed that SIRT7 overexpression was associated with both better RFS and OS. It seemed that SIRT6 and SIRT7 displayed both oncogenic and tumor-suppressive properties in ovarian cancer.
In conclusion, SIRT3 was an independent favorable prognostic factor for OS in serous ovarian cancer, and added prognostic value to the traditional clinicopathological factors used to evaluate patients’ prognosis.
References
Coleman RL, Monk BJ, Sood AK, Herzog TJ. Latest research and treatment of advanced-stage epithelial ovarian cancer. Nat Rev Clin Oncol. 2013;10:211–24.
North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224.
Vassilopoulos A, Fritz KS, Petersen DR, Gius D. The human sirtuin family: evolutionary divergences and functions. Hum Genomics. 2011;5:485–96.
Wu Y, Gao WN, Xue YN, Zhang LC, Zhang JJ, Lu SY, Yan XY, Yu HM, Su J, Sun LK. SIRT3 aggravates metformin-induced energy stress and apoptosis in ovarian cancer cells. Exp Cell Res. 2018;367:137–49.
Jiang W, Jiang P, Yang R, Liu DF. Functional role of SIRT1-induced HMGB1 expression and acetylation in migration, invasion and angiogenesis of ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22:4431–9.
Bae JS, Noh SJ, Kim KM, Park SH, Hussein UK, Park HS, Park BH, Ha SH, Lee H, Chung MJ, Moon WS, Cho DH, Jang KY. SIRT6 is involved in the progression of ovarian carcinomas via beta-catenin-mediated epithelial to mesenchymal transition. Front Oncol. 2018;8:538.
Ray U, Roy SS, Chowdhury SR. Lysophosphatidic acid promotes epithelial to mesenchymal transition in ovarian Cancer cells by repressing SIRT1. Cell Physiol Biochem. 2017;41:795–805.
Mvunta DH, Miyamoto T, Asaka R, Yamada Y, Ando H, Higuchi S, Ida K, Kashima H, Shiozawa T. SIRT1 regulates the Chemoresistance and invasiveness of ovarian carcinoma cells. Transl Oncol. 2017;10:621–31.
Dong XC, Jing LM, Wang WX, Gao YX. Down-regulation of SIRT3 promotes ovarian carcinoma metastasis. Biochem Biophys Res Commun. 2016;475:245–50.
Shuang T, Wang M, Zhou Y, Shi C. Over-expression of Sirt1 contributes to chemoresistance and indicates poor prognosis in serous epithelial ovarian cancer (EOC). Med Oncol. 2015;32:260.
Pinton G, Nilsson S, Moro L. Targeting estrogen receptor beta (ERbeta) for treatment of ovarian cancer: importance of KDM6B and SIRT1 for ERbeta expression and functionality. Oncogenesis. 2018;7:15.
Ansari A, Rahman MS, Saha SK, Saikot FK, Deep A, Kim KH. Function of the SIRT3 mitochondrial deacetylase in cellular physiology, cancer, and neurodegenerative disease. Aging Cell. 2017;16:4–16.
Torrens-Mas M, Oliver J, Roca P, Sastre-Serra J. SIRT3: oncogene and tumor suppressor in Cancer. Cancers (Basel). 2017;9.
George J, Ahmad N. Mitochondrial Sirtuins in Cancer: emerging roles and therapeutic potential. Cancer Res. 2016;76:2500–6.
Lai CC, Lin PM, Lin SF, Hsu CH, Lin HC, Hu ML, Hsu CM, Yang MY. Altered expression of SIRT gene family in head and neck squamous cell carcinoma. Tumour Biol. 2013;34:1847–54.
Yang B, Fu X, Shao L, Ding Y, Zeng D. Aberrant expression of SIRT3 is conversely correlated with the progression and prognosis of human gastric cancer. Biochem Biophys Res Commun. 2014;443:156–60.
Yu W, Denu RA, Krautkramer KA, Grindle KM, Yang DT, Asimakopoulos F, Hematti P, Denu JM. Loss of SIRT3 provides growth advantage for B cell malignancies. J Biol Chem. 2016;291:3268–79.
Yan SM, Han X, Han PJ, Chen HM, Huang LY, Li Y. SIRT3 is a novel prognostic biomarker for esophageal squamous cell carcinoma. Med Oncol. 2014;31:103.
Alhazzazi TY, Kamarajan P, Joo N, Huang JY, Verdin E, D'Silva NJ, Kapila YL. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer. 2011;117:1670–8.
Shackelford R, Hirsh S, Henry K, Abdel-Mageed A, Kandil E, Coppola D. Nicotinamide phosphoribosyltransferase and SIRT3 expression are increased in well-differentiated thyroid carcinomas. Anticancer Res. 2013;33:3047–52.
Ashraf N, Zino S, Macintyre A, Kingsmore D, Payne AP, George WD, Shiels PG. Altered sirtuin expression is associated with node-positive breast cancer. Br J Cancer. 2006;95:1056–61.
Desouki MM, Doubinskaia I, Gius D, Abdulkadir SA. Decreased mitochondrial SIRT3 expression is a potential molecular biomarker associated with poor outcome in breast cancer. Hum Pathol. 2014;45:1071–7.
Igci M, Kalender ME, Borazan E, Bozgeyik I, Bayraktar R, Bozgeyik E, Camci C, Arslan A. High-throughput screening of Sirtuin family of genes in breast cancer. Gene. 2016;586:123–8.
Yang Y, Cao Y, Chen L, Liu F, Qi Z, Cheng X, Wang Z. Cryptotanshinone suppresses cell proliferation and glucose metabolism via STAT3/SIRT3 signaling pathway in ovarian cancer cells. Cancer Med. 2018;7:4610–8.
Yu FY, Xu Q, Wu DD, Lau AT, Xu YM. The prognostic and Clinicopathological roles of Sirtuin-3 in various cancers. PLoS One. 2016;11:e0159801.
Huang S, Chen X, Zheng J, Huang Y, Song L, Yin Y, Xiong J. Low SIRT3 expression contributes to tumor progression, development and poor prognosis in human pancreatic carcinoma. Pathol Res Pract. 2017;213:1419–23.
Torrens-Mas M, Hernandez-Lopez R, Oliver J, Roca P, Sastre-Serra J. Sirtuin 3 silencing improves oxaliplatin efficacy through acetylation of MnSOD in colon cancer. J Cell Physiol. 2018;233:6067–76.
Huang G, Zhu G. Sirtuin-4 (SIRT4), a therapeutic target with oncogenic and tumor-suppressive activity in cancer. Onco Targets Ther. 2018;11:3395–400.
Chen XF, Tian MX, Sun RQ, Zhang ML, Zhou LS, Jin L, Chen LL, Zhou WJ, Duan KL, Chen YJ, Gao C, Cheng ZL, Wang F, Zhang JY, Sun YP, Yu HX, Zhao YZ, Yang Y, Liu WR, Shi YH, Xiong Y, Guan KL, Ye D. SIRT5 inhibits peroxisomal ACOX1 to prevent oxidative damage and is downregulated in liver cancer. EMBO Rep. 2018;19.
Shi L, Yan H, An S, Shen M, Jia W, Zhang R, Zhao L, Huang G, Liu J. SIRT5-mediated deacetylation of LDHB promotes autophagy and tumorigenesis in colorectal cancer. Mol Oncol. 2019;13:358–75.
Zhang G, Liu Z, Qin S, Li K. Decreased expression of SIRT6 promotes tumor cell growth correlates closely with poor prognosis of ovarian cancer. Eur J Gynaecol Oncol. 2015;36:629–32.
Zhang J, Yin XJ, Xu CJ, Ning YX, Chen M, Zhang H, Chen SF, Yao LQ. The histone deacetylase SIRT6 inhibits ovarian cancer cell proliferation via down-regulation of notch 3 expression. Eur Rev Med Pharmacol Sci. 2015;19:818–24.
Wang HL, Lu RQ, Xie SH, Zheng H, Wen XM, Gao X, Guo L. SIRT7 exhibits oncogenic potential in human ovarian Cancer cells. Asian Pac J Cancer Prev. 2015;16:3573–7.
Acknowledgements
We thank all the investigators who shared their high throughput expression data.
Funding
This study was supported by funding from the Shanghai Sailing Program (16YF1401100), National Natural Science Foundation of China (No.81802596), and Natural Science Foundation of Shanghai (17ZR1403500).
Availability of data and materials
All data used in present study are all available in TCGA and GEO databases.
Author information
Authors and Affiliations
Contributions
Conception and design: JL, XX. Collection and assembly of data: JL, HY. Data analysis and interpretation: All authors. Manuscript writing: JL, XX. Final approval of manuscript: All authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All data in present study are obtained from TCGA and GEO databases, which have received ethics approval and consent to participate. The study was performed in accordance with the Declaration of Helsinki.
Consent for publication
All data in present study are obtained from TCGA and GEO databases, which have received consent for publication.
Competing interests
The authors declare that they have no competing interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, J., Yue, H., Yu, H. et al. Development and validation of SIRT3-related nomogram predictive of overall survival in patients with serous ovarian cancer. J Ovarian Res 12, 47 (2019). https://doi.org/10.1186/s13048-019-0524-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13048-019-0524-2