Abstract
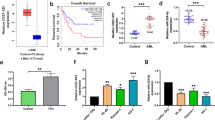
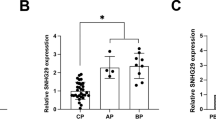
This study aimed to investigate the correlation of long non-coding RNA (lncRNA) taurine-upregulated gene 1 (TUG1) with clinicopathological feature and prognosis, and to explore its effect on cell proliferation and apoptosis as well as the relevant target genes in adult acute myeloid leukemia (AML). LncRNA TUG1 expression was detected in bone marrow samples from 186 AML patients and 62 controls. Blank mimic, lncRNA TUG1 mimic, blank inhibitor, and lncRNA TUG1 inhibitor lentivirus vectors were transfected in KG-1 cells. Rescue experiment was performed by transfection of lncRNA TUG1 inhibitor and aurora kinase A (AURKA) mimic lentivirus vectors. Cell proliferation, apoptosis, RNA, and protein expressions were determined by CKK-8, annexin V-FITC-propidium iodide, quantitative polymerase chain reaction, and western blot assays. LncRNA TUG1 expression was higher in AML patients compared to controls and correlated with higher white blood cell counts, monosomal karyotype, FLT3-ITD mutation, poor-risk stratification, and poor prognosis, which independently predicted worse event-free survival and overall survival. In vitro, lncRNA TUG1 expression was higher in AML cell lines (KG-1, MOLM-14, HL-60, NB-4, and THP-1 cells) compared to controls. LncRNA TUG1 mimic promoted cell proliferation and decreased cell apoptosis rate, while lncRNA TUG1 inhibitor repressed cell proliferation and increased cell apoptosis rate. Rescue experiment showed that AURKA attenuated the influence of lncRNA TUG1 on AML cell proliferation and apoptosis. In conclusion, lncRNA TUG1 associates with advanced disease and worse prognosis in adult AML patients, and it induces AML cell proliferation and represses cell apoptosis via targeting AURKA.









Similar content being viewed by others
References
Dohner H, Weisdorf DJ, Bloomfield CD (2015) Acute myeloid leukemia. N Engl J Med 373(12):1136–1152. https://doi.org/10.1056/NEJMra1406184
Siegel RL, Miller KD, Jemal A (2017) Cancer statistics, 2017. CA Cancer J Clin 67(1):7–30. https://doi.org/10.3322/caac.21387
Guttman M, Rinn JL (2012) Modular regulatory principles of large non-coding RNAs. Nature 482(7385):339–346. https://doi.org/10.1038/nature10887
Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR (2012) Landscape of transcription in human cells. Nature 489(7414):101–108. https://doi.org/10.1038/nature11233
Qiu MT, Hu JW, Yin R, Xu L (2013) Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol 34(2):613–620. https://doi.org/10.1007/s13277-013-0658-6
Wang L, Zhao Z, Feng W, Ye Z, Dai W, Zhang C, Peng J, Wu K (2016) Long non-coding RNA TUG1 promotes colorectal cancer metastasis via EMT pathway. Oncotarget 7(32):51713–51719. https://doi.org/10.18632/oncotarget.10563
Liu L, Chen X, Zhang Y, Hu Y, Shen X, Zhu W (2017) Long non-coding RNA TUG1 promotes endometrial cancer development via inhibiting miR-299 and miR-34a-5p. Oncotarget 8(19):31386–31394. https://doi.org/10.18632/oncotarget.15607
Wang PQ, Wu YX, Zhong XD, Liu B, Qiao G (2017) Prognostic significance of overexpressed long non-coding RNA TUG1 in patients with clear cell renal cell carcinoma. Eur Rev Med Pharmacol Sci 21(1):82–86
Li T, Liu Y, Xiao H, Xu G (2017) Long non-coding RNA TUG1 promotes cell proliferation and metastasis in human breast cancer. Breast Cancer 24(4):535–543. https://doi.org/10.1007/s12282-016-0736-x
Zhai HY, Sui MH, Yu X, Qu Z, Hu JC, Sun HQ, Zheng HT, Zhou K, Jiang LX (2016) Overexpression of long non-coding RNA TUG1 promotes colon cancer progression. Med Sci Monit 22:3281–3287
Wenzel A, Akbasli E, Gorodkin J (2012) RIsearch: fast RNA-RNA interaction search using a simplified nearest-neighbor energy model. Bioinformatics 28(21):2738–2746. https://doi.org/10.1093/bioinformatics/bts519
Tafer H, Hofacker IL (2008) RNAplex: a fast tool for RNA-RNA interaction search. Bioinformatics 24(22):2657–2663. https://doi.org/10.1093/bioinformatics/btn193
Li J, Ma W, Zeng P, Wang J, Geng B, Yang J, Cui Q (2015) LncTar: a tool for predicting the RNA targets of long noncoding RNAs. Brief Bioinform 16(5):806–812. https://doi.org/10.1093/bib/bbu048
Pinero J, Bravo A, Queralt-Rosinach N, Gutierrez-Sacristan A, Deu-Pons J, Centeno E, Garcia-Garcia J, Sanz F, Furlong LI (2017) DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 45(D1):D833–D839. https://doi.org/10.1093/nar/gkw943
Estey E, Dohner H (2006) Acute myeloid leukaemia. Lancet 368(9550):1894–1907. https://doi.org/10.1016/S0140-6736(06)69780-8
Pan JQ, Zhang YQ, Wang JH, Xu P, Wang W (2017) lncRNA co-expression network model for the prognostic analysis of acute myeloid leukemia. Int J Mol Med. https://doi.org/10.3892/ijmm.2017.2888
Hirano T, Yoshikawa R, Harada H, Harada Y, Ishida A, Yamazaki T (2015) Long noncoding RNA, CCDC26, controls myeloid leukemia cell growth through regulation of KIT expression. Mol Cancer 14:90. https://doi.org/10.1186/s12943-015-0364-7
Ou C, Li G (2017) Long non-coding RNA TUG1: a novel therapeutic target in small cell lung cancer. J Thorac Dis 9(7):E644–E645. https://doi.org/10.21037/jtd.2017.06.94
Iliev R, Kleinova R, Juracek J, Dolezel J, Ozanova Z, Fedorko M, Pacik D, Svoboda M, Stanik M, Slaby O (2016) Overexpression of long non-coding RNA TUG1 predicts poor prognosis and promotes cancer cell proliferation and migration in high-grade muscle-invasive bladder cancer. Tumour Biol 37(10):13385–13390. https://doi.org/10.1007/s13277-016-5177-9
Zhang E, He X, Yin D, Han L, Qiu M, Xu T, Xia R, Xu L, Yin R, De W (2016) Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis 7:e2109. https://doi.org/10.1038/cddis.2015.356
Sun J, Ding C, Yang Z, Liu T, Zhang X, Zhao C, Wang J (2016) The long non-coding RNA TUG1 indicates a poor prognosis for colorectal cancer and promotes metastasis by affecting epithelial-mesenchymal transition. J Transl Med 14:42. https://doi.org/10.1186/s12967-016-0786-z
Zeng B, Ye H, Chen J, Cheng D, Cai C, Chen G, Chen X, Xin H, Tang C, Zeng J (2017) LncRNA TUG1 sponges miR-145 to promote cancer progression and regulate glutamine metabolism via Sirt3/GDH axis. Oncotarget 8(69):113650–113661. https://doi.org/10.18632/oncotarget.21922
Guo P, Zhang G, Meng J, He Q, Li Z, Guan Y (2018) Upregulation of long non-coding RNA TUG1 promotes bladder cancer cell 5 proliferation, migration and invasion by inhibiting miR-29c. Oncol Res. https://doi.org/10.3727/096504018X15152085755247
Fang T, Huang H, Li X, Liao J, Yang Z, Hoffman RM, Cheng XI, Liang L, Hu W, Yun S (2018) Effects of siRNA silencing of TUG1 and LCAL6 long non-coding RNAs on patient-derived xenograft of non-small cell lung cancer. Anticancer Res 38(1):179–186. https://doi.org/10.21873/anticanres.12206
Jiang L, Wang W, Li G, Sun C, Ren Z, Sheng H, Gao H, Wang C, Yu H (2016) High TUG1 expression is associated with chemotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. Cancer Chemother Pharmacol 78(2):333–339. https://doi.org/10.1007/s00280-016-3066-y
Kuang D, Zhang X, Hua S, Dong W, Li Z (2016) Long non-coding RNA TUG1 regulates ovarian cancer proliferation and metastasis via affecting epithelial-mesenchymal transition. Exp Mol Pathol 101(2):267–273. https://doi.org/10.1016/j.yexmp.2016.09.008
Zhu J, Shi H, Liu H, Wang X, Li F (2017) Long non-coding RNA TUG1 promotes cervical cancer progression by regulating the miR-138-5p-SIRT1 axis. Oncotarget 8(39):65253–65264. https://doi.org/10.18632/oncotarget.18224
Warner SL, Bearss DJ, Han H, Von Hoff DD (2003) Targeting Aurora-2 kinase in cancer. Mol Cancer Ther 2(6):589–595
Lee EC, Frolov A, Li R, Ayala G, Greenberg NM (2006) Targeting Aurora kinases for the treatment of prostate cancer. Cancer Res 66(10):4996–5002. https://doi.org/10.1158/0008-5472.CAN-05-2796
Lens SM, Voest EE, Medema RH (2010) Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer 10(12):825–841. https://doi.org/10.1038/nrc2964
Chuang TP, Wang JY, Jao SW, Wu CC, Chen JH, Hsiao KH, Lin CY, Chen SH, Su SY, Chen YJ, Chen YT, Wu DC, Li LH (2016) Over-expression of AURKA, SKA3 and DSN1 contributes to colorectal adenoma to carcinoma progression. Oncotarget 7(29):45803–45818. https://doi.org/10.18632/oncotarget.9960
Eterno V, Zambelli A, Villani L, Tuscano A, Manera S, Spitaleri A, Pavesi L, Amato A (2016) AurkA controls self-renewal of breast cancer-initiating cells promoting wnt3a stabilization through suppression of miR-128. Sci Rep 6:28436. https://doi.org/10.1038/srep28436
Wu J, Yang L, Shan Y, Cai C, Wang S, Zhang H (2016) AURKA promotes cell migration and invasion of head and neck squamous cell carcinoma through regulation of the AURKA/Akt/FAK signaling pathway. Oncol Lett 11(3):1889–1894. https://doi.org/10.3892/ol.2016.4110
Lucena-Araujo AR, de Oliveira FM, Leite-Cueva SD, dos Santos GA, Falcao RP, Rego EM (2011) High expression of AURKA and AURKB is associated with unfavorable cytogenetic abnormalities and high white blood cell count in patients with acute myeloid leukemia. Leuk Res 35(2):260–264. https://doi.org/10.1016/j.leukres.2010.07.034
Ye D, Garcia-Manero G, Kantarjian HM, Xiao L, Vadhan-Raj S, Fernandez MH, Nguyen MH, Medeiros LJ, Bueso-Ramos CE (2009) Analysis of Aurora kinase A expression in CD34(+) blast cells isolated from patients with myelodysplastic syndromes and acute myeloid leukemia. J Hematop 2(1):2–8. https://doi.org/10.1007/s12308-008-0019-3
Huang XF, Luo SK, Xu J, Li J, Xu DR, Wang LH, Yan M, Wang XR, Wan XB, Zheng FM, Zeng YX, Liu Q (2008) Aurora kinase inhibitory VX-680 increases Bax/Bcl-2 ratio and induces apoptosis in Aurora-A-high acute myeloid leukemia. Blood 111(5):2854–2865. https://doi.org/10.1182/blood-2007-07-099325
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 81201857) and Nantong Science and Technology Project Foundation, China (No. MS12016012).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics statement
This study was performed under the Institutional Review Board approvals from The Affiliated Hospital of Nantong University Hospital and conducted in accordance with the Declaration of Helsinki. Written informed consents had been obtained from all patients and controls.
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Wang, X., Zhang, L., Zhao, F. et al. Long non-coding RNA taurine-upregulated gene 1 correlates with poor prognosis, induces cell proliferation, and represses cell apoptosis via targeting aurora kinase A in adult acute myeloid leukemia. Ann Hematol 97, 1375–1389 (2018). https://doi.org/10.1007/s00277-018-3315-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-018-3315-8