Abstract
Background
Progression of chronic myelogenous leukemia (CML) is frequently accompanied by cytogenetic evolution, commonly unbalanced chromosomal changes, such as an extra copy of Philadelphia chromosome (Ph), +8, and i(17)(q10). Balanced chromosomal translocations typically found in de novo acute myeloid leukemia occur occasionally in CML, such as inv(3)/t(3;3), t(8;21), t(15;17), and inv(16). Translocations involving the 11q23, a relatively common genetic abnormality in acute leukemia, have been seldom reported in CML. In this study, we explored the prevalence and prognostic role of 11q23 in CML.
Methods
We searched our pathology archives for CML cases diagnosed in our institution from 1998 to present. Cases with 11q23 rearrangements were retrieved. The corresponding clinicopathological data were reviewed.
Results
A total of 2,012 cases of CML with available karyotypes were identified. Ten (0.5%) CML cases had 11q23 rearrangement in Ph-positive cells, including 4 cases of t(9;11), 2 cases of t(11;19), and 1 case each of t(2;11), t(4;11), t(6;11), and t(4;9;11). Eight cases (80%) had other concurrent chromosomal abnormalities. There were 6 men and 4 women with a median age of 50 years (range, 21–70 years) at time of initial diagnosis of CML. 11q23 rearrangement occurred after a median period of 12.5 months (range, 0–172 months): 1 patient in chronic phase, 2 in accelerated phase, and 7 in blast phase. Eight of ten patients died after a median follow-up of 16.5 months (range, 8–186 months) following the initial diagnosis of CML, and a median of 6.7 months (range, 0.8–16.6 months) after the emergence of 11q23 rearrangement. The remaining two patients had complete remission at the last follow-up, 50.2 and 6.9 months, respectively. In addition, we also identified a case with 11q23/t(11;17) in Ph-negative cells in a patient with a history of CML. MLL involvement was tested by fluorescence in situ hybridization in 10 cases, and 7 cases (70%) were positive.
Conclusions
In summary, chromosomal rearrangements involving 11q23 are rare in CML, frequently occurring in blast phase, and are often associated with other cytogenetic abnormalities. These patients had a low response rate to tyrosine kinase inhibitors and a poor prognosis.
Similar content being viewed by others
Background
BCR-ABL1 derived from t(9;22)(q34;q11), its variant translocations, or cryptic fusions is the sole chromosomal abnormality in about 80–90% of chronic mye-logenous leukemia (CML) diagnosed in chronic phase (CP). BCR-ABL1 signaling is believed to be the driving force in CML pathogenesis, leading to disease progression and secondary genetic changes. Continuous BCR-ABL1 activity induces DNA damage and inhibits DNA repair, leading to genetic instability and clonal evolution [1,2]. Clonal evolution, manifested by cytogenetics and mutational changes, occurs in 5–10% of patients diagnosed in chronic phase, approximately 30% of patients in accelerated phase (AP), and 50–80% of patients in blast phase (BP) [2,3]. A recent study demonstrated that the pattern of cytogenetic changes during clonal evolution remains similar in CML patients treated with or without tyrosine kinase inhibitors (TKIs), supporting the idea of genetic instability induced by BCR-ABL1 as the mechanism of clonal evolution[4]. The most common chromosomal aberrations during clonal evolution are unbalanced chromosomal changes, including + Ph, +8, i(17)(q10), and +19 [4], which are the so-called “major route” abnormalities. Reciprocal translocations are much less common and represent “minor route” abnormalities. Several rearrangements typically occurring in de novo acute myeloid leukemia (AML) and conferring prognostic value, such as inv(3)(q21q26)/t(3;3)(q21;q26), t(8;21)(q22,q22), t(15;17)(q22;q21), and inv(16)(p13q22), have been infrequently observed as secondary cytogenetic changes during clonal evolution of CML. Reciprocal rearrangements involving 11q23 are extremely rare in CML and until now only about ten cases in the form of single case reports are available in the literature [5-14]. The clinicopathological characteristics and prognostic value of 11q23 translocations in CML have not been studied systematically.
Rearrangements involving 11q23/MLL locus are frequently encountered in acute leukemia. About 70%–80% of infant acute leukemia has 11q23/MLL abnormalities, associated with a distinct genetic profile and poor prognosis [15,16]. In adults, about 3%–4% of de novo AML carries 11q23 translocations [17]. Rearrangements involving 11q23/MLL are also observed in therapy-related AML, especially after topoisomerase inhibitor treatment. The translocation partners for 11q23 are diverse, and over 70 fusion partners have been characterized at the molecular level to date [18]. The most common translocations involving 11q23/MLL include t(9;11)(p22;q23), t(4;11)(q21;q23), and t(11;19)(q23;p13) [18,19]. Although the mechanisms of MLL-induced leukemogenesis seem diverse and are not fully understood, most MLL gene rearrangements juxtapose the amino terminal portion of MLL with the carboxyl terminal portion of its partners, resulting in chimeric oncoproteins [20]. Gene-expression profiling studies demonstrated that MLL fusion proteins induce leukemic transformation, at least in part, through activation of homeobox genes, such as HOXA5 and HOXA9 [21]. A recent study demonstrated that MLL-AF6 fusion oncogene derived from t(6;11)(q27;q23) can induce aberrant activation of RAS and its downstream targets [22].
In this study, we aimed to explore the incidence, pathology, and clinical features of CML cases with 11q23 rearrangement.
Results
Incidence and clinical characteristics
A total of 2,012 cases of CML with karyotypes were identified in our pathology database and from this group, 10 cases (0.5%) carried translocations involving the 11q23 locus in Ph-positive cells. There were six men and four women with a median age of 50 years (range, 21–70 years) at the time of initial diagnosis of CML. All patients received therapies following CML diagnosis. Depending on the time of CML diagnosed, different regimens were given; historically with IFNa, and more recently with TKIs, including first-generation TKI imatinib and second-generation TKIs dasatinib and nilotinib (Table 1). In contrast to cases 1–10, case 11 was a 48-year-old male with the emergence of 11q23 translocation in Ph-negative cells.
Emergence of 11q23 rearrangement in Ph-positive cells
Rearrangement involving 11q23 developed after a median period of 12.5 months (range, 0–172 months) following the initial diagnosis of CML (Table 1). It was detected in one patient (case 1) in CP, in two patients (cases 2 and 3) in AP, accompanied with other AP features including increased blast counts and thrombocytopenia; both were transformed to BP during targeted therapy with TKIs, after 1.6 and 12 months, respectively. The remaining eight patients (cases 4–10) had 11q23 rearrangement in BP; two patients (cases 5 and 6) presented as BP with 11q23 rearrangements at the initial diagnosis of CML. Both cases showed small hypolobated “dwarf” megakaryocytes. In addition, case 5 presented with basophilia (8%) and eosinophilia (4%). These features support the diagnosis of BP of CML instead of de novo AML with t(9;22). The other six cases developed 11q23 rearrangements during the course of CML treatment.
Emergence of 11q23 rearrangement in Ph-negative cells
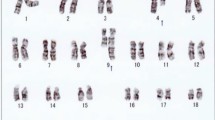
In case 11, the 11q23 translocation developed in Ph-negative cells. The patient was initially diagnosed with CML in CP, with t(9;22)(q34;q11) as a sole abnormality. He was treated with imatinib, nilotinib, and dasatinib. The patient showed a partial response to the treatment. Thirty-two months after the original CML diagnosis, the patient developed AML with the following cytogenetics: 46,XY,t(11;17)(q23;q25) [20]. No Philadelphia chromosome was identified, and fluorescence in situ hybridization (FISH) for BCR-ABL1 was negative.
Emergence of 11q23 rearrangement in a CML case that initially presented as myeloid sarcoma
In case 4, the patient initially presented with a breast mass that was diagnosed as myeloid sarcoma with t(9;22). The status of peripheral blood and bone marrow at that time was unknown. The patient was treated with IFNa. Three months later, bone marrow showed CML in AP, and the patient was treated with idarubicin and ara-C with a brief remission. Another 4 months later, the patient showed massive and diffuse lymphadenopathy with hepatosplenomegaly. A lymph node biopsy showed myeloid sarcoma. Bone marrow biopsy showed acute leukemia with a mixed myeloid/T phenotype (Table 2). The conventional cytogenetics of this bone marrow showed t(2;11)(q32;q23) in Ph-positive cells (Table 3).
Morphology and immunophenotype
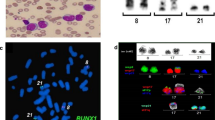
Of ten cases with blast transformation in this study (cases 2–11), nine showed myeloid, and one (case 4) showed a mixed lineage (myeloid/T) immunophenotype (Table 2). Several cases expressed lymphoid markers: case 10 expressed CD10 and CD22 while case 2 had partial CD19. The blast morphology was heterogeneous and some cases have monocytic differentiation (Figure 1 and Table 2). CD64, a monocytic marker, was expressed in six cases.
Cytogenetic data
The translocation partners for 11q23 were diverse and included four cases of t(9;11), two cases of t(11;19), and one case each of t(2;11), t(4;11), t(6;11), t(11;17), and t(4;9;11) (Table 3). In a case of t(9;11) (case 6), t(11;18)(q23;q11.2) and t(9;18)(p22;q11.2) were present, which indicates a three-way translocation (Table 3). Besides 11q23, 8 of 11 cases had additional cytogenetic abnormalities, including − Y, i(17)(q10), inv(3)(q21q26.2), etc. FISH studies to evaluate MLL gene status were performed in ten cases. Seven cases showed MLL rearrangement, whereas three cases had an intact MLL gene using a commercially available breakapart probe.
Treatment and prognosis after emergence of 11q23
Eight patients (case 1–8) received TKI therapy, and three of them (cases 5–7) also received allogeneic stem cell transplant following TKI therapy. The list of TKIs treatment was case 1 with nilotinib, case 2 with nilotinib and bosutinib, case 3 with imatinib, case 4 with imatinib, case 5 with dasatinib and nilotinib, case 6 with dasatinib, case 7 with nilotinib, and case 8 with dasatinib. The remaining patients (case 9–11) received conventional chemotherapy targeting to acute leukemia.
In patients with 11q23 rearrangements in Ph-positive cells, two patients achieved complete cytogenetic response and major molecular response after TKI treatment; one patient (case 1) had 11q23 as the sole secondary abnormality and the other patient (case 5) had 11q23 at initial CML diagnosis. These two patients were alive at the last follow-up, 50.2 and 6.9 months after 11q23 emergence. The rest of the patients (8/10) died at the last follow-up, at a median of 16.5 months (range, 8–186 months) following the initial diagnosis of CML and a median of 6.7 months (range, 0.8–16.6 months) following emergence of the 11q23 translocation. The patient with 11q23 rearrangement in Ph-negative cells (case 11) died 4.1 months following emergence of 11q23 translocation.
Of the three patients who received allogeneic stem cell transplant, one patient (case 5) achieved complete remission after TKI treatment, followed by stem cell transplant. The other two patients (cases 6 and 7) had no response to TKI treatment. Transplant was performed after induction chemotherapy. Both patients had relapsed disease shortly after transplant and died at the last follow-up.
Discussion
Although frequently encountered in AML and acute lymphoid leukemia (ALL), 11q23 rearrangement occurs very rarely in CML, in less than 1% of cases in this study by conventional karyotype. The translocation partners for 11q23 are diverse with t(9;11) (p22;q23) being the most common. Most cases developed 11q23 translocation in BP, and were often associated with other chromosomal changes. Patients had a low response rate to TKI treatment, and the overall survival was poor.
Clonal evolution with additional chromosomal abnormalities besides t(9;22) is considered as a feature of disease progression in CML. The predominant emergence of 11q23 in BP (8/11) suggests that 11q23/MLL may play a direct role in CML blast transformation. In AML and ALL, the role of MLL in leukemic transformation has been extensively studied. Although the fusion partners for MLL are diverse with more than 70 fusion partners described, most MLL rearrangements join the N-terminal region of MLL with C-terminal portion of its partners, forming chimeric oncoproteins [21]. Although the mechanisms of MLL-induced leukemia are complex and not fully understood, the chimeric proteins have been shown to induce HOX expression, which can block hematopoietic differentiation and induce leukemia [20]. Whether MLL plays a similar role in CML blastic transformation needs further confirmation.
The prognostic significance of clonal evolution in CML is variable and depends on many factors, including the particular type of cytogenetic changes, the time and phase of emergence of clonal evolution, other concurrent chromosomal abnormalities, the presence or absence of other AP features, and treatment regimens. In the era of pre-TKI therapy, INFa induced a complete suppression of clonal evolution in 46% of patients with only Ph chromosome and/or diploid karyotype present after resolution of clonal evolution. About 7% of patients with clonal evolution achieved a complete cytogenetic remission on INFa therapy, and the absence of other AP features was associated with a better response [23]. With the first generation of TKI, imatinib, one study showed that clonal evolution had no significant effect on achieving major or complete cytogenetic response, but it was a poor prognostic factor for survival in both CP and AP [24]. Another study showed that clonal evolution was associated with lower cytgenetic response rate in patients in AP with other signs of acceleration [25]. For patients on second generation of TKI therapy, cases with clonal evolution but without other features of AP had similar hematologic/cytogenetic response and overall survival to cases in CP without clonal evolution. However, when other features of AP were present, the presence of clonal evolution adversely affected overall survival [26].
As mentioned above, the prognostic significance of clonal evolution also depends on the specific type of chromosomal changes. Fabarius et al. reported that the major route, not minor route cytogenetic aberrations at initial diagnosis, had a negative prognostic impact on CML patients [27]. Consistently, a recent study indicated that trisomy 8, chromosome 17 abnormalities, and complex cytogenetic changes carried worse prognosis than others [26]. Accordingly, in European LeukemiaNet recommendations, the major routes, not the minor routes of chromosomal abnormalities, are defined as one of criteria for AP [28]. Although 11q23 abnormalities are considered as “minor route” abnormalities, they appear to carry a poor prognosis. In our cohort, nine patients (9/11) died at the last follow-up. The majority of cases had 11q23 abnormalities in BP(8/11). Only one patient (case 1) had 11q23 in CP, in the absence of other AP features. This patient achieved complete cytogenetic response and major molecular response after nilotinib treatment. Two patients (cases 2 and 3) had 11q23 in AP, accompanied with other AP features including increased blast count and thrombocytopenia. These two cases showed poor response to TKI therapy and transformed to blast phase after a period of 1.6 and 12 months.
Interestingly, not all cases with 11q23 rearrangements showed MLL involvement by FISH analysis. In our series, 30% (3/10) cases showed negative MLL rearrangement by FISH study using a commercial MLL breakapart probe. Although not frequent, similar findings had been described in CML and other myeloid neoplasms. Giurliano et al. reported two cases of myeloid neoplasms (AML and atypical CML) with cytogenetic changes involving 11q23 but with intact MLL gene by FISH: one with t(3;11)(q21;q23) and another with t(6;11)(q15;q23) [29]. Cox et al. studied 378 adult acute leukemia cases and found that in 18 cases with rearrangements involving 11q22-25 bands by conventional karyotyping, only 11 had MLL gene involvements [30]. In CML, two cases with 11q23 rearrangements but intact MLL were previously reported. One case had t(2;11)(p21;q23) [12] and the other had t(4;11)(q21;q23) [5]. In our study, three patients showed 11q23 translocations by conventional cytogenetics but were negative for MLL involvement by FISH, including t(2;11)(q32;q23) (case 4), t(4;11)(q21;q23) (case 1), and t(6;11)(p11.2;q23) (case 2). The exact mechanisms and roles of 11q23+/MLL− in leukemia are unknown. We proposed two potential mechanisms here. First, although MLL gene appeared intact in these cases, MLL gene may be controlled and dysregulated by other promoters or enhancers as a result of translocations. Secondly, other genes at the 11q23 locus may play roles in leukemogenesis, such as FLI1 [31,32]. Future molecular study to identify the breakpoints of these 11q23+/MLL− translocations and their fusion partner is needed.
In case 11, although the patient initially presented with chronic phase CML and responded to TKI, he later developed blast transformation with 11q23 translocation. FISH for BCR-ABL1 was negative. The development of acute leukemia in Ph-negative clones is intriguing. The occurrence of chromosomal changes in Ph-negative cells is less common than in Ph-positive cells, and the significance of this phenomenon is not fully understood [33]. Jabbour et al. analyzed 258 CML patients in CP with imatinib therapy [34]. After a median follow-up of 37 months, 9% patients developed chromosomal abnormalities in Ph-negative cells, with − Y and trisomy 8 being the most common. Others included del (7), del (20), t(11;17), etc. Although similar cytogenetic changes are associated with myelodysplastic syndromes, these cytogenetic abnormalities in CML seem transient and most disappeared after a median of 5 months. Development of MDS or acute leukemia is extremely rare in Ph-negative cells. In a study of 1,701 CML patients, Kovitz found that 3 patients developed AML and MDS in Ph-negative cells [35]. Monosomy 7 and complex cytogenetic changes were the most common chromosomal abnormalities in reported cases of MDS/AML developed in Ph-negative cells [36-40]. In our case, the patient developed AML in Ph-negative cells carrying 11q23/MLL abnormality. To our best knowledge, this is the first case of acute leukemia developing in Ph-negative cells with 11q23/MLL involvement. The mechanisms behind the development of chromosomal abnormalities in Ph-negative cells are not clear, but it is less likely due to TKI therapy [34]. A plausible explanation is that t(9;22) is not a random event, but rather develops from a preceding genetic abnormality, which occurs at the level of hematopoietic stem cells. This genetic abnormality not only induces t(9;22) but also other chromosomal abnormalities. During CML treatment, cells with t(9;22) are eradicated and cells with other chromosomal abnormalities emerge [41].
Two patients (cases 5 and 6) presented with acute leukemia with t(9;22) and 11q23 rearrangements at time of initial diagnosis. This timing raised the question of whether they were de novo AML or BP of CML. Features that favor the latter include the presence of i(17)(q10) and “dwarf” megakaryocytes; both are more frequently seen in CML cases. Also in case 5, the patient had basophilia and eosinophilia, a phenomenon more often encountered in CML.
As mentioned above, occasional case reports of CML with 11q23 translocations are available in the literature. The fusion partners previously reported for 11q23 in CML include 1q21 [10], 2p21 [12], 4q21 [5], 6q25 [7], 9p22 [11,8,6], 17q21 [9], 19p13.1 [14], and 19p13.3 [13]. In our study, 9p21-22/AF9 was the most common partner for 11q23. In addition, we identified several new fusion partners for 11q23 in CML: 2q32, 6p11.2, 17q25, and 18q11.2. Of these, t(11;17)(q23;q25) potentially involves the SEPT9 gene on 17q25 and had been reported in AML and myelodysplastic syndrome with a poor prognosis [42], whereas t(2;11)(q32;q23), t(6;11)(p11.2;q23), and t(11;18)(q23;q11.2) translocations have never been previously described in the literature. It will be of interest to explore and characterize the potential genes involved in these reciprocal translocations and their roles in leukemia.
In conclusion, 11q23 translocations are rare events in CML, frequently occurring in blast phase and associated with a poor prognosis. Not all 11q23 translocations involve MLL gene. We described a first case of acute leukemia developed in Ph-negative cells with 11q23/MLL involvement. Several novel genetic loci are identified as fusion partners for 11q23.
Materials and methods
Case selection, histologic and clinical review
We searched the pathology archives of our institution from 1998 to the present for cases of CML with available karyotypes. Conventional cytogenetics and FISH results were reviewed to identify cases with 11q23 translocations. Other genetic changes involving 11q23, such as del(11)q23, were not included in this study. Biopsy and smear slides were retrospectively reviewed. The corresponding medical records were reviewed to obtain clinical data, including age, sex, disease progression, treatment regimens, response to therapy, and follow-up data.
Cytogenetic studies
Conventional chromosomal analysis was performed on G-banded metaphase cells prepared from unstimulated 24- and 48-h BM aspirate culture using standard methods. Twenty metaphases were analyzed and the results were reported using the International System for Human Cytogenetic Nomenclature (ISCN 2013). FISH for MLL gene rearrangement was conducted on BM culture cells or direct aspirate smears using MLL dual color, breakapart probe (Abbott Molecular, Inc.) according to the standard protocols.
Definition of cytogenetic and molecular response
Complete cytogenetic response is defined as 0% Ph-positive metaphases using conventional cytogenetics analysis of at least 20 metaphases. Major molecular response is defined as more than 3-log reduction of BCR-ABL mRNA.
BCR-ABL1 transcript measurement
BCR-ABL1 and ABL1 transcript levels were detected simultaneously and quantitative results expressed as the ratio of BCR-ABL1 to ABL levels.
References
Hehlmann R. How I, treat CML blast crisis. Blood. 2012;120(4):737–47. doi:10.1182/blood-2012-03-380147.
Quintas-Cardama A, Cortes J. Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood. 2009;113(8):1619–30. doi:10.1182/blood-2008-03-144790.
Mu Q, Ma Q, Wang Y, Chen Z, Tong X, Chen FF, et al. Cytogenetic profile of 1,863 Ph/BCR-ABL-positive chronic myelogenous leukemia patients from the Chinese population. Ann Hematol. 2012;91(7):1065–72. doi:10.1007/s00277-012-1421-6.
Haferlach C, Bacher U, Schnittger S, Weiss T, Kern W, Haferlach T. Similar patterns of chromosome abnormalities in CML occur in addition to the Philadelphia chromosome with or without tyrosine kinase inhibitor treatment. Leukemia. 2010;24(3):638–40. doi:10.1038/leu.2009.222.
Aydin C, Cetin Z, Salim O, Yucel OK, Undar L, Berker KS. Previously unreported chromosomal aberrations of t(3;3)(q29;q23), t(4;11)(q21;q23), and t(11;18)(q10;q10) in a patient with accelerated phase Ph + CML. Case reports in genetics. 2014;2014:582016. doi:10.1155/2014/582016.
Dastugue N, Duchayne E, Huguet F, Demur C, Plaisancie H, Calvas P, et al. t(9;11)(p22;q23) translocation in blastic phase of chronic myeloid leukemia. Cancer Genet Cytogenet. 1992;63(1):37–42.
Haus O, Noworolska A, Laskowski M, Kuliszkiewicz-Janus M, Kozlowska J, Harlozinska-Szmyrka A, et al. Prognostic significance of secondary cytogenetic changes and nonspecific cross-reacting antigen (NCA) in patients with Ph-positive chronic myeloid leukemia. Exp Mol Pathol. 1990;52(2):235–42.
Li L, Ritterbach J, Harbott J, Schroyens W, Lohmeyer J, Pralle H, et al. Blastic phase chronic myeloid leukemia with a four-break rearrangement: t(11;9)(9;22)(q23;p22q34;q11). Cancer Genet Cytogenet. 1993;68(2):131–4.
Nishii K, Usui E, Sakakura M, Miyata E, Ridge SA, Ford AM, et al. Additional t(11;17)(q23;q21) in a patient with Philadelphia-positive mixed lineage antigen-expressing leukemia. Cancer Genet Cytogenet. 2001;126(1):8–12.
Otero L, Moellmann AC, Pombo-de-Oliveira MS, Ornellas MH, Pires V, Bouzas LF, et al. Additional t(1;11)(q21;q23) with mixed lineage leukemia rearrangement in T-blastic crisis of a Ph-positive chronic myeloid leukemia. Eur J Haematol. 2007;79(2):179–81. doi:10.1111/j.1600-0609.2007.00884.x.
Rajcan-Separovic E, Bence-Bruckler I, Wells P, Wang H. Fluorescence in situ hybridization analysis of complex translocations in two newly diagnosed Philadelphia chromosome-positive chronic myelogenous leukemia patients. Cancer Genet Cytogenet. 1999;114(1):71–4.
Royer-Pokora B, Hildebrandt B, Redmann A, Herold C, Kronenwett R, Haas R, et al. Simultaneous occurrence of a t(9;22) (Ph) with a t(2;11) in a patient with CML and emergence of a new clone with the t(2;11) alone after imatinib mesylate treatment. Leukemia. 2003;17(4):807–10. doi:10.1038/sj.leu.2402877.
Suzuki K, Sugawara T, Kowata S, Utsugizawa T, Ito S, Murai K, et al. Uncommon karyotypic abnormality, t(11;19)(q23;p13.3), in a patient with blastic phase of chronic myeloid leukemia. Cancer Genet Cytogenet. 2004;150(2):159–63. doi:10.1016/j.cancergencyto.2003.09.005.
Zamecnikova A. Acquisition of mixed lineage leukemia rearrangement in a chronic myeloid leukemia patient while on imatinib. Hematology reports. 2011;3(2):e13.
Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol. 2005;23(26):6306–15. doi:10.1200/JCO.2005.05.047.
Tomizawa D, Koh K, Sato T, Kinukawa N, Morimoto A, Isoyama K, et al. Outcome of risk-based therapy for infant acute lymphoblastic leukemia with or without an MLL gene rearrangement, with emphasis on late effects: a final report of two consecutive studies, MLL96 and MLL98, of the Japan Infant Leukemia Study Group. Leukemia. 2007;21(11):2258–63. doi:10.1038/sj.leu.2404903.
Schoch C, Schnittger S, Klaus M, Kern W, Hiddemann W, Haferlach T. AML with 11q23/MLL abnormalities as defined by the WHO classification: incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood. 2003;102(7):2395–402. doi:10.1182/blood-2003-02-0434.
Meyer C, Hofmann J, Burmeister T, Groger D, Park TS, Emerenciano M, et al. The MLL recombinome of acute leukemias in 2013. Leukemia. 2013;27(11):2165–76. doi:10.1038/leu.2013.135.
Zeisig BB, Schreiner S, Garcia-Cuellar MP, Slany RK. Transcriptional activation is a key function encoded by MLL fusion partners. Leukemia. 2003;17(2):359–65. doi:10.1038/sj.leu.2402804.
Slany RK. The molecular biology of mixed lineage leukemia. Haematologica. 2009;94(7):984–93. doi:10.3324/haematol.2008.002436.
Harper DP, Aplan PD. Chromosomal rearrangements leading to MLL gene fusions: clinical and biological aspects. Cancer Res. 2008;68(24):10024–7. doi:10.1158/0008-5472.CAN-08-2208.
Manara E, Baron E, Tregnago C, Aveic S, Bisio V, Bresolin S, et al. MLL-AF6 fusion oncogene sequesters AF6 into the nucleus to trigger RAS activation in myeloid leukemia. Blood. 2014;124(2):263–72. doi:10.1182/blood-2013-09-525741.
Cortes J, Talpaz M, O’Brien S, Rios MB, Majlis A, Keating M, et al. Suppression of cytogenetic clonal evolution with interferon alfa therapy in patients with Philadelphia chromosome-positive chronic myelogenous leukemia. J Clin Oncol. 1998;16(10):3279–85.
Cortes JE, Talpaz M, Giles F, O’Brien S, Rios MB, Shan J, et al. Prognostic significance of cytogenetic clonal evolution in patients with chronic myelogenous leukemia on imatinib mesylate therapy. Blood. 2003;101(10):3794–800. doi:10.1182/blood-2002-09-2790.
O’Dwyer ME, Mauro MJ, Kurilik G, Mori M, Balleisen S, Olson S, et al. The impact of clonal evolution on response to imatinib mesylate (STI571) in accelerated phase CML. Blood. 2002;100(5):1628–33. doi:10.1182/blood-2002-03-0777.
Verma D, Kantarjian H, Shan J, O’Brien S, Estrov Z, Garcia-Manero G, et al. Survival outcomes for clonal evolution in chronic myeloid leukemia patients on second generation tyrosine kinase inhibitor therapy. Cancer. 2010;116(11):2673–81. doi:10.1002/cncr.25015.
Fabarius A, Leitner A, Hochhaus A, Muller MC, Hanfstein B, Haferlach C, et al. Impact of additional cytogenetic aberrations at diagnosis on prognosis of CML: long-term observation of 1151 patients from the randomized CML Study IV. Blood. 2011;118(26):6760–8. doi:10.1182/blood-2011-08-373902.
Baccarani M, Deininger MW, Rosti G, Hochhaus A, Soverini S, Apperley JF, et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood. 2013;122(6):872–84. doi:10.1182/blood-2013-05-501569.
Giugliano E, Rege-Cambrin G, Scaravaglio P, Wlodarska I, Emanuel B, Stul M, et al. Two new translocations involving the 11q23 region map outside the MLL locus in myeloid leukemias. Haematologica. 2002;87(10):1014–20.
Cox MC, Panetta P, Lo-Coco F, Del Poeta G, Venditti A, Maurillo L, et al. Chromosomal aberration of the 11q23 locus in acute leukemia and frequency of MLL gene translocation: results in 378 adult patients. Am J Clin Pathol. 2004;122(2):298–306. doi:10.1309/RX27-R8GJ-QM33-0C22.
Kornblau SM, Qiu YH, Zhang N, Singh N, Faderl S, Ferrajoli A, et al. Abnormal expression of FLI1 protein is an adverse prognostic factor in acute myeloid leukemia. Blood. 2011;118(20):5604–12. doi:10.1182/blood-2011-04-348052.
Baud V, Lipinski M, Rassart E, Poliquin L, Bergeron D. The human homolog of the mouse common viral integration region, FLI1, maps to 11q23-q24. Genomics. 1991;11(1):223–4.
Loriaux M, Deininger M. Clonal cytogenetic abnormalities in Philadelphia chromosome negative cells in chronic myeloid leukemia patients treated with imatinib. Leuk Lymphoma. 2004;45(11):2197–203. doi:10.1080/10428190410001723278.
Jabbour E, Kantarjian HM, Abruzzo LV, O’Brien S, Garcia-Manero G, Verstovsek S, et al. Chromosomal abnormalities in Philadelphia chromosome negative metaphases appearing during imatinib mesylate therapy in patients with newly diagnosed chronic myeloid leukemia in chronic phase. Blood. 2007;110(8):2991–5. doi:10.1182/blood-2007-01-070045.
Kovitz C, Kantarjian H, Garcia-Manero G, Abruzzo LV, Cortes J. Myelodysplastic syndromes and acute leukemia developing after imatinib mesylate therapy for chronic myeloid leukemia. Blood. 2006;108(8):2811–3. doi:10.1182/blood-2006-04-017400.
Perel JM, McCarthy C, Walker O, Irving I, Williams B, Kennedy GA. Clinical significance of development of Philadelphia-chromosome negative clones in patients with chronic myeloid leukemia treated with imatinib mesylate. Haematologica. 2005;90 Suppl:ECR25.
Bacher U, Hochhaus A, Berger U, Hiddemann W, Hehlmann R, Haferlach T, et al. Clonal aberrations in Philadelphia chromosome negative hematopoiesis in patients with chronic myeloid leukemia treated with imatinib or interferon alpha. Leukemia. 2005;19(3):460–3. doi:10.1038/sj.leu.2403607.
Andersen MK, Pedersen-Bjergaard J, Kjeldsen L, Dufva IH, Brondum-Nielsen K. Clonal Ph-negative hematopoiesis in CML after therapy with imatinib mesylate is frequently characterized by trisomy 8. Leukemia. 2002;16(7):1390–3. doi:10.1038/sj.leu.2402634.
Chee YL, Vickers MA, Stevenson D, Holyoake TL, Culligan DJ. Fatal myelodysplastic syndrome developing during therapy with imatinib mesylate and characterised by the emergence of complex Philadelphia negative clones. Leukemia. 2003;17(3):634–5. doi:10.1038/sj.leu.2402842.
Quintas-Cardama A, Kantarjian H, Abruzzo LV, Cortes J. Extramedullary BCR-ABL1-negative myeloid leukemia in a patient with chronic myeloid leukemia and synchronous cytogenetic abnormalities in Philadelphia-positive and -negative clones during imatinib therapy. Leukemia. 2007;21(11):2394–6. doi:10.1038/sj.leu.2404865.
Perrotti D, Jamieson C, Goldman J, Skorski T. Chronic myeloid leukemia: mechanisms of blastic transformation. J Clin Invest. 2010;120(7):2254–64. doi:10.1172/JCI41246.
Lee SG, Park TS, Oh SH, Park JC, Yang YJ, Marschalek R, et al. De novo acute myeloid leukemia associated with t(11;17)(q23;q25) and MLL-SEPT9 rearrangement in an elderly patient: a case study and review of the literature. Acta Haematol. 2011;126(4):195–8. doi:10.1159/000329389.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
WW wrote the manuscript. WW and SH designed the study. WW, DA, and SH collected the data and GT performed FISH studies. All authors read and approved the final manuscript.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Wang, W., Tang, G., Cortes, J.E. et al. Chromosomal rearrangement involving 11q23 locus in chronic myelogenous leukemia: a rare phenomenon frequently associated with disease progression and poor prognosis. J Hematol Oncol 8, 32 (2015). https://doi.org/10.1186/s13045-015-0128-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13045-015-0128-2