Abstract
Background
Recently, we showed a >60% difference in 5-year survival for patients with tubo-ovarian high-grade serous carcinoma (HGSC) when stratified by a 101-gene mRNA expression prognostic signature. Given the varied patient outcomes, this study aimed to translate prognostic mRNA markers into protein expression assays by immunohistochemistry and validate their survival association in HGSC.
Methods
Two prognostic genes, FOXJ1 and GMNN, were selected based on high-quality antibodies, correlation with protein expression and variation in immunohistochemical scores in a preliminary cohort (n = 134 and n = 80, respectively). Six thousand four hundred and thirty-four (FOXJ1) and 5470 (GMNN) formalin-fixed, paraffin-embedded ovarian neoplasms (4634 and 4185 HGSC, respectively) represented on tissue microarrays from the Ovarian Tumor Tissue Analysis consortium underwent immunohistochemical staining and scoring, then univariate and multivariate survival analysis.
Results
Consistent with mRNA, FOXJ1 protein expression exhibited a linear, increasing association with improved overall survival in HGSC patients. Women with >50% expression had the most favourable outcomes (HR = 0.78, 95% CI 0.67–0.91, p < 0.0001). GMNN protein expression was not significantly associated with overall HSGC patient survival. However, HGSCs with >35% GMNN expression showed a trend for better outcomes, though this was not significant.
Conclusion
We provide foundational evidence for the prognostic value of FOXJ1 in HGSC, validating the prior mRNA-based prognostic association by immunohistochemistry.
Similar content being viewed by others
Background
Ovarian carcinoma (OC) is a heterogenous disease that can be better understood through its five main histotypes being tubo-ovarian high-grade serous (HGSC), low-grade serous (LGSC), mucinous (MC), endometrioid (EC) and clear-cell (CCC) carcinoma. HGSCs are the most commonly diagnosed histotype and are typically diagnosed at a high stage. Patients diagnosed with this disease have a 5-year survival of about 40% in most Western countries [1]. Based on a prognostic signature of 101 genes, we have recently shown a dramatic difference (>60%) in median 5-year overall survival when stratified by gene expression score quintiles [2]. While the median survival time for the most favourable quintile reached 9.5 years, it was only 2.3 years in the least favourable quintile. This effect size outperforms any individual validated prognostic marker to date and further supports a remarkable biological heterogeneity within HGSC [3,4,5,6].
Historically, a blanket approach was applied to the treatment of women diagnosed with HGSC, though it is now undergoing a dramatic shift due to high response rates to poly (ADP-ribose) polymerase (PARP) inhibitors in BRCA1/2-deficient cases, allowing them to be added as maintenance to the first line of therapy [7, 8]. However, this targeted therapy does not show the same efficacy across all patients with HGSC, and resistance may occur[9]. Therefore, other biomarkers related to prognosis may help to further delineate the biology of this aggressive disease. Candidate biomarkers that were discovered by mRNA expression profiling could be validated by immunohistochemistry (IHC). IHC is utilised routinely in clinical diagnostics across the globe, thus supporting its use as a tool for fast-tracked, statistically powered biological validation, particularly if large existing cohorts of tissue microarrays (TMAs) are also employed [10, 11].
The purpose of this study was to translate previously identified prognostic mRNA targets into clinically applicable IHC assays and to validate their prognostic association at the protein level in HGSC and the other main OC histotypes.
Methods
IHC of candidate markers
Five candidate markers: geminin (GMNN), small nuclear ribonucleoprotein polypeptide A (SNRPA1), flap structure-specific endonuclease 1 (FEN1), histone cluster 1 H2B family member D (HIST1H2BD) and forkhead box J1 (FOXJ1), were assessed by IHC in a training cohort (n = 80 (GMNN, SNRPA1, FEN1 and HIST12BD) or 134 (FOXJ1). IHC was automated on the DAKO Omnis platform (Agilent, Santa Clara, CA, USA) and performed centrally at the Department of Pathology and Laboratory Medicine of the University of Calgary, Canada. TMA sections were subjected to heat-induced epitope retrieval. (Supplementary Table 1). Slides were scanned using an Aperio CS2 up to ×400 magnification (Leica Biosystems, Wetzlar, Hessen, Germany), and viewed using ImageScope v12.2.2.5015.
Study design and participants
Sections of archival ovarian tumour tissue microarrays (TMA) were submitted by 21 studies from the Ovarian Tumor Tissue Analysis (OTTA) consortium (Supplementary Table 2) for analysis of GMNN and FOXJ1. Ethics approval was obtained for this project (University of New South Wales Human Research Ethics Advisory panel, #HC16299) and for participants through written informed consent or an Institutional Review Board ethics approval waiver (Supplementary Table 2). A retrospective cohort of 8798 and 7662 patient samples represented by individual or replicate TMA cores with a primary diagnosis of OC was assembled for FOXJ1 and GMNN staining, respectively. Contributing studies provided clinical covariates, including patient age at diagnosis, the time from diagnosis to OTTA study admission, histotype, stage of disease (FIGO (International Federation of Gynecology and Obstetrics) Stage I-IV, or localised/regional/distant), final vital status and overall survival (OS), defined as the time from diagnosis to death or last follow-up.
Immunohistochemistry of FOXJ1 and GMNN
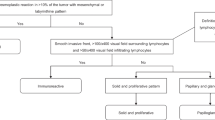
Positive and negative on-slide controls for FOXJ1 and GMNN protein expression were used to assess stain specificity (Supplementary Table 3). Healthy pre-menopausal and peri-menopausal fallopian tube samples (n = 1 case each), were also examined through haematoxylin and eosin staining, and IHC for FOXJ1 and GMNN (Supplementary Fig. 1).
IHC scoring was performed by two observers (AW and EYK). Observers were blinded to the clinicopathological data; one observer scored cases for FOXJ1 using a Nikon Eclipse 80i microscope (Nikon Inc., Chicago, IL, USA) at ×200 and another observer scored cases for GMNN using ImageScope v12.2.2.5015. Nuclear staining in each tumour core was semi-quantified by scores representing the percentage of immunopositive tumour cells in the core, in 5% intervals. Where cores were absent or tumour cells represented <25% of the core, no score was given. The maximum score was taken in cases with multiple cores on the TMAs. Intratumoural heterogeneity was assessed by a comparison of duplicate cores in n = 3401 (FOXJ1) and n = 3057 (GMNN) HGSC cases. Interobserver variability in FOXJ1 and GMNN expression scoring was evaluated in n = 221 and n = 311 cases, respectively.
FOXJ1 and GMNN expression score stratification
IHC scores were visualised through frequency distributions of FOXJ1and GMNN scores in OC, and by histotype. Score stratification models simplified IHC scores. Thresholds used in stratification were selected to preserve the shape of distribution in the frequency histogram of HGSC scores. As FOXJ1 and GMNN expression differed in the distribution of their IHC scores, different score stratification models were applied to each data set, being FOXJ1 0%, 5%, 10–15%, 20–45% and 50–100%, and GMNN 0%, 5%, 10–15%, 20–25%, 30% and 35–100%. Score stratification was required to assess the association between expression and OS.
Correlation analysis of mRNA and protein expression
The correlation between mRNA expression data [2] and IHC protein expression score was assessed. To ensure score stratification models would not alter this relationship, the correlation between mRNA and stratified FOXJ1 and GMNN protein was also assessed.
FOXJ1 and GMNN protein expression in only chemotherapy-naive samples
Sensitivity univariate and multivariate survival analyses in HGSC patient where treatment by primary debulking surgery was confirmed (n = 4440 (FOXJ1) and n = 4009 (GMNN)) were performed. This was to ensure the effects of neoadjuvant chemotherapy (NACT) did not impact the trends detected in the main survival analysis. Paired tissue samples taken before and after NACT were also assessed for FOXJ1 and GMNN protein expression (n = 23 and n = 21 respectively) to determine whether there were changes in expression in the tissue following exposure to chemotherapy.
Statistical analysis
The correlation between mRNA and protein expression was measured for each marker by Pearson correlation coefficients; the explainable variance was examined through the coefficient of determination. Clinical variables in FOXJ1 and GMNN cohorts were assessed through Chi-square testing of proportions. Kruskal–Wallis testing assessed IHC score differences by histotype. Squared-weighted Cohen’s kappa statistics and frequency count matrices were used to assess intratumoural heterogeneity across duplicate TMA cores from the same case. Squared-weighted Cohen’s kappa statistics were used to estimate concordance between multiple observers. A two-tailed Wilcoxon–Pratt matched-pairs signed rank test was used to assess protein expression differences in matched pairs of HGSC tumour samples taken before and after NACT.
Histotype-specific univariate and multivariate analysis of the survival associations of stratified FOXJ1 and GMNN expression were performed, with death by any cause being the primary end point. Right censoring at 10 years was applied to account for deaths not related to OC. Left truncation mitigated against survival bias introduced by time between diagnosis and enrolment into a study. Kaplan–Meier curves with corresponding log-rank testing and calculation of year-specific survival, were used to compare prognosis between strata of the score stratification model. Multivariate Cox proportional hazards regression modelling estimated hazard ratios (HRs) and corresponding 95% confidence intervals (CIs). Models were adjusted for age, stage and strata of score and stratified by OTTA study site. Assumptions of proportional hazards were tested. In cases of non-proportional covariate hazards or large sample sizes, scaled Schoenfeld residuals were plotted to assess the violation. Adjusted HRs for each strata of the score model were visualised through forest plots, and used to identify a linear relationship between expression and the hazard of death. All statistical analyses were carried out using RStudio v1.1.463 or GraphPad Prism v7.02. R packages “survival” (version 3.3-1) and “survminer” (version 0.4.9) were employed in univariate and multivariate survival analysis. Statistical significance was defined by p < 0.05.
Results
Selection of FOXJ1 and GMNN for analysis by IHC
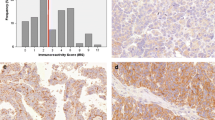
mRNA expression analysis by NanoString was previously performed on 3769 HGSC cases from the Ovarian Tumor Tissue Analysis (OTTA) consortium and 276 genes significantly associated with patient survival were identified [2]. The 90 genes of the greatest significance were examined for suitability for IHC analysis (Supplementary Table 4). We focussed on nuclear markers (avoiding stromal or immune markers due to challenges in interpretation of staining) with high-quality antibodies suitable for IHC in formalin-fixed paraffin-embedded tissues. Five genes met these criteria: GMNN, SNRPA1, FEN1, HIST1H2BD and FOXJ1. The previously identified HRs for one standard deviation change in gene expression, 95% CIs and adjusted q-values for these genes were: GMNN: 0.85 (95% CI 0.82–0.89) q = 1.89 × 10−11, SNRPA1: 0.87 (95% CI 0.83–0.91) q = 1.84×10−10, FEN1: 0.89 (95% CI 0.85–0.92) q = 1.44 × 10−7, HIST1H2BD: 0.89 (95% CI 0.85–0.93) q = 1.44 × 10−7, FOXJ1: 0.90 (95% CI 0.86–0.94) q = 2.56 × 10−6 [2]. Protein expression of these markers was assessed by IHC in a small cohort of 80–134 cases. The distribution of IHC scores were examined; FOXJ1 and GMNN expression showed the greatest variation in scores (Supplementary Fig. 2). Expression of these markers was positively skewed with median expression scores of 5 and 20%, respectively. The correlation between mRNA expression data from NanoString analysis and protein expression was determined (Supplementary Fig. 2). A significant correlation between mRNA and protein expression was identified for FEN1, FOXJ1 and GMNN (Supplementary Fig. 3). The levels of mRNA were able to explain some variance in FOXJ1 r2 = 0.283 and GMNN r2 = 0.23 protein expression. Both FOXJ1 and GMNN were selected for further analysis as they showed prognostic significance at the mRNA level, demonstrated a significant correlation between mRNA and protein, and had variation in IHC scores, suggesting they could be suitable for prognostic stratification of patients.
As a marker of ciliogenesis, FOXJ1 protein expression was observed specifically in ciliated cells in pre- and peri-menopausal fallopian tube epithelium (Supplementary Fig. 1). Both pre- and peri-menopausal fallopian tube epithelium did not express GMNN (Supplementary Fig. 1). The interobserver concordance of the stratified IHC scoring systems (FOXJ1 0%, 5%, 10–15%, 20–45% and 50–100%, and GMNN 0%, 5%, 10–15%, 20–25%, 30% and 35–100%) for both FOXJ1 and GMNN expression, between 2 observers, was evaluated in a subset of 221 and 311 cases, respectively, and achieved a weighted Cohen’s kappa of 0.957 and 0.673, respectively.
Protein expression of GMNN and FOXJ1 across OC histotypes, and the relationship with mRNA expression
Of the 8798 (FOXJ1) and 7662 (GMNN) cases, 2364 and 2192 cases respectively were excluded due to diagnosis with a histotype other than HGSC, LGSC, MC, EC or CCC, no survival data, uninterpretable or insufficient tumour tissue (<25% of the tumour core), overlapping cases between TMAs or withdrawn consent. The clinicopathological parameters by histotype for 6434 FOXJ1 and 5470 GMNN cases that met the inclusion criteria were evaluated (Table 1 and Supplementary Table 5). Subsets of HGSC cases had 80% power to detect a HR of 1.3, with a type-1 error rate of 0.001. OC histotypes had significantly different FOXJ1 and GMNN expression patterns (Fig. 1). FOXJ1 protein expression varied more than GMNN expression across histotypes with greater interquartile ranges of expression scores in all histotypes (apart from MC). Intratumoural heterogeneity was assessed by weighted Cohen’s kappa statistics in duplicate HGSC TMA cores when FOXJ1 and GMNN expression were given as 5% intervals (Cohen’s kappa 0.41 and 0.40 respectively) and stratified IHC scores (Cohen’s kappa 0.74 and 0.72 respectively; Supplementary Fig. 4). 3111 FOXJ1 and 2900 GMNN HGSC cases also had mRNA expression data collected through NanoString nCounter analysis [2]. The correlation between mRNA and protein expression was preserved in a HGSC subset of the final cohort to undergo survival analysis (Fig. 1). Stratification of FOXJ1 and GMNN protein expression did not appear to alter this correlation (Fig. 1).
Boxplots comparing the distribution of a FOXJ1 (n = 6434) and b GMNN (n = 5470) expression scores between histotypes. Unfilled boxes are defined by the 25th and 75th percentile, giving the interquartile range. Median scores are indicated by vertical lines within the interquartile range. If a vertical line is not shown, the median score is at the minimum or maximum value on the plot (0 or 100%, respectively). Tails indicate the minimum and maximum score given. Kruskal–Wallis testing evaluated the differences between histotypes; p values represent a comparison between IHC expression scores of each marker in OC histotypes. *p < 0.001. Relationship between normalised mRNA expression and protein expression in HGSC cases stained for c FOXJ1 (n = 3111) and d GMNN (n = 2900) and e FOXJ1 and f GMNN expression simplified by IHC score stratification models. Pearson’s correlation analysis given by r and the coefficient of determination by R2. *p < 0.0001
Association of stratified GMNN and FOXJ1 protein expression and OS, by histotype
Stratified FOXJ1 protein expression was significantly associated with OS in univariate (p < 0.0001) and multivariate (p = 0.0002) analysis of HGSC survival (Fig. 2 and Table 2). Higher FOXJ1 expression was significantly associated with a higher probability of survival, relative to cases with absent or lower expression (Table 2). Prognosis was most favourable in tumours with >50% of cells expressing FOXJ1 (HR 0.78, 95% CI 0.67–0.91, p < 0.05), with the 5-year survival being >10% higher than those without expression.
Univariate and multivariate analysis of stratified FOXJ1 protein expression in CCC indicated a significant association with improved survival (p = 0.0096 and p = 0.02, respectively; Supplementary Fig. 5 and Table 2). Multivariate analysis indicated that 10–15% (HR 0.50, 95% CI 0.30–0.83, p < 0.05) and >50% expression (HR 0.64, 95% CI 0.44–0.93, p < 0.05) had a significant survival association. However, review of the Kaplan–Meier curve (Supplementary Fig. 5) did not indicate a linear relationship between FOXJ1 expression and CCC survival, whereby the probability of survival increased alongside increasing FOXJ1 expression. Similarly, no threshold effects were observed, such as any expression of FOXJ1 conferring better outcomes in comparison to tumours having no expression of the protein. No survival associations with stratified FOXJ1 expression were observed in univariate or adjusted multivariate models of LGSC, MC and EC cases (Table 2).
Stratified GMNN protein expression was not significantly associated with OS in HGSC (p = 0.14), though tumours with >35% GMNN expression appeared to be associated with an improved OS (Fig. 2). This trend was not supported by multivariate survival analysis adjusting for age and stage, although the overall p value for GMNN was significant (p = 0.001) (Table 3). Univariate analysis of stratified GMNN expression in EC and CCC indicated a significant association with survival (Supplementary Fig. 6), where higher expression in both cases conferred improved outcomes. However, adjusted analysis indicated that stratified GMNN expression was not significantly associated with EC or CCC patient survival (p = 0.09 and p = 0.65, respectively; Table 3). Adjusted analysis of GMNN expression in the two LGSC cases with >30% expression in tumours had significantly poorer outcomes (HR 8.28, 95% CI 1.36–50.43, p < 0.05; Table 3).
The proportional hazards assumption of Cox regression modelling was not violated by age, stage or OTTA site, in any histotypes, for both markers. To account for age and stage artefacts in these results, induced by the large sample sizes of HGSC-specific analyses, Schoenfeld residuals were plotted and assessed (Supplementary Fig. 7). No significant deviations from the line of best fit were detected.
The univariate and multivariate survival analysis results were replicated in a sensitivity analysis of a subset of the HGSC cohort, using only known chemotherapy naïve tissue, from patients where primary debulking surgery was the confirmed first line of therapy (Supplementary Fig. 8 and Supplementary Table 6).
GMNN and not FOXJ1 protein expression was significantly different before and after NACT
FOXJ1 and GMNN protein expression in pairs of HGSC samples taken before and after NACT was assessed (Supplementary Fig. 9). While FOXJ1 expression was not significantly different in these matched pairs (n = 23, p = 0.93), GMNN expression did differ significantly after NACT (n = 21, p = 0.04).
Internal validation of further stratified FOXJ1 protein expression in HGSC
Forest plots were used to visualise the relationship between patient survival and FOXJ1 and GMNN score stratification models (Fig. 3). A linear relationship between FOXJ1 protein expression and the risk of death was observed in HGSC patients (Fig. 3). As FOXJ1 expression increased the probability of HGSC patient survival also increased. The HR for GMNN expression was not linear but was suggestive of a threshold-effect (Fig. 3). Cases with >35% of tumour cells expressing GMNN were less likely to succumb to disease, than those with lower expression.
Discussion
Our study validates the linear, increasing prognostic association of FOXJ1 protein expression in HGSC in a large, international cohort. While our analysis of FOXJ1 exemplifies the power of IHC for rapid biological validation of prognostic biomarkers that were originally discovered as mRNA candidates, the lack of translation of other markers also illustrates the challenges.
As FOXJ1 protein expression increased, the probability of survival in HGSC increased, implying expression has a linear association with outcome. Patients with tumours with >50% of cells expressing FOXJ1 had the longest survival. Sui and colleagues have already suggested that high FOXJ1 expression is associated with favourable OC prognosis [12]. The present analysis uses a statistically powered, histotype-specific approach to differentiate that FOXJ1 is only associated with prognosis in HGSC and does not indicate the prognosis of other histotypes, perhaps except for CCC. High expressing CCC were associated with favourable survival as were cases with lower expression, contrasting the observed linear association with survival in HGSC. Sui and colleagues proposed that the stem cell-related transcription factor NANOG is a negative regulator of FOXJ1-mediated OC migration and invasion [12]. This suggests that NANOG high/FOXJ1 low are more akin to cancer stem cells, which may be more likely chemo-resistant and thereby less susceptible to chemotherapy, leading to poor prognosis. However, we were unable to address the response to chemotherapy in the current data set due to lack of data, e.g. the Response Evaluation Criteria in Solid Tumors (RECIST) data. Yet this generates an interesting hypothesis for future studies.
The biological significance and prognostic association of FOXJ1 in cancer is varied [13, 14]. Survival associations consistent with our HGSC results have been observed in gastric cancer, as well as ependymomas and choroid plexus tumours, where high expression of FOXJ1 is thought to be a marker of better tumour differentiation and a more favourable prognosis [15, 16]. It may be appealing to conclude that FOXJ1 is a marker for well-differentiated HGSCs. FOXJ1’s characteristic role is in motile ciliogenesis, and we confirmed FOXJ1 expression in ciliated, but not secretory cells of the fallopian tube. However, this would somewhat contradict the prevailing concept that HGSC arises from the secretory cells, as opposed to ciliated cells, of the fallopian tube via precursor serous tubal intraepithelial carcinoma (STIC) lesions and diffuse PAX8 staining, a marker of secretory cells, is present in virtually all HGSCs, supporting the STIC hypothesis [17,18,19,20,21]. As motile cilia are neither a feature of STIC nor HGSC, the expression of FOXJ1 does not suggest that HGSC arise from terminally differentiated ciliated cells of the fallopian tube or that a cell lineage switch occurs [22]. Instead, the expression of the ciliated marker FOXJ1 in HGSC can be considered as a marker of enigmatic differentiation, which does not result in morphologically apparent cilia and is associated with better outcome; a phenomenon that has been previously suggested for endometrial cancers [23]. “Aberrant” FOXJ1 expression in HGSC may reflect epigenetic reprogramming of secretory cell-derived tumour cells or a unique differentiation state of the cell of origin. A recent study proposed that HGSC are composed of a mixture of cancer cell states inherited from its cell of origin in the fallopian tube including a ciliated state [24]. An enigmatic ciliated differentiation is further supported by expression of other ciliated markers (such as Ezrin) in OC confirming that FOXJ1 expression is not an isolated, aberrantly expressed marker of ciliogenesis, but represents a more general differentiation programme [25, 26]. In addition to FOXJ1’s potential in HGSC prognostication, future studies may use FOXJ1 to gain further insights into HGSC development.
Contrary to results in FOXJ1, the univariate survival analysis with GMNN expression indicated that there was no significant prognostic association in HGSC. Contrastingly, multivariate survival analysis did indicate that GMNN expression was significantly associated with HGSC survival, although there were no observable trends in HRs across protein expression groups that suggested this association was linear. In a threshold effect, individuals with >35% of tumour tissue expressing GMNN did demonstrate a trend towards a more favourable prognosis, although this was not supported by the 95%CI in the multivariate analysis. As GMNN’s significant mRNA association with survival did not translate into protein expression, GMNN is not currently suitable for prognostic assessment through IHC. Various explanations of this result can be drawn. Large-scale studies have shown the correlation between mRNA and protein expression is varied and difficult to predict for a given biomarker [27, 28]. In the final cohort, the correlation between mRNA and GMNN protein expression was weaker, relative to FOXJ1 expression. While this difference was slight, it may have contributed to the result. Dissimilar mRNA and protein abundances may be reflective of the translation rate, its modulation, delays in protein synthesis and transport, and the modulation of the proteins’ turnover-rate [29,30,31]. As a surrogate marker of proliferation, GMNN’s expression is highly regulated during the cell cycle [32]. Protein translation and degradation may be uncoupled from mRNA transcription and stability, and may explain the lack of association with survival for GMNN protein levels. Another explanation might be that the FOXJ1 IHC assay was superior in quality and therefore easier to interpret as shown by the better interobserver reproducibility. Perhaps another reason that GMNN did not translate into a suitable prognostic IHC marker is the less variable expression within HGSC, when compared to FOXJ1. While prognostic significance was not shown in LGSC, multivariate analysis indicated that individuals with >35% of tumour tissue expressing GMNN were associated with a more unfavourable prognosis. However, as this group only included two individuals this should only be considered an interesting observation highlighting the challenges of biomarker studies in rare histotypes even in a consortium type approach. No associations were seen in the other histotypes. GMNN expression did however differ significantly in samples taken before and after NACT in small cohort of HGSC cases. This difference can potentially be attributed to the role of GMNN in cell division alongside the impact of chemotherapy on this process [33]. However, trends that would suggest GMNN expression increased or decreased following NACT were not observed. Furthermore, the potential clinical relevance of this result is limited by the small sample size, which would not completely capture disease heterogeneity. HGSC cases known to be chemotherapy naïve underwent univariate and multivariate survival analysis to confirm the prognostic associations revealed in this study.
The potential for FOXJ1 and GMNN expression stratification into broader score-groupings was observed in our HGSC cohort. As 0% and 5%, as well as 10%-15% and 20%-45% exhibited consistently similar survival associations in the cohort, analysis of FOXJ1 expression by 0%-5%, 10-45% and 50%-100% IHC score groupings may prove more-appropriate for clinical assessment. Similarly, the trend observed in tumours with high GMNN expression suggested that stratification into 0%-35% and 40%-100% may clarify the relationship with survival, though the lack of significant results in this study, does render the value of further GMNN investigation dubious. We acknowledge that the translational potential of our findings is limited by heterogeneity within HGSC. Future work may take important molecular alterations within HGSC, such BRCA1/2 mutation or homologous repair deficiency status, into account. Further, we recognise that the immediate translational potential of FOXJ1 as an individual prognostic biomarker is limited given the observed survival increase of ~10% in a small subset (less than 10%) of HGSC patients. Yet we believe that due to robust prognostic associations FOXJ1 should be evaluated in prognostic multimarker models.
Several variables can interfere with the translation of mRNA targets into IHC biomarkers. Our marker selection was severely limited by the number of genes with available high-quality monoclonal antibodies, which were necessary as they ensured increased stain reproducibility across FFPE TMA sections. We avoided proteins of the microenvironment and focussed on those with nuclear expression as they are often the most robust markers for established clinical tests. When selecting markers, we observed only moderate correlations between mRNA and protein expression for the five initially selected markers. While gene expression or protein stability can influence these correlations, we believe that a main factor is the use of conventional IHC, which is not quantitative in nature [34,35,36,37]. Modern IHC detection systems greatly amplify the signal leading to a quick saturation. This might explain why some of the initial markers showed ubiquitous expression by IHC in comparison to variable mRNA expression levels. In addition, markers were assessed on TMAs with an intrinsic limitation in assessing for intratumoural heterogeneity. However, our assessment of intratumoural heterogeneity in cases with duplicate TMA cores demonstrated strong agreement in FOXJ1 and GMNN protein expression scores. While this may not capture all variation in protein expression across each tumour, it does support that the scores used in our analysis were representative of the tumour profile.
Overall, our study has provided foundational evidence that the prognostic mRNA signature of FOXJ1 can be translated into an IHC biomarker, to stratify prognosis in HGSC. Survival improved with stratified, increasing FOXJ1 protein expression. It indicated this relationship could be further stratified, to improve clinical utility, which is intriguing regarding its unexpected expression in HGSC. We also demonstrate the challenges of validating prognostic IHC markers. The careful study design and power has ensured these results provide much-needed insights into the likely outcomes of HGSC patients.
Data availability
Participants of this study did not agree to their data being shared publicly; accordingly, the data used in this research will not be made available.
References
Arnold M, Rutherford MJ, Bardot A, Ferlay J, Andersson TM, Myklebust TA, et al. Progress in cancer survival, mortality, and incidence in seven high-income countries 1995-2014 (ICBP SURVMARK-2): a population-based study. Lancet Oncol. 2019;20:1493–505.
Millstein J, Budden T, Goode EL, Anglesio MS, Talhouk A, Intermaggio MP, et al. Prognostic gene expression signature for high-grade serous ovarian cancer. Ann Oncol. 2020;31:1240–50.
Bolton KL, Chenevix-Trench G, Goh C, Sadetzki S, Ramus SJ, Karlan BY, et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. 2012;307:382–90.
Ovarian Tumor Tissue Analysis C, Goode EL, Block MS, Kalli KR, Vierkant RA, Chen W, et al. Dose-response association of CD8+ tumor-infiltrating lymphocytes and survival time in high-grade serous ovarian cancer. JAMA Oncol. 2017;3:e173290.
Sieh W, Kobel M, Longacre TA, Bowtell DD, deFazio A, Goodman MT, et al. Hormone-receptor expression and ovarian cancer survival: an Ovarian Tumor Tissue Analysis consortium study. Lancet Oncol. 2013;14:853–62.
Chan AM, Enwere E, McIntyre JB, Wilson H, Nwaroh C, Wiebe N, et al. Combined CCNE1 high-level amplification and overexpression is associated with unfavourable outcome in tubo-ovarian high-grade serous carcinoma. J Pathol Clin Res. 2020;6:252–62.
Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–8.
Coleman RL, Fleming GF, Brady MF, Swisher EM, Steffensen KD, Friedlander M, et al. Veliparib with first-line chemotherapy and as maintenance therapy in ovarian cancer. N Engl J Med. 2019;381:2403–15.
Virani S, Baiocchi G, Bowtell D, Cabasag CJ, Cho KR, Fortner RT, et al. Joint IARC/NCI International Cancer Seminar Series Report: expert consensus on future directions for ovarian carcinoma research. Carcinogenesis. 2021;42:785–93.
Magaki S, Hojat SA, Wei B, So A, Yong WH. An introduction to the performance of immunohistochemistry. Methods Mol Biol. 2019;1897:289–98.
Kobel M, Anglesio MS, Brenton JD. You won’t believe this old test… that does cheap single-cell mutation detection. J Pathol Clin Res. 2018;4:149–53.
Siu MK, Wong ES, Kong DS, Chan HY, Jiang L, Wong OG, et al. Stem cell transcription factor NANOG controls cell migration and invasion via dysregulation of E-cadherin and FoxJ1 and contributes to adverse clinical outcome in ovarian cancers. Oncogene. 2013;32:3500–9.
Liu K, Fan J, Wu J. Forkhead box protein J1 (FOXJ1) is overexpressed in colorectal cancer and promotes nuclear translocation of beta-catenin in SW620 cells. Med Sci Monit. 2017;23:856–66.
Xian S, Shang D, Kong G, Tian Y. FOXJ1 promotes bladder cancer cell growth and regulates Warburg effect. Biochem Biophys Res Commun. 2018;495:988–94.
Abedalthagafi MS, Wu MP, Merrill PH, Du Z, Woo T, Sheu SH, et al. Decreased FOXJ1 expression and its ciliogenesis programme in aggressive ependymoma and choroid plexus tumours. J Pathol. 2016;238:584–97.
Wang J, Cai X, Xia L, Zhou J, Xin J, Liu M, et al. Decreased expression of FOXJ1 is a potential prognostic predictor for progression and poor survival of gastric cancer. Ann Surg Oncol. 2015;22:685–92.
Gilks CB, Irving J, Kobel M, Lee C, Singh N, Wilkinson N, et al. Incidental nonuterine high-grade serous carcinomas arise in the fallopian tube in most cases: further evidence for the tubal origin of high-grade serous carcinomas. Am J Surg Pathol. 2015;39:357–64.
Perets R, Wyant GA, Muto KW, Bijron JG, Poole BB, Chin KT, et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer Cell. 2013;24:751–65.
Mehrad M, Ning G, Chen EY, Mehra KK, Crum CP. A pathologist’s road map to benign, precancerous, and malignant intraepithelial proliferations in the fallopian tube. Adv Anat Pathol. 2010;17:293–302.
Munakata S, Yamamoto T. Incidence of serous tubal intraepithelial carcinoma (STIC) by algorithm classification in serous ovarian tumor associated with PAX8 expression in tubal epithelia: a study of single institution in Japan. Int J Gynecol Pathol. 2015;34:9–18.
Laury AR, Perets R, Piao H, Krane JF, Barletta JA, French C, et al. A comprehensive analysis of PAX8 expression in human epithelial tumors. Am J Surg Pathol. 2011;35:816–26.
Abdelhamed ZA, Ryan TA, Fuller M, Coulson-Gilmer C, Abdelmottaleb DI, Wang TL, et al. Characterization of primary cilia in normal fallopian tube epithelium and serous tubal intraepithelial carcinoma. Int J Gynecol Cancer. 2018;28:1535–44.
Cochrane DR, Campbell KR, Greening K, Ho GC, Hopkins J, Bui M, et al. Single cell transcriptomes of normal endometrial derived organoids uncover novel cell type markers and cryptic differentiation of primary tumours. J Pathol. 2020;252:201–14.
Hu Z, Artibani M, Alsaadi A, Wietek N, Morotti M, Shi T, et al. The repertoire of serous ovarian cancer non-genetic heterogeneity revealed by single-cell sequencing of normal fallopian tube epithelial cells. Cancer Cell. 2020;37:226.e7–42.e7.
Horwitz V, Davidson B, Stern D, Trope CG, Tavor Re’em T, Reich R. Ezrin is associated with disease progression in ovarian carcinoma. PLoS ONE. 2016;11:e0162502.
Kobel M, Gradhand E, Zeng K, Schmitt WD, Kriese K, Lantzsch T, et al. Ezrin promotes ovarian carcinoma cell invasion and its retained expression predicts poor prognosis in ovarian carcinoma. Int J Gynecol Pathol. 2006;25:121–30.
Payne SH. The utility of protein and mRNA correlation. Trends Biochem Sci. 2015;40:1–3.
Papatheodorou I, Fonseca NA, Keays M, Tang YA, Barrera E, Bazant W, et al. Expression Atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 2018;46:D246–51.
Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–50.
Kays I, Chen BE. Protein and RNA quantification of multiple genes in single cells. Biotechniques. 2019;66:15–21.
Li JJ, Chew GL, Biggin MD. Quantitating translational control: mRNA abundance-dependent and independent contributions and the mRNA sequences that specify them. Nucleic Acids Res. 2017;45:11821–36.
Ballabeni A, Zamponi R, Moore JK, Helin K, Kirschner MW. Geminin deploys multiple mechanisms to regulate Cdt1 before cell division thus ensuring the proper execution of DNA replication. Proc Natl Acad Sci USA. 2013;110:E2848–53.
Kang EY, Millstein J, Popovic G, Meagher NS, Bolithon A, Talhouk A, et al. MCM3 is a novel proliferation marker associated with longer survival for patients with tubo-ovarian high-grade serous carcinoma. Virchows Arch. 2021;480:855–71.
Buccitelli C, Selbach M. mRNAs, proteins and the emerging principles of gene expression control. Nat Rev Genet. 2020;21:630–44.
Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42.
Mauger DM, Cabral BJ, Presnyak V, Su SV, Reid DW, Goodman B, et al. mRNA structure regulates protein expression through changes in functional half-life. Proc Natl Acad Sci USA. 2019;116:24075–83.
Taube JM, Akturk G, Angelo M, Engle EL, Gnjatic S, Greenbaum S, et al. The Society for Immunotherapy of Cancer statement on best practices for multiplex immunohistochemistry (IHC) and immunofluorescence (IF) staining and validation. J Immunother Cancer. 2020;8:e000155.
Acknowledgements
We thank all the study participants who contributed to this study and all the researchers, clinicians, and technical and administrative staff who have made this work possible. We thank Shuhong Liu and Young Ou for performing immunohistochemistry (Anatomical Pathology Research Laboratory, Department of Pathology and Laboratory Medicine, University of Calgary). The AOCS gratefully acknowledges additional support from S. Boldeman, the Agar family, Ovarian Cancer Action (UK), Ovarian Cancer Australia and the Peter MacCallum Foundation. The AOCS also acknowledges the cooperation of the participating institutions in Australia and acknowledges the contribution of the study nurses, research assistants, and all clinical and scientific collaborators to the study. The complete AOCS Study Group can be found at http://www.aocstudy.org. We thank Mie Konno, Michelle Darago, Faye Chambers, SEARCH team, Craig Luccarini, Caroline Baynes, Don Conroy, I. Jacobs, S. Gayther, E. Wozniak, J. Ford, and N. Balogun. We thank the Genetic Pathology Evaluation Centre (GPEC) for technical support in construction of tissue micro-arrays; and the OVCARE clinical informatics core, Gynecological tissue bank (GTB), and Cheryl Brown Gynecologic Cancer Outcomes Unit for facilitating recruitment of patients, banking biospecimens, and maintaining clinical data. We thank the Gynaecological Oncology Biobank at Westmead, a member of the Australasian Biospecimen Network‐Oncology group. We thank all patients who participated in and donated tissue samples to this study. We acknowledge funding and support from Cancer Research UK and the Cancer Research UK Cambridge Centre (A22905, A29580, A25117).
Funding
This study was funded by NCI/NIH R01CA172404 to SJR. This research was supported by the NIHR Cambridge Biomedical Research Centre (BRC‐1215‐20014). The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care. AOC: The Australian Ovarian Cancer Study was supported by the U.S. Army Medical Research and Materiel Command under DAMD17‐01‐1‐0729, The Cancer Council Victoria, Queensland Cancer Fund, The Cancer Council New South Wales, The Cancer Council South Australia, The Cancer Foundation of Western Australia, The Cancer Council Tasmania and the National Health and Medical Research Council of Australia (NHMRC; ID199600; ID400413 and ID400281); AOV: Canadian Institutes of Health Research (MOP‐86727).; BAV: ELAN Funds of the University of Erlangen‐Nuremberg; BRZ: Brazilian National Council for Scientific and Technological Development, grant No. 478416/2009-1; CAL: Cancer Research Society (19319) and CLS Internal Research support RS14‐5008; CNI: CNI funded by Instituto de Salud Carlos Tercero (AES grant PI19/01730) and Fondo Europeo de Desarrollo Regional, FEDER. Instituto de Salud Carlos III (PI 12/01319); Ministerio de Economía y Competitividad (SAF2012); DOV: NCI/NIH R01CA168758, NCI/NIH R01CA112523 and NCI/NIH R01CA087538; HAW: US National Institutes of Health (R01‐CA58598, N01‐CN‐55424 and N01‐PC‐67001); HOP: Department of Defense (DAMD17‐02‐1‐0669) and NCI (K07‐CA080668, R01‐CA95023, MO1‐RR000056). This project used the UPMC Hillman Cancer Center and Tissue and Research Pathology/Pitt Biospecimen Core shared resource which is supported in part by award P30CA047904. LAX: American Cancer Society Early Detection Professorship (SIOP‐06‐258‐01‐COUN) and the National Center for Advancing Translational Sciences (NCATS), Grant UL1TR000124; MAY: National Institutes of Health (R01‐CA122443, R01-CA243483, P30‐CA15083, P50‐CA136393); Mayo Foundation; Minnesota Ovarian Cancer Alliance; Fred C. and Katherine B. Andersen Foundation; NCT: Parts of the study at the NCT were funded by “Heuer Stiftung für medizinische Forschung”; POC: Pomeranian Medical University; SEA: Cancer Research UK C490/A16561, the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge, Cambridge Cancer Centre. The University of Cambridge has received salary support for PDPP from the NHS in the East of England through the Clinical Academic Reserve. TVA: This work was supported by Canadian Institutes of Health Research grant (MOP‐86727) and by NIH/NCI 1 R01CA160669‐01A1; UKO: The UKOPS study was funded by The Eve Appeal (The Oak Foundation) with contribution to author’s salary through MRC core funding MC_UU_00004/01 and the National Institute for Health Research University College London Hospitals Biomedical Research Centre; VAN: BC’s Gynecological Cancer Research Team (OVCARE) receives core funding from The BC Cancer Foundation and the VGH and UBC Hospital Foundation; WMH: National Health and Medical Research Council of Australia, Enabling Grants ID 310670 and ID 628903. Cancer Institute NSW Grants 12/RIG/1‐17 and 15/RIG/1‐16. The Westmead GynBiobank acknowledges financial support from the Sydney West Translational Cancer Research Centre, funded by the Cancer Institute NSW. NSM is supported by the NSW Ministry of Health and UNSW Sydney under the NSW Health PhD Scholarship Program, and the Translational Cancer Research Network, a translational cancer research centre programme funded by the Cancer Institute NSW. DB is supported by the National Health and Medical Research Council of Australia (NHMRC Fellowship (APP1117044) and Program Grant APP1092856) and the United States Department of Defence (DoD) Ovarian Cancer Transitional Leverage Award (W81XWH‐12‐1‐0104); FJCdR is funded by the Brazilian National Council for Scientific and Technological Development – CNPq (427983/2016-9, 303210/2018-4). MK received support through the Calgary Laboratory Services research support fund (RS19-612). AB received support through the Ovarian Cancer Research Fund (OCRF). BYK is funded by the American Cancer Society Early Detection Professorship (SIOP-06-258-01-COUN) and the National Center for Advancing Translational Sciences (NCATS), grant UL1TR000124. SH received grants from University of Heidelberg Medical Faculty, Novartis Oncology, Stiftung für Krebs- und Scharlachforschung Mannheim, and Deutsche Forschungsgemeinschaft. JDB acknowledges funding and support from Cancer Research UK (grant numbers A22905, A15601 and A17197). UKO: MW is funded by the European Union’s Horizon 2020 European Research Council Program, H2020 BRCA-ERC under Grant Agreement No. 742432. AT is funded through a Michael Smith Foundation for Health Research Scholar Award. MA is funded through a Michael Smith Foundation for Health Research Scholar Award and the Janet D. Cottrelle Foundation Scholars programme managed by the BC Cancer Foundation. DGH receives support from the Dr. Chew Wei Memorial Professorship in Gynecologic Oncology and the Canada Research Chairs programme (Research Chair in Molecular and Genomic Pathology). Funding for ADF is funded by Cancer Institute NSW grant 15/TRC/1-01. RE was supported by the Interdisciplinary Center for Clinical Research (IZKF; Clinician Scientist Program) of the Medical Faculty FAU Erlangen-Nürnberg. The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript and decision to submit the manuscript for publication. Open Access funding enabled and organized by CAUL and its Member Institutions.
Author information
Authors and Affiliations
Consortia
Contributions
SJR and MK conceived, designed, and supervised the study. AW and EYK collected immunohistochemical protein scores, performed analysis, drafted the manuscript and revised the manuscript. PDPP provided statistical advice. All authors contributed through collection, curation and maintenance of respective consortia based, or local institution, collections of patient samples including recruitment and consenting of patients, clinical care, abstraction of clinical data, and updating of outcome and follow-up data. All authors revised the manuscript and approved submission of the final version.
Corresponding author
Ethics declarations
Competing interests
ADF, SF, and NT have received a research grant from AstraZeneca, not directly related to this work. DDB receives funding from Astra Zeneca and Genentech Roche for the conduct of research studies and clinical trials unrelated to the work described in this article. DGH is a founder and Chief Medical Officer of Contextual Genomics, a somatic mutation testing laboratory; the company’s work and interests do not overlap with the subject of and methodologies used in this article. UM had stock ownership in Abcodia until October 2021, and UCL had a license agreement (2011–2021) with Abcodia not related to this work. PDPP is a member of the British Journal Cancer Editorial Board. MW is a shareholder of Sola Diagnostics GmbH and BreOva Health Ltd. GSN received speaker/advisory fees from 3M, GSK, Medtronic, and Abbott outside of the submitted work. AH has received honoraria from BMS, MSD, Roche, AstraZeneca, Boehringer Ingelheim, Abbvie, Jansen-Cilag, and Ipsen. RE has received honoraria from Roche, Eisai, Pfizer, and Novartis and travel grants from BioNTech. The institution of AH and RE conducts research for AstraZeneca, Roche, Janssen-Cilag, NanoString Technologies, Novartis, Cepheid, and BioNTech. All other authors declare no competing interests.
Ethics approval and consent to participate
The majority of the participants provided informed consent. Three of the 21 sites had a waiver of consent, listed in Supplementary Table 2. The study was approved by the University of New South Wales, Sydney, Human Research Ethics Application (HREA) Panel D: Biomedical, HC16299. The ethics committees that approved the individual participating sites are listed in Supplementary Table 2. The study was performed in accordance with the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Weir, A., Kang, EY., Meagher, N.S. et al. Increased FOXJ1 protein expression is associated with improved overall survival in high-grade serous ovarian carcinoma: an Ovarian Tumor Tissue Analysis Consortium Study. Br J Cancer 128, 137–147 (2023). https://doi.org/10.1038/s41416-022-02014-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-022-02014-y
- Springer Nature Limited