Abstract
Acute myeloid leukemia (AML) is a malignant tumor of the immature myeloid hematopoietic cells in the bone marrow (BM). It is a highly heterogeneous disease, with rising morbidity and mortality in older patients. Although researches over the past decades have improved our understanding of AML, its pathogenesis has not yet been fully elucidated. Long noncoding RNAs (lncRNAs), microRNAs (miRNAs), and circular RNAs (circRNAs) are three noncoding RNA (ncRNA) molecules that regulate DNA transcription and translation. With the development of RNA-Seq technology, more and more ncRNAs that are closely related to AML leukemogenesis have been discovered. Numerous studies have found that these ncRNAs play an important role in leukemia cell proliferation, differentiation, and apoptosis. Some may potentially be used as prognostic biomarkers. In this systematic review, we briefly described the characteristics and molecular functions of three groups of ncRNAs, including lncRNAs, miRNAs, and circRNAs, and discussed their relationships with AML in detail.
Similar content being viewed by others
Background
Acute myeloid leukemia (AML) is an aggressive hematological malignancy characterized by abnormal proliferation and differentiation of the immature myeloid cells [1]. Despite a growing list of treatment options, most patients still relapse and die after remission, and the prognosis remains unideal [2]. It is necessary to explore new biomarkers for diagnosis, prognostication, and therapeutic targets of AML so as to develop more effective surveillance and treatment programs.
The discovery of noncoding RNAs (ncRNAs) opens up new prospects for AML diagnosis, prognosis and treatment. ncRNAs are functional small RNA molecules that are not translated into a protein [3]. The DNA molecules that make up the human genome are about 3 billion base pairs, of which about 5–10% are stably transcribed, but protein-coding genes account for less than 2% of the human genome. The remaining 3–8% of the genome are transcribed into non-coding transcripts, i.e., ncRNAs [4,5,6]. ncRNAs are divided into two categories based on their functions: housekeeping and regulatory, the latter includes miRNAs, circRNAs, and lncRNAs. Regulatory ncRNAs are extensively involved in gene transcription and translation. They are key players in physiological and pathological processes such as cell differentiation, ontogenesis, inflammation, and angiogenesis. There is emerging evidence that miRNAs, circRNAs, and lncRNAs actively participate in the pathogenesis of major hematological malignancies including AML [7]. In this review, we aimed to provide a comprehensive summary of the roles of miRNAs, circRNAs, and lncRNAs in AML, and to illustrate their diagnostic and prognosticating potentials in this disease.
MicroRNA
MicroRNAs (miRNAs) are small RNA molecules of approximately 22 nucleotides that bind to the 3′-untranslated region (3′-UTR) of the target mRNA and negatively regulate the expression of the target gene at the transcriptional level [8]. miRNAs mainly participate in the pathogenesis of AML through the following five mechanisms: copy number alterations, change in the proximity to the oncogenic genomic region due to chromosomal translocation, epigenetic changes, aberrant targeting of miRNA promoter regions by altered transcription factors or oncoproteins, and finally, dysregulated miRNAs processing [9].
Abnormal miRNA expression and function in acute myeloid leukemia
The molecular and cytogenetic criteria currently defined by 2016 WHO is the most widely used diagnostic tool for AML [10]. Each AML subtype seems to exhibit a unique miRNA signature that distinguishes it from others. For example, Chen et al. reported miR-9, an oncogenic miRNA, was overexpressed in the mixed lineage leukemia (MLL)-rearranged AML patients. Inhibition of miR-9 expression could significantly reduce cell growth/viability and promote apoptosis [11]. Emmrich et al. found miR-9, significantly downregulated in pediatric AML with t(8;21), was characterized by its tumor-suppressive property. Upregulation of miR-9 decreased leukemic growth and induced monocytic differentiation of t(8;21) AML cell lines in vitro and in vivo. Functionally, miR-9 exerted its effects by binding to let-7 to suppress the oncogenic LIN28B/HMGA2 axis [12]. In another study, miR-9-1 was observed to be downregulated in t(8;21) AML. Besides, overexpressed miR-9-1 induced differentiation and inhibited proliferation in t(8;21) AML cell lines [13]. MiR-10a/b was significantly increased in AML patients with t(8;21), t(9;11), NPM1 mutation, and particularly M1, M2, and M3 subtype. Abnormal high expression in those patients led to unlimited proliferation of immature blood progenitors and repressed differentiation and maturation of mature blood cell [14]. Another study showed that miR-10a overexpression was significantly associated with French-American-British(FAB)-M3/t(15;17) subtypes and NPM1 mutation, leading to the lower percentage of bone marrow (BM) blasts, while overexpression of miR-10b was correlated with NPM1 and DNMT3A mutations, resulting in higher percentage of BM blasts [15]. Some studies observed overexpression of the miR-181 in cytogenetic normal AML (CN-AML) patients with CEBPA mutations, FLT3-ITD, and/or wild-type NPM1 and t(15;17) [16,17,18,19]. MiR-155 was upregulated in FLT3-ITD-associated AML and targeted the myeloid transcription factor PU.1. Knockdown of miR-155 could repress proliferation and induce apoptosis of FLT3-ITD-associated leukemic cells [20].
MiRNA expression is also associated with morphologic sub-types of AML. MiR-122 expression, as an oncogene, was decreased in BM samples from pediatric patients with FAB subtype M7, and the forced expression of miR-122 in AML cell lines significantly inhibited cell proliferation and reduced the ratio of S-phase cells [21]. Xu et al. recently reported higher expression of miR-196b was observed in pediatric AML with M4/5 subtypes and predicted a poor outcome [22]. Another study compared M1 with M5 samples and noted that expressions of miR-146a/b, miR-181a/b/d, miR-130a, miR-663, and miR-135b were higher in M1, whereas expressions of miR-21, miR-193a, and miR-370 were higher in M5 [23]. Interestingly, in normal BM, miR-181a was enriched in B cells, T cells, monocytes, and granulocytes [24], but its overexpression was less common in monocytic lineage AML subtypes M4 or M5, but more so in M1 or M2 subtypes [25]. The expression levels of miR-195 in both BM and serum were significantly decreased, and pediatric patients with low serum miR-195 level more often had FAB-M7, unfavorable karyotypes, and shorter relapse-free and overall survival (OS) [26].
Changes in miRNA expression levels alter the expressions of downstream genes, promoting AML leukemogenesis [27]. For example, miR-155, acting as an oncogenic miRNA, may participate in the pathogenesis of AML by targeting SHIP1 and downregulating transcription factor PU.1 expression [28, 29]. This miRNA was regulated by NF-κB, whose activity was partly controlled by the NEDD8-dependent ubiquitin ligases [30, 31]. Schneider et al. reported that miR-155 expression was positively correlated with Meis1 expression level in MLL-rearranged AML and first indicated that the transforming efficacy of MLL-fusions remained unaltered in the absence of miR-155, while knocking out miR-155 did not affect in vitro leukemia formation or progression [32]. Other studies demonstrated that miR-9/9* was aberrantly expressed in myeloid progenitors of most AML cases to inhibit neutrophil differentiation by regulating EGN post-transcriptional level. Moreover, miR-9 could promote proliferation of leukemia cells in adult CD34+ AML with normal karyotype by suppressing Hes1 expression and knockdown of miR-9 could reduce circulating leukemic cell counts in peripheral blood (PB) and BM, attenuate splenomegaly and prolong survival in a xenotransplant mouse model [33, 34]. Li et al. showed that miR-193a expression was downregulated in AML1/ETO-positive leukemia cells because AML1/ETO triggered the heterochromatic silencing of miR-193a by binding at AML1-binding sites and recruiting chromatin-remodeling enzymes. Then the epigenetic silencing of tumor suppressor gene miR-193a led to leukemogenesis in AML with t(8;21) by activating the PTEN/PI3K signal pathway [35]. The latest study found that Erbin was the target of miR-183-5p that negatively regulated the Erbin expression, resulting in enhanced cell proliferation of AML cells via activation of RAS/RAF/MEK/ERK and PI3K/AKT/FoxO3a pathways [36]. MiR-125b, as an oncogenic miRNA, frequently overexpressed in human AML, could promote MLL-AF9-driven murine AML by TET2-VEGFA pathway. Zhang et al. reported that miR-203 downregulation frequently occurred in CD34 + AML cells in relation to CD34− cells isolated from patients. Additionally, re-expression of miR-203 inhibited cell proliferation, self-renewal, and sphere formation in LSCs by targeting survivin and Bmi-1 [37].
MicroRNAs are associated with chemoresistance of AML
Chemoresistance is commonly seen in refractory and recurrent AML. Studies have shown that miRNAs are involved in AML chemotherapy resistance in many ways, such as apoptosis, cell cycle and ATP-binding cassette (ABC) transporter-mediated multidrug resistance.
Li et al. reported that miR-181a expression level was lower in the K562/A02 cells than in the K562 cells and could reduce doxorubicin resistance of K562/A02 cells by directly targeting the 3′-UTR of BCL-2 and MCL-1 mRNAs [38]. Similarly, miR-181a was underexpressed in the HL-60/Ara-C cell line compared with HL-60 cell line, while upregulated miR-181a in HL-60/Ara-C cells sensitized the cells to Ara-C treatment and promoted apoptosis by releasing cytochrome C and activating caspase-9/caspase-3 pathway. Functionally, BCL-2 was confirmed as a direct miR-181a target [39]. MiR-182-5p expression levels were higher in blood samples of AML patients than the normal samples. Cellular function indicated miR-182-5p inhibition in AML cells could decrease cell proliferation, promote AML cell apoptosis, and reverse cisplatin (DDP) resistance via targeting BCL2L12 and BCL2 expression [40].
Clinical chemotherapy drugs mainly interfere with cell cycle by inhibiting cellular DNA and RNA synthesis. FoxM1, an established oncogenic factor promoting cell cycle progression, plays a role in this process. MiR-370 expression was decreased in both leukemia cell lines (K562 and HL-60) and primary leukemic cells from patients BM with de novo AML. Ectopic expression of miR-370 in HL60 and K562 cells arrested cell growth and led senescence, while knockout of miR-370 expression promoted the proliferation of those leukemic cells. Mechanistically, miR-370 played a tumor suppressive role by targeting FoxM1. Moreover, when AML cells were treated with 5-aza-2′-deoxycytidine (a DNA methylation inhibitor), upregulation of miR-370 expression was observed, suggesting epigenetic silencing of miR-370 in leukemic cells [41]. Cyclin D1 is a target protein of PTEN signaling pathway. PTEN mainly negatively regulates PI3K/AKT pathway through lipid phosphatase activity, then degrades Cyclin D1, leading to cell cycle organization in G1 phase. MiR-21 may desensitize leukemia cells to chemotherapy by interfering PTEN expression. Bai et al. reported high miR-21 expression in daunorubicin (DNR) resistant cell line K562/DNR. K562/DNR cell line stable transfected with miR-21 inhibitor was induced drug resistance, while inhibition of miR-21 enhanced cell sensitivity to cytotoxicity. Drug resistance mechanism of miR-21 was associated with regulating PTEN protein expression [42].
Chemotherapy drug resistance is also associated with efflux of hydrophobic drugs out of cells. ABC transporter and P-glycoprotein (P-gp), encoded by the MDR1 gene, play pivotal roles in this process [43, 44]. MiR-381 and miR-495 were strongly underexpressed in K562/ADM cells. Restoring expression of miR-381 or miR-495 reduced expression of the MDR1 gene and its protein product P-gp, and increased drug uptake via targeting the 3′-UTR of the MDR1 gene [45]. In the drug-resistant cell line HL-60/VCR, miR-138 was significantly downregulated. Enhanced miR-138 expression significantly downregulated P-gp expression level and MRP1 transcription to promote doxorubicin-induced apoptosis and reversed HL-60/VCR resistance to P-gp dependent and P-gp independent to drug delivery [46]. Besides, Feng et al. found that the expression of miR-331-5p and miR-27a was negatively correlated with MDR1 expression, and the upregulation of miR-331-5p and miR-27a decreased MDR1 expression and increased the sensitivity of K562-resistant cell line to doxorubicin [47].
MicroRNAs and DNA methylation
Aberrant DNA methylation is an important epigenetic modification in the pathogenesis of AML. DNA methyltransferases are mainly divided into two types: DNMT1 and DNMT3. The former maintains methylation, and the latter performs de novo methylation [48]. Garzon et al. demonstrated that miR-29b directly targeted DNMT3A and DNMT3B and indirectly targeted DNMT1, leading to DNA hypomethylation and tumor suppressor gene reactivation [49]. The indirect inhibition of DNMT1 was mediated by a zinc finger-like structural transcription factor SP1, which bound directly to the DNMT1 promoter region to start transcription [50]. MiR-29b downregulates SP1 expression, thereby disrupting SP1-dependent DNMT1 transcription [11]. Another example of DNMTs inhibition was hypomethylating tumor suppressor P115INK4b which could reduce susceptibility to myeloid leukemia in mouse model [51]. Phase 2 data of decitabine in elderly AML patients confirmed that miR-29b upregulation in BM cells could reduce the expression of DNMTs, enhance the effect of DNA hypomethylating agents, and therefore improve the remission rate [52].
MiR-29b could, however, be downregulated by SP1, as well as KIT. KIT overexpression has been observed in various tumors, including AML, and it promotes malignant cell proliferation [53]. Liu et al. identified that aberrant activation of KIT resulted in decreased MYC-dependent miR-29b expression and increased SP1 expression, the latter then interacted with the NF-κB/HDAC complex to further inhibit miR-29b expression and transactivate KIT [54].
Contrary to miR-29b, which suppressed leukemogenesis, miR-221 was able to contribute to the aggressive nature of AML via the NCL/miR-221/NF-κB/DNMT1 network. A group in China designed a nanoparticle that delivered anti-miR-221 antisense RNA in to leukemia cells. The nanoparticle could directly reactivate tumor suppressor gene p27Kip1 by annihilating miR-221 and upregulate other tumor suppressor gene expressions by downregulating DNMT1. In mouse model, the nanoparticle showed promising therapeutic outcome [55].
Gene targets of miRNA may overcome the suppression or even downregulate the respective miRNA by DNA hypermethylation. For example, miR-375 could suppress HOXB3 expression and cause AML cell proliferation arrest and colony reduction. In return, HOXB3 enhanced DNMT3B’s binding to the promoter of miR-375, leading to DNA hypermethylation and lower expression of miR-375 [56].
The role of exosomal microRNAs in acute myeloid leukemia
Exosomes are cell-derived, biologically active membrane-bound vesicles. The role of exosomes in hematopoiesis is receiving increasing attention. In 2015, Hornick et al. identified a set of miRNAs enriched in AML exosomes from the NOD/SCID/IL-2rγnull (NSG) mice serum, such as let-7a, miR-99b, miR-146a, miR-150, miR-155, miR-191, and miR-1246. These serum exosomal miRNAs could potentially be used for early detection of AML [57]. Barrera-Ramirez et al. later sequenced miRNAs from exosomes isolated from AML patients’ marrow samples and from healthy controls. Of the five candidate miRNAs identified by differential packaging in exosomes, miR-26a-5p and miR-101-3p were significantly increased in AML, while miR-23b-5p, miR-339-3p, and miR-425-5p were significantly decreased, but the role and target genes of these exosomal miRNAs were still unknown [58]. Some of them might be AML tumor suppressors. Another study found that exosomes isolated from cultured AML cells or AML mice plasma were enriched with miR-150 and miR-155. Hematopoietic stem/progenitor cells (HSPCs) co-cultured with either of the two exosomes experienced impaired clonogenicity through the miR-150- or miR-155-mediated suppression of c-MYB, a transcription factor involved in HSPC differentiation and proliferation [59]. Moreover, Huan et al. found that the Molm-14 exosome was also enriched in miR-150. This exosome was responsible for decreasing migration of AML cell lines and reducing the surface expression of CXCR4 [60].
Some exosomal miRNAs may promote AML leukemogenesis. In a recent study, miR-7977 was found to have higher levels in AML exosomes than those from normal CD34+ cells. It might be a critical player in disrupting normal hematopoiesis via suppression of poly(rC)-binding protein. It also induced aberrant production of hematopoietic growth factors in mesenchymal stem cells, resulting in a hostile microenvironment for the normal stem cells [61].
Leukemia stem cells (LSCs) are believed to be the primary source of exosomes. Shedding harmful miRNAs via exosomes might be a mechanism of LSCs’ self-protection. Peng et al. discovered that miR-34c-5p was significantly downregulated in AML (excluding APL) stem cells compared to normal HSPCs. Increased expression of miR-34c-5p could induce LSC senescence ex vivo via both p53-dependent and independent CKD/Cyclin pathways. LSC could generate miR-34c-5p deficiency by actively packing and transporting miR-34c-5p out of the cells in exosomes. In return, miR-34c-5p could suppress exosome-mediated transfer via a positive feedback loop through RAB27B, a molecule that promotes exosome shedding. By targeting RAB27B, miR-34c-5p could enrich its intracellular level and induce LSC senescence [62].
MicroRNAs as biomarkers for prognosis in acute myeloid leukemia
miRNAs have many properties of good AML prognostic biomarkers, such as wide presence in various tissues, highly conserved sequences, and easy and sensitive detection, as well as stability under extreme conditions [63, 64]. Mounting studies have shown that miRNAs can be used to predict outcome in CN-AML. Zhang et al. reported miR-216b overexpression as an independently poor prognostic factor for CN-AML and may provide a valuable biomarker associated with disease recurrence in AML [65]. In 224 patients with CN-AML, high miR-362-5p expression was associated with older age and shorter OS compared with low expressers [66]. Diaz-Beya et al. reported that high miR-3151 expression was commonly found in AML patients and obtained shorter disease-free, OS, lower CR rate and higher cumulative incidence of relapse compared with low expressers [67]. The underexpression of miR-328 in AML patients had poor clinical outcome and may provide a diagnostic and prognostic biomarker [68]. MiR-34a expression was negatively correlated with aggressive clinical variable. Patients with low miR-34a expression showed shorter overall and recurrence-free survival [69]. Xu et al. reported miR-135a as an independent prognostic factor for outcome in AML and a tumor suppressor in AML by inversely regulating HOXA10 expression [70]. Moreover, patients with high expression levels of miR-146a and miR-3667 tended to have more favorable prognoses than their low expression counterparts [71], while underexpression of miR-122, miR-192, miR-193b-3p, miR-204, and miR-217, as well as miR-340 had been well studied to be unfavorable prognostic predictors of AML [72,73,74,75,76,77].
Some polymorphic miRNAs only had prognostic impact in certain subtypes. MiR-204 has two sites of variations: one is the upstream flanking region (rs718447 A > G), and the other is the gene itself (rs112062096 A > G). Butrym et al. demonstrated that miR-204 rs718447 GG homozygosity was a risk factor and associated with short survival [78].
Some miRNAs biomarkers might be helpful in selecting patients for allogenic hematopoietic stem cell transplant (allo-HSCT). High miR-425 level was associated with significantly longer OS and event-free survival (EFS) in non-transplant patients, but this association was not observed in post allo-HSCT patients. Instead, patients with downregulated miR-425 did better if they had allo-HSCT, suggesting that low miR-425 level might be an indication for transplant [79]. Overexpression of miR-99a predicted adverse prognosis in AML patients irrespective of transplant status, necessitating the investigation of novel alternative treatment in miR-99a overexpressors [80]. Moreover, high expression of miR-98 correlated with good clinical outcome in AML patients treated with chemotherapy alone [81].
miRNAs have potential prognostic value complementing information gained from gene mutations. MiR-181 family, which has been associated with CEBPA mutations and FLT3-ITD and/or NPM1 wild-type in CN-AML, did demonstrate prognostic value [17]. Marcucci et al. reported favorable clinical outcomes in CN-AML patients with miR-181 overexpression and CEBPA mutations or miR-181 overexpression with FLT3-ITD [82]. In BM mononuclear cells of 113 de novo AML patients, miR-19b overexpression had more frequently occurred and high miR-19b expression had a higher frequency of mutations of U2AF1 and IDH1/2 genes and associated with poor prognosis and disease recurrence in AML [83]. AML patients with low miR-186 expression were frequently observed, and harbored lower complete remission rate and shorter OS, while miR-186high patients had a significantly higher frequency of CEBPA mutation [84]. These findings suggested that measuring miRNA may have potential advantages for predicting prognosis of AML compared to assessed gene mutations such as DMNT3A, FLT3-ITD, NPM1, and CEBPA. In published studies, univariate and multivariate analysis showed that miR-98, miR-99a, miR-340, miR-216b, and miR-34c had independent stronger prognostic impact on EFS and OS (P < 0.05) than gene mutations in FLT3-ITD, NPM1, DMNT3A, RUNX1, CEBPA, and TP53 [80, 81, 85, 86].
To summarize, miRNA researches in AML have yielded important results. The major miRNAs and their roles in AML were listed in Table 1.
Circular RNAs
Circular RNAs (circRNAs) are ubiquitous, stable, and conserved non-coding RNAs. They are closed circular RNA molecules and lack the 3′- and 5′-ends, different from the linear RNAs [141]. This structure was first described in viroids but later was also found in eukaryotic cells [142]. There are four types of circRNAs, namely exonic circRNAs (ecircRNAs), circRNAs from introns, exon-intron circRNAs (EIciRNAs), and intergenic circRNAs [143].
Aberrant circRNA expression levels in acute myeloid leukemia
With the help of sequencing technology, more than 10,000 circRNAs in human have been identified [144, 145]. Aiming to pinpoint circRNAs that correlated with AML, Li et al. [146] used circRNAs microarray and characterized the expression profile of circRNAs in CN-AML, in which 147 circRNAs were upregulated and 317 circRNAs were downregulated compared with healthy control. An interesting phenomenon was that while hsa_circ_0004277 was one of the most significantly downregulated circRNAs in AML, its expression level was restored in patients who achieved complete remission, and the level post-remission was the same as healthy control, but it significantly dropped if the patient became relapse-refractory. Their findings suggested that hsa_circ_0004277 could be a potential diagnostic biomarker in detecting early relapse. Another circRNA, circPVT1, was overexpressed in AML harboring oncogene MYC amplification [147], and this association could hint that circPVT1 might impact the survival of AML patients.
In vitro and in vivo experiments have confirmed that the fusion circRNAs are derived from a fusion gene produced by chromosomal translocation. The study by Guarnerio et al. discovered PML/RARα-derivative f-circPR, and MLL/AF9-derivative f-circM9, and both promoted malignant transformation, chemoresistance, and leukemia cell survival [148]. AML1 transcription factor complex is the most common target for leukemia-associated chromosomal translocations. HIPK2 is part of the AML1 complex and activates AML1-mediated transcription. Li et al. screened mutations of the HIPK2 gene in 50 cases of AML and found two missense mutations (R868W and N958I) of HIPK2 that are localized to nuclear regions with conical or ring shapes [149]. Hirsch et al. detected circular RNAs of NPM1. They found that the circular NPM1 transcript, i.e., has_circ_0075001, had lower expression in healthy volunteers than in AML cell lines, and its expression was positively correlated with total NPM1 expression, but not with the status of NPM1 mutation [150]. Nevertheless, none of the current studies have elucidated the role of circRNA in AML pathogenesis.
The AML-related circRNAs and their roles in AML have been summarized in Table 2.
Long noncoding RNA
Long noncoding RNAs (lncRNAs) are noncoding RNAs that are more than 200 nucleotides in length and lack a meaningful open reading frame [159]. lncRNAs are classified into intergenic lncRNAs, intron lncRNAs, sense lncRNAs, and antisense lncRNAs [160]. In cells, different lncRNAs may act as (1) a signal molecule, expressed at specific time and in specific tissues, regulating the expression of certain genes; (2) a miRNA sponge; (3) a leader molecule, directing RNAs that bind to RNA-binding proteins to reach regulatory sites, and regulating the expression of the relevant gene; and (4) a scaffold molecule, being a central platform for the assembly of other molecules.
lncRNAs involved in acute myeloid leukemia pathogenesis
lncRNAs play an important role in BM cell differentiation and are subjected to differentiation-inducing therapies. HOTAIRM1 and NEAT1 are two important examples. HOTAIRM1 is a myeloid-specific lncRNA that is transcribed from the locus between the HOXA1 and HOXA2 genes. In the initial studies of lncRNAs in AML, HOTAIRM1 was found to be a regulator of myeloid differentiation and maturation by affecting the expression levels of integrin genes such as ITGA4(CD49d) and ITGAX(CD11c). Knocking down HOTAIRM1 would prohibit all-trans retinoic acid (ATRA)-induced granulocyte differentiation [161]. The fact that HOTAIRM1 came from the HOXA cluster might imply that it could regulate nearby genes in the HOXA cluster, although this warranted further investigation. The other lncRNA, NEAT1, was significantly downregulated by PML-RARα in de novo APL samples compared with those of healthy donors. In NB4 cells, silencing NEAT1 could block ATRA-induced differentiation [162]. The roles of HOTAIRM1 and NEAT1 in normal hematopoiesis and leukemogenesis are awaiting further elucidation.
Other lncRNAs participate in regulating AML cell proliferation, cell cycle, and apoptosis. A typical example is lncRNA PVT1 [163]. The coding sequence of PVT1 on the chromosome is adjacent to MYC. Functional acquisition of MYC and PVT1 due to amplification of 8q24.21 is observed in approximately 10% of AML patients [164]. In AML cell lines, overexpression of PVT1 could induce apoptosis and necrosis, probably through downregulating c-MYC expression [165, 166]. UCA1 is another lncRNA that might have the capability to modulate AML cell proliferation; silencing of UCA1 by short hairpin RNA would result in a significantly slower cell proliferation and G1 cell cycle arrest. UCA1 could promote proliferation by inhibiting the expression of the cell cycle regulator p27kip1 [167]. Similarly, CRNDE could coordinate the proliferation and differentiation of AML cells as demonstrated by Wang et al. in their experiment with the U937 cell line [168]. At present, most of the lncRNA studies in AML are ex vivo, and the detailed mechanisms of lncRNA regulating cell proliferation remain to be investigated.
LncRNA expression in AML with recurrent genetic mutations
Distinct lncRNA expression patterns have been observed in different AML subtypes, reflecting the heterogeneity of this disease. AML is most common in older patients (age ≥ 60) although they often have a worse prognosis [169, 170]. Numerous studies have identified characteristic lncRNA profiles in age ≥ 60 CN-AML patients with recurrent genetic mutations such as FLT3-ITD, NPM1, CEBPA, and RUNX1 mutations.
FLT3-ITD-related lncRNAs
Wilms’ tumor 1(WT1) expression positively correlates with FLT3-ITD in patients with AML [171]. Benetatos et al. identified that lncRNA MEG3 could be activated by WT1 and TET2 and it acted as a cofactor of WT1, enhancing leukemogenesis [172].
CEBPA mutation-related lncRNAs
CCAAT/enhancer-binding protein-α (CEBPA) is a critical regulator of myeloid differentiation and 10% of AML have mutations in CEBPA, which may lead to the expression of a 30-kDa dominant negative isoform (C/EBPα-p30) [173]. Hughes et al. identified a C/EBPα-p30 target lncRNA UCA1. It was increased in CN-AML patients with biallelic CEBPA mutations and could promote cell proliferation [167]. Another study reported that HOXB-AS3 was the most downregulated lncRNA in CEBPA-mutated AML while it was upregulated in NPM1-mutated AML [174].
NPM1 mutation-related lncRNAs
Besides the aforementioned HOXB-AS3 [175], the coiled-coil domain containing 26 (CCD26) is also upregulated in the NPM1-mutated AML and is a retinoic acid-dependent modulator of myeloid cell differentiation and death [176]. Apart from them, a recent study employing RNA-sequencing identified another NPM1 mutation-associated lncRNA XLOC_109948 whose high expression predicted a poor prognosis [177].
RUNX1 mutation-related lncRNAs
Fernando et al. first characterized CASC15, a conserved lncRNA upregulated in pediatric AML with RUNX1 mutation. High expression of CASC15 led to myeloid-predominant BM development, decreased engraftment, and colony formation. Researchers also found that CASC15 positively regulated YY1-mediated SOX4 promoter [178].
Prognostic value of lncRNAs in acute myeloid leukemia
LncRNA expression level could predict AML clinical features and outcomes. A published study has confirmed that lncRNAs can assist to predict clinical outcome in older patients with CN-AML. In the basic of 148 CN-older (age > 60 years) AML patients, Garzon et al. evaluated the associations of lncRNA expression with clinical characteristics, gene mutations, and outcome and built a lncRNA score including 48 lncRNAs for independently outcome prognosis [179]. Li et al. reported that SNHG5 overexpression was frequently observed in AML patients with advanced FAB classification and unfavorable cytogenetics. Furthermore, a higher SNHG5 expression level was also associated with shorter OS [180]. Yang et al. have determined the PANDAR expression level and its clinical significance in 119 de novo AML patients. AML patients expressing a higher level of PANDAR were associated with low complete remission rate and adverse prognosis in comparison with those with lower expression of PANDAR [181]. Moreover, high HOTAIR expression was associated with adverse clinical outcomes [182]. Based on 64 de novo non-M3 AML patients, Pashaiefar et al. found that low expression of IRAIN was independently associated with adverse prognosis: higher white blood cell count and blast counts and shorter OS and relapse-free survival. Besides, patients with refractory response to chemotherapies and those with subsequent relapse were more likely to show a lower initial IRAIN expression [183].
TUG1 has been in the spotlight of AML research. Higher TUG1 expression level occurred in AML patients with monosomal karyotype, FLT3-ITD mutation, and poor-risk and correlated with higher white blood cell counts and worse event-free survival and overall survival [184]. Luo et al. investigated the correlation of TUG1 expression with clinicopathological features and its predictive value for treatment response and survival profiles in refractory or relapsed AML patients age ≥ 60 years. They demonstrated that AML patients with higher TUG1 expression had shorter OS, and a lower rate of complete response and overall response than those with lower TUG1 expression [185].
Overall, there are only a few published reports of lncRNAs’ prognostic value in AML; thus, more profound works are required to investigate the association of lncRNAs, clinical characteristics, mutations, and outcome. The researches on AML-related lncRNAs are summarized in Table 3.
lncRNAs and circRNAs can interfere with miRNA function in AML
It has recently been learned that aberrant expression of lncRNAs and circRNAs in AML can change the function of specific miRNAs contributing to initiation, maintenance, and development of leukemogenesis.
In 2011, Salmena et al. proposed a competing endogenous (ceRNA) hypothesis that lncRNAs competitively binds to endogenous miRNAs in AML. A lncRNA, H19, for example, was found overexpressed in BM samples from patients with AML-M2; it promoted AML cell proliferation by sequestering miR-19a/b [186]. The lncRNA NEAT1 that competitively binds miR-23a-3p, an oncogenic miRNA, thus modulating the expression of SMC1A in AML cells, which affected myeloid leukemia cell proliferation and apoptosis [187]. UCA1 is a functional lncRNA that promoted cell proliferation, migration, and invasion of human AML cells via binding miR-126 [188]. In accord with Zhang et al.’s study, its expression was abnormally upregulated following doxorubicin-based chemotherapy and knockdown of UCA1 helped overcome chemoresistance in pediatric AML by suppressing glycolysis via binding miR-125a [189]. FTX is another lncRNA involved in chemoresistance, and it controlled the expression of ALG3 by binding miR-342 [193]. HOXA cluster antisense RNA 2 (HOXA-AS2) was significantly upregulated in BM samples from AML patients after treatment with adriamycin-based chemotherapy and sponged miR-520c-3p to contribute to chemoresistance in AML [195]. An oncogenic activity of lncRNA was also shown by HOTAIR that regulating the expression of c-Kit in AML cells through competitively binding miR-193a, an important tumor-suppressor miRNA to predict a poor clinical outcome [190]. HOTAIRM1, a lncRNA located in the HOXA genomic region, is related to myeloid differentiation which sequestered miR-20a, miR-106b and miR-125b, all of which targets autophagy-associated genes, leading to the degradation of oncoprotein PML-RARA. Moreover, Chen et al. showed that CCAT1 is an oncogenic lncRNA that upregulated c-Myc via its ceRNA activity on miR-155 to repress monocytic differentiation and promote cell growth [192]. The host non-coding transcript of miR-223 of linc-223, found downregulated in AML, is a functional lncRNA which regulated proliferation and differentiation of AML cells by binding miR-125-5p [203].
In recent years, the research of circRNAs, as one of ncRNAs, is focused on their function as “miRNA sponges” in the complex endogenous RNA networks. A circRNA HIPK2, for example, sponged miR-124-3p to regulate the differentiation of all-trans retinoic acid (ATRA)-induced NB4 cells [155]. Chen et al. [151] reported that circANAPC7 was significantly upregulated in AML and used an Arraystar human circRNAs microarray and bioinformatics analysis to predict when ANAPC7 might bind miR-181 family to participate in AML pathogenesis. An oncogenic activity of circRNA was also shown by DLEU2, which was highly expressed in AML, that inhibited miR-496 expression to promote cell proliferation and inhibit cell apoptosis [157]. A circular RNA 100290, which as an oncogenic circRNA was upregulated in AML, showed that it sponged miR-203 to control AML cell proliferation and apoptosis [152]. Moreover, Shang et al. demonstrated the circRNA PAN3 controlled AML chemoresistance by sequestering miR-153-5p and miR-183-5p, [153]. Moreover, Ping et al. showed that circ_0009910, upregulated in AML BM and predicting adverse outcome of AML patients, sponged miR-20a-5p to promote cell proliferation and inhibit [154].
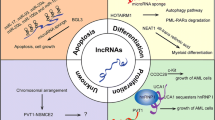
In combination, lncRNAs and circRNAs introduce a complex layer in the miRNA target network, respectively, while lncRNA HOTAIRM1 and circ_0009910 can bind with the same miRNA, miR-20a, to play a different function in AML. The connections of these three ncRNAs involved in AML is shown in Fig. 1. But how lncRNAs and circRNAs compete with each other to bind with the same miRNAs remains unclear, thus making it necessary to further explore the relationship between lncRNAs and circRNAs in AML, to illustrate AML pathogenesis and therapy.
Conclusion
ncRNAs are widely recognized as critical participators in AML pathogenesis. Indeed, specific ncRNA expression could assist clinicians to classify subtypes, to evaluate prognosis, and to predict the response of drug treatment in AML. In this review, we discussed miRNAs, circRNAs, and lncRNAs, involving in subtypes, molecular function, chemoresistance and prognosis in AML, and the interactions between three major ncRNAs. Currently, the role of miRNAs in AML is most studied, but the mechanisms of miRNAs in AML still remain complex and unclear owing to miRNAs target genes ranging from tens to hundreds and involving different signaling pathways. In recent years, lncRNAs and circRNAs are introduced into miRNA network one after another and can be used as ceRNA of miRNAs and miRNAs sponge to regulate miRNA expression in AML. In our review, we reported that some lncRNAs such as UCA1 and linc223 could target the same miRNA, miR-125, to control proliferation, apoptosis, and differentiation, and lncRNA HOTAIRM1 participated in autophagy pathway by binding with miR-125. MiR-125 has been reported to promote MLL-AF9-driven murine AML by TET2-VEGFA pathway and target autophagy-associated genes, leading to the degradation of oncoprotein PML-RARA. CircRNA_0009910 could also bind miR-20 via competing with lncRNA HOTAIRM1 to regulate proliferation and apoptosis. However, whether these three lncRNAs directly affect MLL-AF9-driven AML and autophagy, the target genes of miR-20 are not clear. Thus it is important to find the crossover miRNAs of the three ncRNAs to help illustrate the connections among these three ncRNAs. However, currently, there is very little literature on this subject and the connection networks of the three ncRNAs are required for further study. Subsequently, we will also trace relative studies and update the interaction networks of miRNAs, lncRNAs, and circRNAs.
Abbreviations
- 3′-UTR:
-
3′-untranslated region
- ABC:
-
ATP-binding cassette
- allo-HSCT:
-
Allogenic hematopoietic stem cell transplant
- AML:
-
Acute myeloid leukemia
- ATRA:
-
All-trans retinoic acid
- BM:
-
Bone marrow
- C/EBPα-p30:
-
30-kDa dominant negative isoform
- CCD26:
-
Coiled-coil domain containing 26
- CEBPA:
-
CAAT/enhancer-binding protein-α
- ceRNA:
-
Competing endogenous RNA
- circRNA:
-
Circular RNA
- CN-AML:
-
Cytogenetic normal AML
- DNR:
-
Daunorubicin
- EFS:
-
Event-free survival
- EZH2:
-
Zeste homolog 2
- FAB:
-
French-American-British
- HOXA-AS2:
-
HOXA cluster antisense RNA 2.
- HSPCs:
-
Hematopoietic stem/progenitor cells
- LncRNA:
-
Long noncoding RNA
- LSCs:
-
Leukemia stem cells
- miRNA:
-
MicroRNA
- MLL:
-
Mixed lineage leukemia
- ncRNA:
-
Noncoding RNA
- OS:
-
Overall survival
- PB:
-
Peripheral blood
- P-gp:
-
P-glycoprotein
- WT1:
-
Wilms' tumor 1
References
Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136–52.
Burnett A, Wetzler M, Löwenberg B. Therapeutic advances in acute myeloid leukemia. J Clin Oncol. 2011;29(5):487–94.
Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297(5589):2053–6.
International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–45.
Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Bailey DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, Gingeras TR. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308(5725):1149–54.
Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–8.
Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–74.
Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–4.
Wallace JA, O'Connell RM. MicroRNAs and acute myeloid leukemia: therapeutic implications and emerging concepts. Blood. 2017;130(11):1290–301.
Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–405.
Chen P, Price C, Li Z, Li Y, Cao D, Wiley A, He C, Gurbuxani S, Kunjamma RB, Huang H, Jiang X, Arnovitz S, Xu M, Hong GM, Elkahloun AG, Neilly MB, Wunderlich M, Larson RA, Le Beau MM, Mulloy JC, Liu PP, Rowley JD, Chen J. miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia-rearranged leukemia. Proc Natl Acad Sci U S A. 2013;110(28):11511–6.
Emmrich S, Katsman-Kuipers JE, Henke K, Khatib ME, Jammal R, Engeland F, Dasci F, Zwaan CM, den Boer ML, Verboon L, Stary J, Baruchel A, de Haas V, Danen-van Oorschot AA, Fornerod M, Pieters R, Reinhardt D, Klusmann JH, van den Heuvel-Eibrink MM. miR-9 is a tumor suppressor in pediatric AML with t(8;21). Leukemia. 2014;28(5):1022–32.
Fu L, Shi J, Liu A, Zhou L, Jiang M, Fu H, Xu K, Li D, Deng A, Zhang Q, Pang Y, Guo Y, Hu K, Zhou J, Wang Y, Huang W, Jing Y, Dou L, Wang L, Xu K, Ke X, Nervi C, Li Y, Yu L. A minicircuitry of microRNA-9-1 and RUNX1-RUNX1T1 contributes to leukemogenesis in t(8;21) acute myeloid leukemia. Int J Cancer. 2017;140(3):653–61.
Bi L, Sun L, Jin Z, Zhang S, Shen Z. MicroRNA-10a/b are regulators of myeloid differentiation and acute myeloid leukemia. Oncol Lett. 2018;15(4):5611–9.
Zhang TJ, Guo H, Zhou JD, Li XX, Zhang W, Ma JC, Wen XM, Yao XY, Lin J, Qian J. Bone marrow miR-10a overexpression is associated with genetic events but not affects clinical outcome in acute myeloid leukemia. Pathol Res Pract. 2018;214(1):169–73.
Marcucci G, Maharry K, Radmacher MD, Mrózek K, Vukosavljevic T, Paschka P, Whitman SP, Langer C, Baldus CD, Liu CG, Ruppert AS, Powell BL, Carroll AJ, Caligiuri MA, Kolitz JE, Larson RA, Bloomfield CD. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B Study. J Clin Oncol. 2008;26(31):5078–87.
Marcucci G, Radmacher MD, Maharry K, Mrózek K, Ruppert AS, Paschka P, Vukosavljevic T, Whitman SP, Baldus CD, Langer C, Liu CG, Carroll AJ, Powell BL, Garzon R, Croce CM, Kolitz JE, Caligiuri MA, Larson RA, Bloomfield CD. MicroRNA expression in cytogenetically normal acute myeloid leukemia. New Engl J Med. 2008;358(18):1919–28.
Li ZLJ, Sun M, Mi S, Zhang H, Luo RT, Chen P, Wang Y, Yan M, Qian Z, Neilly MB, Jin J, Zhang Y, Bohlander SK, Zhang DE, Larson RA, Le Beau MM, Thirman MJ, Golub TR, Rowley JD, Chen J. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci U S A. 2008;105(40):15535–40.
Li Z, Huang H, Li Y, Jiang X, Chen P, Arnovitz S, Radmacher MD, Maharry K, Elkahloun A, Yang X, He C, He M, Zhang Z, Dohner K, Neilly MB, Price C, Lussier YA, Zhang Y, Larson RA, Le Beau MM, Caligiuri MA, Bullinger L, Valk PJ, Delwel R, Lowenberg B, Liu PP, Marcucci G, Bloomfield CD, Rowley JD, Chen J. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. 2012;119(10):2314–24.
Gerloff D, Grundler R, Wurm AA, Brauer-Hartmann D, Katzerke C, Hartmann JU, Madan V, Muller-Tidow C, Duyster J, Tenen DG, Niederwieser D, Behre G. NF-kappaB/STAT5/miR-155 network targets PU.1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia. 2015;29(3):535–47.
Yang J, Yuan Y, Yang X, Hong Z, Yang L. Decreased expression of microRNA-122 is associated with an unfavorable prognosis in childhood acute myeloid leukemia and function analysis indicates a therapeutic potential. Pathol Res Pract. 2017;213(9):1166–72.
Xu L, Guo Y, Yan W, Cen J, Niu Y, Yan Q, He H, Chen CS, Hu S. High level of miR-196b at newly diagnosed pediatric acute myeloid leukemia predicts a poor outcome. EXCLI J. 2017;16:197–209.
Lutherborrow M, Bryant A, Jayaswal V, Agapiou D, Palma C, Yang YH, Ma DD. Expression profiling of cytogenetically normal acute myeloid leukemia identifies microRNAs that target genes involved in monocytic differentiation. Am J Hematol. 2011;86(1):2–11.
Ramkissoon SH, Mainwaring LA, Ogasawara Y, Keyvanfar K, McCoy JP Jr, Sloand EM, Kajigaya S, Young NS. Hematopoietic-specific microRNA expression in human cells. Leuk Res. 2006;30(5):643–7.
Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-McIver A, Young BD. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21(5):912–6.
Hong Z, Zhang R, Qi H. Diagnostic and prognostic relevance of serum miR-195 in pediatric acute myeloid leukemia. Cancer Biomark. 2018;21(2):269–75.
Liao Q, Wang B, Li X, Jiang G. miRNAs in acute myeloid leukemia. Oncotarget. 2017;8(2):3666–82.
Xue H, Hua LM, Guo M, Luo JM. SHIP1 is targeted by miR-155 in acute myeloid leukemia. Oncol Rep. 2014;32(5):2253–9.
Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, Smith KG, Rada C, Enright AJ, Toellner KM, Maclennan IC, Turner M. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27(6):847–59.
Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, Mallardo M. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 2008;36(20):6608–19.
Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, Manfredi M, Narayanan U, Rolfe M, Staudt LM, Soucy TA, Yu J, Zhang J, Bolen JB, Smith PG. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood. 2010;116(9):1515–23.
Schneider E, Staffas A, Röhner L, Krowiorz K, Heuser M, Döhner K, Bullinger L, Döhner H, Fogelstrand L, Rouhi A, Kuchenbauer F, Palmqvist L. miR-155 is also upregulated in MLL-rearranged AML but its absence does not affect leukemia development. Exp Hematol. 2016;44(12):1166–71.
Nowek K, Sun SM, Bullinger L, Bindels EM, Exalto C, Dijkstra MK, van Lom K, Döhner H, Erkeland SJ, Löwenberg B, Jongen-Lavrencic M. Aberrant expression of miR-9/9* in myeloid progenitors inhibits neutrophil differentiation by post-transcriptional regulation of ERG. Leukemia. 2016;30(1):229–37.
Tian C, You MJ, Yu Y, Zhu L, Zheng G, Zhang Y. MicroRNA-9 promotes proliferation of leukemia cells in adult CD34-positive acute myeloid leukemia with normal karyotype by downregulation of Hes1. Tumour Biol. 2016;37(6):7461–71.
Li Y, Gao L, Luo X, Wang L, Gao X, Wang W, Sun J, Dou L, Li J, Xu C, Wang L, Zhou M, Jiang M, Zhou J, Caligiuri MA, Nervi C, Bloomfield CD, Marcucci G, Yu L. Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t(8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood. 2013;121(3):499–509.
Zheng Z, Zheng X, Zhu Y, Gu X, Gu W, Xie X, Hu W, Jiang J. miR-183-5p inhibits occurrence and progression of acute myeloid leukemia via targeting erbin. Mol Ther. 2019;27(3):542–58.
Zhang Y, Zhou SY, Yan HZ, Xu DD, Chen HX, Wang XY, Wang X, Liu YT, Zhang L, Wang S, Zhou PJ, Fu WY, Ruan BB, Ma DL, Wang Y, Liu QY, Ren Z, Liu Z, Zhang R, Wang YF. miR-203 inhibits proliferation and self-renewal of leukemia stem cells by targeting survivin and Bmi-1. Sci Rep. 2016;6:19995.
Li H, Hui L, Xu W. miR-181a sensitizes a multidrug-resistant leukemia cell line K562/A02 to daunorubicin by targeting BCL-2. Acta Biochim Biophys Sin (Shanghai). 2012;44(3):269–77.
Bai H, Cao Z, Deng C, Zhou L, Wang C. miR-181a sensitizes resistant leukaemia HL-60/Ara-C cells to Ara-C by inducing apoptosis. J Cancer Res Clin Oncol. 2012;138(4):595–602.
Zhang S, Zhang Q, Shi G, Yin J. MiR-182-5p regulates BCL2L12 and BCL2 expression in acute myeloid leukemia as a potential therapeutic target. Biomed Pharmacother. 2018;97:1189–94.
Zhang X, Zeng J, Zhou M, Li B, Zhang Y, Huang T, Wang L, Jia J, Chen C. The tumor suppressive role of miRNA-370 by targeting FoxM1 in acute myeloid leukemia. Mol Cancer. 2012;11:56.
Bai H, Xu R, Cao Z, Wei D, Wang C. Involvement of miR-21 in resistance to daunorubicin by regulating PTEN expression in the leukaemia K562 cell line. FEBS Lett. 2011;585(2):402-8.
Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5(3):219–34.
Xu Y, Zhi F, Xu G, Tang X, Lu S, Wu J, Hu Y. Overcoming multidrug-resistance in vitro and in vivo using the novel P-glycoprotein inhibitor 1416. Biosci Rep. 2012;32(6):559–66.
Xu Y, Ohms SJ, Li Z, Wang Q, Gong G, Hu Y, Mao Z, Shannon MF, Fan JY. Changes in the expression of miR-381 and miR-495 are inversely associated with the expression of the MDR1 gene and development of multi-drug resistance. PLoS One. 2013;8(11):e82062.
Zhao X, Yang L, Hu J, Ruan J. miR-138 might reverse multidrug resistance of leukemia cells. Leuk Res. 2010;34(8):1078–82.
Feng DD, Zhang H, Zhang P, Zheng YS, Zhang XJ, Han BW, Luo XQ, Xu L, Zhou H, Qu LH, Chen YQ. Down-regulated miR-331–5p and miR-27a are associated with chemotherapy resistance and relapse in leukaemia. J Cell Mol Med. 2011;15(10):2164–75.
Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature reviews Genetics. 2002;3(6):415–28.
Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, Havelange V, Volinia S, Blum W, Rush LJ, Perrotti D, Andreeff M, Bloomfield CD, Byrd JC, Chan K, Wu LC, Croce CM, Marcucci G. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113(25):6411–8.
Liu S, Liu Z, Xie Z, Pang J, Yu J, Lehmann E, Huynh L, Vukosavljevic T, Takeki M, Klisovic RB, Baiocchi RA, Blum W, Porcu P, Garzon R, Byrd JC, Perrotti D, Caligiuri MA, Chan KK, Wu LC, Marcucci G. Bortezomib induces DNA hypomethylation and silenced gene transcription by interfering with Sp1/NF-kappaB-dependent DNA methyltransferase activity in acute myeloid leukemia. Blood. 2008;111(4):2364–73.
Wolff L, Garin MT, Koller R, Bies J, Liao W, Malumbres M, Tessarollo L, Powell D, Perella C. Hypermethylation of the Ink4b locus in murine myeloid leukemia and increased susceptibility to leukemia in p15(Ink4b)-deficient mice. Oncogene. 2003;22(58):9265–74.
Blum W, Garzon R, Klisovic RB, Schwind S, Walker A, Geyer S, Liu S, Havelange V, Becker H, Schaaf L, Mickle J, Devine H, Kefauver C, Devine SM, Chan KK, Heerema NA, Bloomfield CD, Grever MR, Byrd JC, Villalona-Calero M, Croce CM, Marcucci G. Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. Proc Natl Acad Sci U S A. 2010;107(16):7473–8.
Mark A. Lemmon, Joseph Schlessinger. Cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–34.
Liu S, Wu LC, Pang J, Santhanam R, Schwind S, Wu YZ, Hickey CJ, Yu J, Becker H, Maharry K, Radmacher MD, Li C, Whitman SP, Mishra A, Stauffer N, Eiring AM, Briesewitz R, Baiocchi RA, Chan KK, Paschka P, Caligiuri MA, Byrd JC, Croce CM, Bloomfield CD, Perrotti D, Garzon R, Marcucci G. Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell. 2010;17(4):333–47.
Deng R, Shen N, Yang Y, Yu H, Xu S, Yang YW, Liu S, Meguellati K, Yan F. Targeting epigenetic pathway with gold nanoparticles for acute myeloid leukemia therapy. Biomaterials. 2018;167:80–90.
Bi L, Zhou B, Li H, He L, Wang C, Wang Z, Zhu L, Chen M, Gao S. A novel miR-375-HOXB3-CDCA3/ DNMT3B regulatory circuitry contributes to leukemogenesis in acute myeloid leukemia. BMC Cancer. 2018;18(1):182.
Hornick NI, Huan J, Doron B, Goloviznina NA, Lapidus J, Chang BH, Kurre P. Serum exosome icroRNA as a minimally-invasive early biomarker of AML. Sci Rep. 2015;5:11295.
Barrera-Ramirez J, Lavoie JR, Maganti HB, Stanford WL, Ito C, Sabloff M, Brand M, Rosu-Myles M, Le Y, Allan DS. Micro-RNA profiling of exosomes from marrow-derived mesenchymal stromal cells in patients with acute myeloid leukemia: implications in leukemogenesis. Stem Cell Rev. 2017;13(6):817–25.
Hornick NI, Doron B, Abdelhamed S, Huan J, Harrington CA, Shen R, Cambronne XA, Chakkaramakkil Verghese S, Kurre P. AML suppresses hematopoiesis by releasing exosomes that contain microRNAs targeting c-MYB. Sci Signal. 2016;9(444):ra88.
Huan J, Hornick NI, Shurtleff MJ, Skinner AM, Goloviznina NA, Roberts CT Jr, Kurre P. RNA trafficking by acute myelogenous leukemia exosomes. Cancer Res. 2013;73(2):918–29.
Horiguchi H, Kobune M, Kikuchi S, Yoshida M, Murata M, Murase K, Iyama S, Takada K, Sato T, Ono K, Hashimoto A, Tatekoshi A, Kamihara Y, Kawano Y, Miyanishi K, Sawada N, Kato J. Extracellular vesicle miR-7977 is involved in hematopoietic dysfunction of mesenchymal stromal cells via poly(rC) binding protein 1 reduction in myeloid neoplasms. Haematologica. 2016;101(4):437–47.
Peng D, Wang H, Li L, Ma X, Chen Y, Zhou H, Luo Y, Xiao Y, Liu L. miR-34c-5p promotes eradication of acute myeloid leukemia stem cells by inducing senescence through selective RAB27B targeting to inhibit exosome shedding. Leukemia. 2018;32(5):1180–8.
He Y, Lin J, Kong D, Huang M, Xu C, Kim TK, Etheridge A, Luo Y, Ding Y, Wang K. Current state of circulating MicroRNAs as cancer biomarkers. Clin Chem. 2015;61(9):1138–55.
Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Circulating miRNAs: cell-cell communication function. Front Genet. 2013;4:119.
Zhang TJ, Wu DH, Zhou JD, Li XX, Zhang W, Guo H, Ma JC, Deng ZQ, Lin J, Qian J. Overexpression of miR-216b: prognostic and predictive value in acute myeloid leukemia. J Cell Physiol. 2018;233(4):3274–81.
Ma QL, Wang JH, Yang M, Wang HP, Jin J. MiR-362-5p as a novel prognostic predictor of cytogenetically normal acute myeloid leukemia. J Transl Med. 2018;16(1):68.
Diaz-Beya M, Brunet S, Nomdedeu J, Cordeiro A, Tormo M, Escoda L, Ribera JM, Arnan M, Heras I, Gallardo D, Bargay J, Queipo de Llano MP, Salamero O, Marti JM, Sampol A, Pedro C, Hoyos M, Pratcorona M, Castellano JJ, Nomdedeu M, Risueno RM, Sierra J, Monzo M, Navarro A, Esteve J. The expression level of BAALC-associated microRNA miR-3151 is an independent prognostic factor in younger patients with cytogenetic intermediate-risk acute myeloid leukemia. Blood Cancer J. 2015;5:e352.
Liu L, Chen R, Zhang Y, Fan W, Xiao F, Yan X. Low expression of circulating microRNA-328 is associated with poor prognosis in patients with acute myeloid leukemia. Diagn Pathol. 2015;10:109.
Huang Y, Zou Y, Lin L, Ma X, Chen H. Identification of serum miR-34a as a potential biomarker in acute myeloid leukemia. Cancer Biomark. 2018;22(4):799–805.
Xu H, Wen Q. Downregulation of miR-135a predicts poor prognosis in acute myeloid leukemia and regulates leukemia progression via modulating HOXA10 expression. Mol Med Rep. 2018;18(1):1134–40.
Zhu R, Lin W, Zhao W, Fan F, Tang L, Hu Y. A 4-microRNA signature for survival prognosis in pediatric and adolescent acute myeloid leukemia. J Cell Biochem. 2019;120(3):3958–68.
Zhang TJ, Qian Z, Wen XM, Zhou JD, Li XX, Xu ZJ, Ma JC, Zhang ZH, Lin J, Qian J. Lower expression of bone marrow miR-122 is an independent risk factor for overall survival in cytogenetically normal acute myeloid leukemia. Pathol Res Pract. 2018;214(6):896–901.
Tian C, Zhang L, Li X, Zhang Y, Li J, Chen L. Low miR-192 expression predicts poor prognosis in pediatric acute myeloid leukemia. Cancer Biomark. 2018;22(2):209–15.
Bhayadia R, Krowiorz K, Haetscher N, Jammal R, Emmrich S, Obulkasim A, Fiedler J, Schwarzer A, Rouhi A, Heuser M, Wingert S, Bothur S, Döhner K, Mätzig T, Ng M, Reinhardt D, Döhner H, Zwaan CM, van den Heuvel Eibrink M, Heckl D, Fornerod M, Thum T, Humphries RK, Rieger MA, Kuchenbauer F, Klusmann JH. Endogenous tumor suppressor microRNA-193b: Therapeutic and prognostic value in acute myeloid leukemia. J Clin Oncol. 2018;36(10):1007–16.
Butrym A, Rybka J, Baczyńska D, Tukiendorf A, Kuliczkowski K, Mazur G. Low expression of microRNA-204 (miR-204) is associated with poor clinical outcome of acute myeloid leukemia (AML) patients. J Exp Clin Cancer Res. 2015;34:68.
Yan J, Wu G, Chen J, Xiong L, Chen G, Li P. Downregulated miR-217 expression predicts a poor outcome in acute myeloid leukemia. Cancer Biomark. 2018;22(1):73–8.
Wang Q, Feng T, Xu J, Miao MH, Ji XQ, Zhu H, Shao XJ. Low expression of microRNA-340 confers adverse clinical outcome in patients with acute myeloid leukemia. J Cell Physiol. 2019;234(4):4200–5.
Butrym A, Łacina P, Kuliczkowski K, Bogunia-Kubik K, Mazur G. Genetic variation of the gene coding for microRNA-204 (miR-204) is a risk factor in acute myeloid leukaemia. BMC Cancer. 2018;18(1):107.
Yang C, Shao T, Zhang H, Zhang N, Shi X, Liu X, Yao Y, Xu L, Zhu S, Cao J, Cheng H, Yan Z, Li Z, Niu M, Xu K. MiR-425 expression profiling in acute myeloid leukemia might guide the treatment choice between allogeneic transplantation and chemotherapy. J Transl Med. 2018;16(1):267.
Cheng Z, Zhou L, Hu K, Dai Y, Pang Y, Zhao H, Wu S, Qin T, Han Y, Hu N, Chen L, Wang C, Zhang Y, Wu D, Ke X, Shi J, Fu L. Prognostic significance of microRNA-99a in acute myeloid leukemia patients undergoing allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant. 2018;53(9):1089–95.
Hu N, Cheng Z, Pang Y, Zhao H, Chen L, Wang C, Qin T, Li Q, Han Y, Shi J, Fu L. High expression of MiR-98 is a good prognostic factor in acute myeloid leukemia patients treated with chemotherapy alone. J Cancer. 2019;10(1):178–85.
Marcucci G, Maharry K, Radmacher MD, Mrózek K, Vukosavljevic T, Paschka P, Whitman SP, Langer C, Baldus CD, Liu CG, Ruppert AS, Powell BL, Carroll AJ, Caligiuri MA, Kolitz JE, Larson RA. CD. B. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B Study. J Clin Oncol. 2008;26(31):5078–87.
Zhang TJ, Lin J, Zhou JD, Li XX, Zhang W, Guo H, Xu ZJ, Yan Y, Ma JC, Qian J. High bone marrow miR-19b level predicts poor prognosis and disease recurrence in de novo acute myeloid leukemia. Gene. 2018;640:79–85.
Zhang TJ, Wang YX, Yang DQ, Yao DM, Yang L, Zhou JD, Deng ZQ, Wen XM, Guo H, Ma JC, Lin J, Qian J. Down-regulation of miR-186 correlates with poor survival in de novo acute myeloid leukemia. Clin Lab. 2016;62(1-2):113–20.
Zhang TJ, Wu DH, Zhou JD. Overexpression of miR-216b: prognostic and predictive value in acute myeloid leukemia. J Cell Physiol. 2018;233(4):3274–81.
Yang DQ, Zhou JD, Wang YX, Deng ZQ, Yang J, Yao DM, Qian Z, Yang L, Lin J, Qian J. Low miR-34c expression is associated with poor outcome in de novo acute myeloid leukemia. Int J Lab Hematol. 2017;39(1):42–50.
Zhou L, Fu L, Lv N, Chen XS, Liu J, Li Y, Xu QY, Huang S, Zhang XD, Dou LP, Wang LL, Li YH, Yu L. A minicircuitry comprised of microRNA-9 and SIRT1 contributes to leukemogenesis in t(8;21) acute myeloid leukemia. Eur Rev Med Pharmacol Sci. 2017;21(4):786–94.
Senyuk V, Zhang Y, Liu Y, Ming M, Premanand K, Zhou L, Chen P, Chen J, Rowley JD, Nucifora G, Qian Z. Critical role of miR-9 in myelopoiesis and EVI1-induced leukemogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(14):5594–9.
Li G, Song Y, Li G, Ren J, Xie J, Zhang Y, Gao F, Mu J, Dai J. Downregulation of microRNA-21 expression inhibits proliferation, and induces G1 arrest and apoptosis via the PTEN/AKT pathway in SKM-1cells. Mol Med Rep. 2018;18(3):2771–9.
Jiang X, Hu C, Arnovitz S, Bugno J, Yu M, Zuo Z, Chen P, Huang H, Ulrich B, Gurbuxani S, Weng H, Strong J, Wang Y, Li Y, Salat J, Li S, Elkahloun AG, Yang Y, Neilly MB, Larson RA, Le Beau MM, Herold T, Bohlander SK, Liu PP, Zhang J, Li Z, He C, Jin J, Hong S, Chen J. miR-22 has a potent anti-tumour role with therapeutic potential in acute myeloid leukaemia. Nat Commun. 2016;7:11452.
Gong JN, Yu J, Lin HS, Zhang XH, Yin XL, Xiao Z, Wang F, Wang XS, Su R, Shen C, Zhao HL, Ma YN, Zhang JW. The role, mechanism and potentially therapeutic application of microRNA-29 family in acute myeloid leukemia. Cell Death Differ. 2014;21(1):100–12.
Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T, Zanesi N, Kornblau SM, Marcucci G, Calin GA, Andreeff M, Croce CM. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;18(3):E597.
Eyholzer M, Schmid S, Wilkens L, Mueller BU, Pabst T. The tumour-suppressive miR-29a/b1 cluster is regulated by CEBPA and blocked in human AML. Br J Cancer. 2010;103(2):275–84.
Scoville SD, Nalin AP, Chen L, Chen L, Zhang MH, McConnell K, Beceiro Casas S, Ernst G, Traboulsi AA, Hashi N, Williams M, Zhang X, Hughes T, Mishra A, Benson DM, Saultz JN, Yu J, Freud AG, Caligiuri MA, Mundy-Bosse BL. Human AML activates the AHR pathway to impair NK cell development and function. Blood. 2018;132(17):1792–804.
Pulikkan JA, Peramangalam PS, Dengler V, Ho PA, Preudhomme C, Meshinchi S, Christopeit M, Nibourel O, Müller-Tidow C, Bohlander SK, Tenen DG, Behre G. C/EBPα regulated microRNA-34a targets E2F3 during granulopoiesis and is down-regulated in AML with CEBPA mutations. Blood. 2010;116(25):5638–49.
Pyzer AR, Stroopinsky D, Rosenblatt J, Anastasiadou E, Rajabi H, Washington A, Tagde A, Chu JH, Coll M, Jiao AL, Tsai LTT, enen DE, Cole L, Palmer K, Ephraim A, Leaf RK, Nahas M, Apel A, Bar-Natan M, Jain S, McMasters M, Mendez L, Arnason J, Raby BA, Slack F, Kufe D, Avigan D. MUC1 inhibition leads to decrease in PD-L1 levels via upregulation of miRNAs. Leukemia. 2017;31(12):2780–90.
Liu L, Ren W, Chen K. MiR-34a promotes apoptosis and inhibits autophagy by targeting HMGB1 in acute myeloid leukemia cells. Cell Physiol Biochem. 2017;41(5):1981–92.
Pigazzi M, Manara E, Baron E, Basso G. MiR-34b targets cyclic AMP-responsive element binding protein in acute myeloid leukemia. Cancer Res. 2009;69(6):2471–8.
Si X, Zhang X, Hao X, Li Y, Chen Z, Ding Y, Shi H, Bai J, Gao Y, Cheng T, Yang FC, Zhou Y. Upregulation of miR-99a is associated with poor prognosis of acute myeloid leukemia and promotes myeloid leukemia cell expansion. Oncotarget. 2016;7(4):78095–109.
Khalaj M, Woolthuis CM, Hu W, Durham BH, Chu SH, Qamar S, Armstrong SA, Park CY. miR-99 regulates normal and malignant hematopoietic stem cell self-renewal. J Exp Med. 2017;8:126. https://doi.org/10.1186/s13045-015-0223-4.
Zhang L, Li X, Ke Z, Huang L, Liang Y, Wu J, Zhang X, Chen Y, Zhang H, Luo X. MiR-99a may serve as a potential oncogene in pediatric myeloid leukemia. Cancer Cell Int. 2013;13(1):110.
Wan L, Tian Y, Zhang R, Peng Z, Sun J, Zhang W. MicroRNA-103 confers the resistance to long-treatment of driamycin to human leukemia cells by regulation of COP1. J Cell Biochem. 2018;119(5):3843–52.
Bousquet M, Quelen C, Rosati R, Mansat-De Mas V, La Starza R, Bastard C, Lippert E, Talmant P, Lafage-Pochitaloff M, Leroux D, Gervais C, Viguié F, Lai JL, Terre C, Beverlo B, Sambani C, Hagemeijer A, Marynen P, Delsol G, Dastugue N, Mecucci C, Brousset P. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J Exp Med. 2008;205(11):2499–506.
Chaudhuri AA, So AY, Mehta A, Minisandram A, Sinha N, Jonsson VD, Rao DS, O'Connell RM, Baltimore D. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin28A. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(11):4233–8.
So AY, Sookram R, Chaudhuri AA, Minisandram A, Cheng D, Xie C, Lim EL, Flores YG, Jiang S, Kim JT, Keown C, Ramakrishnan P, Baltimore D. Dual mechanisms by which miR-125b represses IRF4 to induce myeloid and B-cell leukemias. Blood. 2014;124(9):1502–12.
Hu J, Zheng L, Shen X, Zhang Y, Li C, Xi T. MicroRNA-125b inhibits AML cells differentiation by directly targeting Fes. Gene. 2017;620:1–9.
Wang Y, Tang P, Chen Y, Chen J, Ma R, Sun L. Overexpression of microRNA-125b inhibits human acute myeloid leukemia cells invasion, proliferation and promotes cells apoptosis by targeting NF-κB signaling pathway. Biochem Biophys Res Commun. 2017;488(1):60–6.
Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, Chen P, Wang Y, Yan M, Qian Z, Neilly MB, Jin J, Zhang Y, Bohlander SK, Zhang DE, Larson RA, Le Beau MM, Thirman MJ, Golub TR, Rowley JD, Chen J. Distinct microRNA expression profiles in acute myeloid leukemia with common translocation. Proc Natl Acad Sci USA. 2008;105(40):15535–40.
de Leeuw DC, Denkers F, Olthof MC, Rutten AP, Pouwels W, Schuurhuis GJ, Ossenkoppele GJ, Smit L. Attenuation of microRNA-126 expression that drives CD34+38- stem/progenitor cells in acute myeloid leukemia leads to tumor eradication. Cancer Res. 2014;74(7):2094–105.
Li Z, Chen P, Su R, Li Y, Hu C, Wang Y, Arnovitz S, He M, Gurbuxani S, Zuo Z, Elkahloun AG, Li S, Weng H, Huang H, Neilly MB, Wang S, Olson EN, Larson RA, Le Beau MM, Zhang J, Jiang X, Wei M, Jin J, Liu PP, Chen J. Overexpression and knockout of miR-126 both promote leukemogenesis. Blood. 2015;126(17):2005–15.
Dorrance AM, Neviani P, Ferenchak GJ, Huang X, Nicolet D, Maharry KS, Ozer HG, Hoellarbauer P, Khalife J, Hill EB, Yadav M, Bolon BN, Lee RJ, Lee LJ, Croce CM, Garzon R, Caligiuri MA, Bloomfield CD, Marcucci G. Targeting leukemia stem cells in vivo with antagomiR-126 nanoparticles in acute myeloid leukemia. Leukemia. 2015;29(11):2143–53.
Lechman ER, Gentner B, Ng SW, Schoof EM, van Galen P, Kennedy JA, Nucera S, Ciceri F, Kaufmann KB, Takayama N, Dobson SM, Trotman-Grant A, Krivdova G, Elzinga J, Mitchell A, Nilsson B, Hermans KG, Eppert K, Marke R, Isserlin R, Voisin V, Bader GD, Zandstra PW, Golub TR, Ebert BL, Lu J, Minden M, Wang JC, Naldini L, Dick JE. miR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell. 2016;29(2):214–28.
Ding Q, Wang Q, Ren Y, Zhu HQ, Huang Z. MicroRNA-126 attenuates cell apoptosis by targeting TRAF7 in acute myeloid leukemia cells. Biochem Cell Biol. 2018.
Emmrich S, Engeland F, El-Khatib M, Henke K, Obulkasim A, Schöning J, Katsman-Kuipers JE, Michel Zwaan C, Pich A, Stary J, Baruchel A, de Haas V, Reinhardt D, Fornerod M, van den Heuvel-Eibrink MM, Klusmann JH. miR-139-5p controls translation in myeloid leukemia through EIF4G2. Oncogene. 2016;35(14):1822–31.
Hartmann JU, Bräuer-Hartmann D, Kardosova M, Wurm AA, Wilke F, Schödel C, Gerloff D, Katzerke C, Krakowsky R, Namasu CY, Bill M, Schwind S, Müller-Tidow C, Niederwieser D, Alberich-Jorda M, Behre G. MicroRNA-143 targets ERK5 in granulopoiesis and predicts outcome of patients with acute myeloid leukemia. Cell Death Dis. 2018;9(8):814.
Sun X, Liu D, Xue Y, Hu X. Enforced miR-144-3p expression as a non-invasive biomarker for the acute myeloid leukemia patients mainly by targeting NRF2. Clin Lab. 2017;63(4):679–87.
Starczynowski DT, Kuchenbauer F, Argiropoulos B, Sung S, Morin R, Muranyi A, Hirst M, Hogge D, Marra M, Wells RA, Buckstein R, Lam W, Humphries RK, Karsan A. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16(1):49–58.
Fang J, Barker B, Bolanos L, Liu X, Jerez A, Makishima H, Christie S, Chen X, Rao DS, Grimes HL, Komurov K, Weirauch MT, Cancelas JA, Maciejewski JP, Starczynowski DT. Myeloid malignancies with chromosome 5q deletions acquire a dependency on an intrachromosomal NF-κB gene network. Cell Rep. 2014;8(5):1328–38.
Zhao JL, Rao DS, Boldin MP, Taganov KD, O'Connell RM, Baltimore D. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc Natl Acad Sci U S A. 2011;108(22):9184–9.
Varney ME, Niederkorn M, Konno H, Matsumura T, Gohda J, Yoshida N, Akiyama T, Christie S, Fang J, Miller D, Jerez A, Karsan A, Maciejewski JP, Meetei RA, Inoue J, Starczynowski DT. Loss of Tifab, a del(5q) MDS gene, alters hematopoiesis through derepression of Toll-like receptor-TRAF6 signaling. J Exp Med. 2015;212(11):1967–85.
Magilnick N, Reyes EY, Wang WL, Vonderfecht SL, Gohda J, Inoue JI, Boldin MP. miR-146a-Traf6 regulatory axis controls autoimmunity and myelopoiesis, but is dispensable for hematopoietic stem cell homeostasis and tumor suppression. Proc Natl Acad Sci USA. 2017;114(34):E7140–E9.
Li X, Xu L, Sheng X, Cai J, Liu J, Yin T, Xiao F, Chen F, Zhong H. Upregulated microRNA-146a expression induced by granulocyte colony-stimulating factor enhanced low-dosage chemotherapy response in aged acute myeloid leukemia patients. Exp Hematol. 2018;68:66–79.
Tian P, Yan L. Inhibition of MicroRNA-149-5p induces apoptosis of acute myeloid leukemia cell line THP-1 by targeting fas ligand (FASLG). Med Sci Monit. 2016;22:5116–23.
Xu DD, Zhou PJ, Wang Y, Zhang Y, Zhang R, Zhang L, Chen SH, Fu WY, Ruan BB, Xu HP, Hu CZ, Tian L, Qin JH, Wang S, Wang X, Liu QY, Ren Z, Gu XK, Li YH, Liu Z, Wang YF. miR-150 Suppresses the proliferation and tumorigenicity of leukemia stem cells by targeting the nanog signaling pathway. Front Pharmacol. 2016;7:439.
Fang ZH, Wang SL, Zhao JT, Lin ZJ, Chen LY, Su R, Xie ST, Carter BZ, Xu B. miR-150 exerts antileukemia activity in vitro and in vivo through regulating genes in multiple pathways. Cell Death Dis. 2016;7(9):e2371.
Huang X, Schwind S, Santhanam R, Eisfeld AK, Chiang CL, Lankenau M, Yu B, Hoellerbauer P, Jin Y, Tarighat SS, Khalife J, Walker A, Perrotti D, Bloomfield CD, Wang H, Lee RJ, Lee LJ, Marcucci G. Targeting the RAS/MAPK pathway with miR-181a in acute myeloid leukemia. Oncotarget. 2016;7(37):59273–86.
Sharifi M, Fasihi-Ramandi M, Sheikhi A, Moridnia A, Saneipour M. Apoptosis induction in acute promyelocytic leukemia cells through upregulation of CEBPα by miR-182 blockage. Mol Biol Res Commun. 2018;7(1):25–33.
Ke S, Li RC, Lu J, Meng FK, Feng YK, Fang MH. MicroRNA-192 regulates cell proliferation and cell cycle transition in acute myeloid leukemia via interaction with CCNT2. Int J Hematol. 2017;106(2):258–65.
Popovic R, Riesbeck LE, Velu CS, Chaubey A, Zhang J, Achille NJ, Erfurth FE, Eaton K, Lu J, Grimes HL, Chen J, Rowley JD, Zeleznik-Le NJ. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113(14):3314–22.
Li Z, Huang H, Chen P, He M, Li Y, Arnovitz S, Jiang X, He C, Hyjek E, Zhang J, Zhang Z, Elkahloun A, Cao D, Shen C, Wunderlich M, Wang Y, Neilly MB, Jin J, Wei M, Lu J, Valk PJ, Delwel R, Lowenberg B, Le Beau MM, Vardiman J, Mulloy JC, Zeleznik-Le NJ, Liu PP, Zhang J, Chen J. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun. 2012;3:688.
Wang Z, Luo HFZ, Fan Y, Liu X, Zhang Y, Rui S, Chen Y, Hong L, Gao J, Zhang M. MiR-204 acts as a potential therapeutic target in acute myeloid leukemia by increasing BIRC6-mediated apoptosis. BMB Rep. 2018;51(9):444–9.
Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, Volinia S, Liu CG, Schnittger S, Haferlach T, Liso A, Diverio D, Mancini M, Meloni G, Foa R, Martelli MF, Mecucci C, Croce CM, Falini B. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci U S A. 2008;105(10):3945–50.
Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L, Diverio D, Ammatuna E, Cimino G, Lo-Coco F, Grignani F, Nervi C. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12(5):457–66.
Pulikkan JA, Dengler V, Peramangalam PS, Peer Zada AA, Müller-Tidow C, Bohlander SK, Tenen DG, Behre G. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115(9):1768–78.
Gentner B, Pochert N, Rouhi A, Boccalatte F, Plati T, Berg T, Sun SM, Mah SM, Mirkovic-Hösle M, Ruschmann J, Muranyi A, Leierseder S, Argiropoulos B, Starczynowski DT, Karsan AHM, Hogge D, Camargo FD, Engelhardt S, Döhner H, Buske C, Jongen-Lavrencic M, Naldini L, Humphries RK, Kuchenbauer F. MicroRNA-223 dose levels fine tune proliferation and differentiation in human cord blood progenitors and acute myeloid leukemia. Exp Hematol. 2015;38(20):e00259-18.
Xiao Y, Su C, Deng T. miR-223 decreases cell proliferation and enhances cell apoptosis in acute myeloid leukemia via targeting FBXW7. Oncol Lett. 2016;12(5):3531–6.
Xiao Y, Su C, Deng T. MicroRNA-339-5p inhibits cell proliferation of acute myeloid leukaemia by directly targeting SOX4. Oncol Lett. 2016;12(5):3531–6.
Ying X, Zhang W, Fang MZW, Wang C, Han L. miR-345-5p regulates proliferation, cell cycle, and apoptosis of acute myeloid leukemia cells by targeting AKT2. J Cell Biochem. 2018.
García-Ortí L, Cristóbal I, Cirauqui C, Guruceaga E, Marcotegui N, Calasanz MJ, Castello-Cros R, Odero MD. Integration of SNP and mRNA arrays with microRNA profiling reveals that MiR-370 is upregulated and targets NF1 in acute myeloid leukemia. PLoS One. 2012;7(10):e47717.
Pelosi A, Careccia S, Lulli V, Romania P, Marziali G, Testa U, Lavorgna S, Lo-Coco F, Petti MC, Calabretta B, Levrero M, Piaggio G, Rizzo MG. MiRNA let-7c promotes granulocytic differentiation in acute myeloid leukemia. Oncogene. 2013;32(31):3648–54.
Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–42.
Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340(6131):440–1.
Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res. 2016;6(6):1167–76.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8.
Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15(3):611–24.
Li W, Zhong C, Jiao J, Li P, Cui B, Ji C, Ma D. Characterization of hsa_circ_0004277 as a new biomarker for acute myeloid leukemia via circular RNA profile and bioinformatics analysis. Int J Mol Sci. 2017;18(3).
L Abbate A, Tolomeo D, Cifola I, Severgnini M, Turchiano A, Augello B, Squeo G, D Addabbo P, Traversa D, Daniele G, Lonoce A, Pafundi M, Carella M, Palumbo O, Dolnik A, Muehlematter D, Schoumans J, Van Roy N, De Bellis G, Martinelli G, Merla G, Bullinger L, Haferlach C, Storlazzi CT. MYC-containing amplicons in acute myeloid leukemia: genomic structures, evolution, and transcriptional consequences. Leukemia. 2018;32(10):2152–66.
Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;166(4):1055–6.
Li XL, Arai Y, Harada H, Shima Y, Yoshida H, Rokudai S, Aikawa Y, Kimura A, Kitabayashi I. Mutations of the HIPK2 gene in acute myeloid leukemia and myelodysplastic syndrome impair AML1- and p53-mediated transcription. Oncogene. 2007;26(51):7231–9.
Hirsch S, Blätte TJ, Grasedieck S, Cocciardi S, Rouhi A, Jongen-Lavrencic M, Paschka P, Krönke J, Gaidzik VI, Döhner H, Schlenk RF, Kuchenbauer F, Döhner K, Dolnik A, Bullinger L. Circular RNAs of the nucleophosmin (NPM1) gene in acute myeloid leukemia. Haematologica. 2017;102(12):2039–47.
Chen H, Liu T, Liu J, Feng Y, Wang B, Wang J, Bai J, Zhao W, Shen Y, Wang X, Yang J, Ji Y, He A, Yang Y. Circ-ANAPC7 is upregulated in acute myeloid leukemia and appears to target the MiR-181 family. Cell Physiol Biochem. 2018;47(5):1998–2007.
Fan H, Li Y, Liu C, Liu Y, Bai J, Li W. Circular RNA-100290 promotes cell proliferation and inhibits apoptosis in acute myeloid leukemia cells via sponging miR-203. Biochem Biophys Res Commun. 2018;507(1-4):178–84.
Shang J, Chen WM, Wang ZH, Wei TN, Chen ZZ, Wu WB. CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. Exp Hematol. 2018;70:42–54. https://doi.org/10.1084/jem.20161595.
Ping L, Jian-Jun C, Chu-Shu L, Guang-Hua L, Ming Z. Silencing of circ_0009910 inhibits acute myeloid leukemia cell growth through increasing miR-20a-5p. Blood Cells Mol Dis. 2019;75:41–7.
Li S, Ma Y, Tan Y, Ma X, Zhao M, Chen B, Zhang R, Chen Z, Wang K. Profiling and functional analysis of circular RNAs in acute promyelocytic leukemia and their dynamic regulation during all-trans retinoic acid treatment. Cell Death Dis. 2018;9(6):651.
Morenos L, Chatterton Z, Ng JL, Halemba MS, Parkinson-Bates M, Mechinaud F, Elwood N, Saffery R, Wong NC. Hypermethylation and down-regulation of DLEU2 in paediatric acute myeloid leukaemia independent of embedded tumour suppressor miR-15a/16-1. Mol Cancer. 2014;13(123).
Wu DM, Wen X, Han XR, Wang S, Wang YJ, Shen M, Fan SH, Zhang ZF, Shan Q, Li MQ, Hu B, Chen GQ, Lu J, Zheng YL. Role of circular RNA DLEU2 in human acute myeloid leukemia. Mol Cell Biol. 2018;38(20).
Li W, Zhong C, Jiao J, Li P, Cui B, Ji C, Ma D. Characterization of hsa_circ_0004277as a new biomarker for acute myeloid leukemia via circular RNA profile and bioinformatics analysis. Int J Mol Sci. 2017;18(3).
Chen LL, Carmichael GG. Long noncoding RNAs in mammalian cells: what, where, and why? Wiley Interdiscip Rev RNA. 2010;1(1):2–21.
Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407.
Zhang X, Weissman SM, Newburger PE. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. 2014;11(6):777–87.
Zeng C, Xu Y, Xu L, Yu X, Cheng J, Yang L, Chen S, Li Y. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer. 2014;14:693.
Zeng C, Yu X, Lai J, Yang L, Chen S, Li Y. Overexpression of the long non-coding RNA PVT1 is correlated with leukemic cell proliferation in acute promyelocytic leukemia. J Hematol Oncol. 2015;8(126).
Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell TC, Essig J, Otto GM, O'Sullivan MG, Largaespada DA, Schwertfeger KL, Marahrens Y, Kawakami Y, Bagchi A. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512(7512):82–6.
Salehi M, Sharifi M. Induction of apoptosis and necrosis in human acute erythroleukemia cells by inhibition of long non-coding RNA PVT1. Mol Biol Res Commun. 2018;7(2):89–96.
Salehi M, Sharifi M, Bagheri M. Knockdown of long noncoding RNA plasmacytoma variant translocation 1 with antisense locked nucleic acid GapmeRs exerts tumor-suppressive functions in human acute erythroleukemia cells through downregulation of C-MYC expression. Cancer Biother Radiopharm. 2018.
Hughes JM, Legnini I, Salvatori B, Masciarelli S, Marchioni M, Fazi F, Morlando M, Bozzoni I, Fatica A. C/EBPα-p30 protein induces expression of the oncogenic long non-coding RNA UCA1 in acute myeloid leukemia. Oncotarget. 2015;2015(6):21.
Wang Y, Zhou Q, Ma JJ. High expression of lnc-CRNDE presents as a biomarker for acute myeloid leukemia and promotes the malignant progression in acute myeloid leukemia cell line U937. Eur Rev Med Pharmacol Sci. 2018;22(3):763–70.
Leith CP, Kopecky KJ, Godwin J, McConnell T, Slovak ML, Chen IM, Head DR, Appelbaum FR, Willman CL. Acute myeloid leukemia in the elderly: assessment of multidrug resistance (MDR1) and cytogenetics distinguishes biologic subgroups with remarkably distinct responses to standard chemotherapy. A Southwest Oncology Group study. Blood. 1997;89(9):3323–9.
Cancer and Leukemia Group B 8461, Farag SS, MK AKJ, CA RAS, Vardiman JW, Pettenati MJ, Baer MR, Qumsiyeh MB, Koduru PR, Ning Y, Mayer RJ, Stone RM, Larson RA, Bloomfield CD. Pretreatment cytogenetics add to other prognostic factors predicting complete remission and long-term outcome in patients 60 years of age or older with acute myeloid leukemia: results from Cancer and Leukemia Group. Blood. 2006;108(1):63–73.
Spassov BV, Stoimenov AS, Balatzenko GN, Genova ML, Peichev DB, Konstantinov SM. Wilms' tumor protein and FLT3-internal tandem duplication expression in patients with de novo acute myeloid leukemia. Hematology. 2011;16(1):37–42.
Lyu Y, Lou J, Yang Y, Feng J, Hao Y, Huang S, Yin L, Xu J, Huang D, Ma B, Zou D, Wang Y, Zhang Y, Zhang B, Chen P, Yu K, Lam EW, Wang X, Liu Q, Yan J, Jin B. Dysfunction of the WT1-MEG3 signaling promotes AML leukemogenesis via p53-dependent and -independent pathways. Leukemia. 2017;31(12):2543–51.
Nerlov C. C/EBPalpha mutations in acute myeloid leukaemias. Nat Rev Cancer. 2004;4(5):394–400.
Schlenk RF, Döhner K, Krauter J, Fröhling S, Corbacioglu A, Bullinger L, Habdank M, Späth D, Morgan M, Benner A, Schlegelberger B, Heil G, Ganser A. H; D, German-Austrian Acute Myeloid Leukemia Study Group. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. The New England journal of medicine. 2008;358(18):1909–18.
Verhaak RG, Goudswaard CS, van Putten W, Bijl MA, Sanders MA, Hugens W, Uitterlinden AG, Erpelinck CA, Delwel R, Löwenberg B, Valk PJ. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106(12):3747–54.
Hirano T, Yoshikawa R, Harada H, Harada YI, Shida A, Yamazaki T. Long noncoding RNA, CCDC26, controls myeloid leukemia cell growth through regulation of KIT expression. Mol Cancer. 2015;14:90.
De Clara E, Gourvest M, Ma H, Vergez F, Tosolini M, Dejean S, Demur C, Delabesse E, Recher C, Touriol C, Martelli MP, Falini B, Brousset P, Bousquet M. Long non-coding RNA expression profile in cytogenetically normal acute myeloid leukemia identifies a distinct signature and a new biomarker in NPM1-mutated patients. Haematologica. 2017;102(10):1718–26.
Fernando TR, Contreras JR, Zampini M, Rodriguez-Malave NI, Alberti MO, Anguiano J, Tran TM, Palanichamy JK, Gajeton J, Ung NM, Aros CJ, Waters EV, Casero D, Basso G, Pigazzi M, Rao DS. The lncRNA CASC15 regulates SOX4 expression in RUNX1-rearranged acute leukemia. Mol Cancer. 2017;16(1):126.
Garzon R, Volinia S, Papaioannou D, Nicolet D, Kohlschmidt J, Yan PS, Mrozek K, Bucci D, Carroll AJ, Baer MR, Wetzler M, Carter TH, Powell BL, Kolitz JE, Moore JO, Eisfeld AK, Blachly JS, Blum W, Caligiuri MA, Stone RM, Marcucci G, Croce CM, Byrd JC, Bloomfield CD. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2014;111(52):18679–84.
Li J, Sun CK. Long noncoding RNA SNHG5 is up-regulated and serves as a potential prognostic biomarker in acute myeloid leukemia. Eur Rev Med Pharmacol Sci. 2018;22(11):3342–7.
Yang L, Zhou JD, Zhang TJ, Ma JC, Xiao GF, Chen Q, Deng ZQ, Lin J, Qian J, Yao DM. Overexpression of lncRNA PANDAR predicts adverse prognosis in acute myeloid leukemia. Cancer Manag Res. 2018;10:4999–5007.
Hao S, Shao Z. HOTAIR is upregulated in acute myeloid leukemia and that indicates a poor prognosis. Int J Clin Exp Pathol. 2015;8(6):7223–8.
Pashaiefar H, Izadifard M, Yaghmaie M, Montazeri M, Gheisari E, Ahmadvand M, Momeny M, Ghaffari SH, Kasaeian A, Alimoghaddam K, Ghavamzadeh A. Low expression of long noncoding RNA IRAIN is associated with poor prognosis in non-M3 acute myeloid leukemia patients. Genet Test Mol Biomarkers. 2018;22(5):288–94.
Wang X, Zhang L, Zhao F, Xu R, Jiang J, Zhang C, Liu H, Huang H. Long non-coding RNA taurine-upregulated gene 1 correlates with poor prognosis, induces cell proliferation, and represses cell apoptosis via targeting aurora kinase A in adult acute myeloid leukemia. Ann Hematol. 2018;97(8):1375–89.
Luo W, Yu H, Zou X, Ni X, Wei J. Long non-coding RNA taurine-upregulated gene 1 correlates with unfavorable prognosis in patients with refractory or relapsed acute myeloid leukemia treated by purine analogue based chemotherapy regimens. Cancer Biomark. 2018;23(4):485–94.
Zhao TF, Jia HZ, Zhang ZZ, Zhao XS, Zou YF, Zhang W, Wan J, Chen XF. LncRNA H19 regulates ID2 expression through competitive binding to hsa-miR-19a/b in acute myelocytic leukemia. Mol Med Rep. 2017;16(3):3687–93.
Zhao C, Wang S, Zhao Y, Du F, Wang W, Lv P, Qi L. Long noncoding RNA NEAT1 modulates cell proliferation and apoptosis by regulating miR-23a-3p/SMC1A in acute myeloid leukemia. J Cell Physiol. 2018.
Sun MD, Zheng YQ, Wang LP, Zhao HT, Yang S. Long noncoding RNA UCA1 promotes cell proliferation, migration and invasion of human leukemia cells via sponging miR-126. Eur Rev Med Pharmacol Sci. 2018;22(8):2233–45.
Zhang Y, Liu Y, Xu X. Knockdown of LncRNA-UCA1 suppresses chemoresistance of pediatric AML by inhibiting glycolysis through the microRNA-125a/hexokinase 2 pathway. J Cell Biochem. 2018;119(7):6296–308.
Xing CY, Hu XQ, Xie FY, Yu ZJ, Li HY. Bin-Zhou, Wu JB, Tang LY, Gao SM. Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Lett. 2015;589(15):1981–7. https://doi.org/10.1002/jcb.27461.
Gao S, Zhou B, Li H, Huang X, Wu Y, Xing C, Yu X, Ji Y. Long noncoding RNA HOTAIR promotes the self-renewal of leukemia stem cells through epigenetic silencing of p15. Exp Hematol. 2018;67:32–40.
Chen L, Wang W, Cao L, Li Z, Wang X. Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol Cells. 2016;39(4):330–6.
Liu B, Ma X, Liu Q, Xiao Y, Pan S, Jia L. Aberrant mannosylation profile and FTX/miR-342/ALG3-axis contribute to development of drug resistance in acute myeloid leukemia. Cell Death Dis. 2018;9(6):688.
Zhao H, Zhang X, Frazão JB, Condino-Neto A, Newburger PE. HOX antisense lincRNA HOXA-AS2 is an apoptosis repressor in all trans retinoic acid treated NB4 promyelocytic leukemia cells. J Cell Biochem. 2013;13:123. https://doi.org/10.1186/1476-4598-13-123.
Dong X, Fang Z, Yu M, Zhang L, Xiao R, Li X, Pan G, Liu J. Knockdown of long noncoding RNA HOXA-AS2 suppresses chemoresistance of acute myeloid leukemia via the miR-520c-3p/S100A4 axis. Cell Physiol Biochem. 2018;51(2):886–96.
Chen ZH, Wang WT, Huang W, Fang K, Sun YM, Liu SR, Luo XQ, Chen YQ. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 2017;24(2):212–24.
Wei S, Zhao M, Wang X, Li Y, Wang K. PU.1 controls the expression of long noncoding RNA HOTAIRM1 during granulocytic differentiation. J Hematol Oncol. 2016;9(1):44.
Díaz-Beyá M, Brunet S, Nomdedéu J, Pratcorona M, Cordeiro A, Gallardo D, Escoda L, Tormo M, Heras I, Ribera JM, Duarte R, de Llano MP, Bargay J, Sampol A, Nomdedeu M, Risueño RM, Hoyos M, Sierra J, Monzo M, Navarro A, Esteve J, Cooperative AML group CETLAM. The lincRNA HOTAIRM1, located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget. 2015;6(31):31613–27.
Sun J, Li W, Sun Y, Yu D, Wen X, Wang H, Cui J, Wang G, Hoffman AR, Hu JF. A novel antisense long noncoding RNA within the IGF1R gene locus is imprinted in hematopoietic malignancies. Nucleic Acids Res. 2014;42(15):9588–601.
Wang H, Li W, Guo R, Sun J, Cui J, Wang G, Hoffman AR, Hu JF. An intragenic long noncoding RNA interacts epigenetically with the RUNX1 promoter and enhancer chromatin DNA in hematopoietic malignancies. Int J Cancer. 2014;135(12):2783–94.
Sun LY, Li XJ, Sun YM, Huang W, Fang K, Han C, Chen ZH, Luo XQ, Chen YQ, Wang WT. LncRNA ANRIL regulates AML development through modulating the glucose metabolism pathway of AdipoR1/AMPK/SIRT1. Mol Cancer. 2018;17(1):127.
Treppendahl MB, Qiu X, Søgaard A, Yang X, Nandrup-Bus C, Hother C, Andersen MK, Kjeldsen L, Möllgård L, Hellström-Lindberg E, Jendholm J, Porse BT, Jones PA, Liang G, Grønbæk K. Allelic methylation levels of the noncoding VTRNA2-1 located on chromosome 5q31.1 predict outcome in AML. Blood. 2012;119(1):206–16.
Mangiavacchi A, Sorci M, Masciarelli S, Larivera S, Legnini I, Iosue I, Bozzoni I, Fazi F, Fatica A. The miR-223 host non-coding transcript linc-223 induces IRF4 expression in acute myeloid leukemia by acting as a competing endogenous RNA. Oncotarget. 2016;7(37):60155–68.
Wang Y, Li Y, Song HQ, Sun GW. Long non-coding RNA LINC00899 as a novel serum biomarker for diagnosis and prognosis prediction of acute myeloid leukemia. Eur Rev Med Pharmacol Sci. 2018;22(21):7364–70.
Acknowledgements
Not applicable
Funding
This work was supported by grants from the National Natural Science Foundation of China (81500118, 61501519), the China Postdoctoral Science Foundation funded project (Project No.2016 M600443), Jiangsu Province Postdoctoral Science Foundation funded project (Project No.1701184B).
Availability of data and materials
Not applicable
Author information
Authors and Affiliations
Contributions
LF and JLS conceptualized the review. YL wrote the manuscript. YL and LZC prepared the figure and tables.YFP, TTQ, LQ and HYZ revised the review. ZHC, XYK and LF critically reviewed and edited the manuscript. All aythors read and approved the fnal manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liu, Y., Cheng, Z., Pang, Y. et al. Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia. J Hematol Oncol 12, 51 (2019). https://doi.org/10.1186/s13045-019-0734-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13045-019-0734-5