Abstract
Fear dysregulation is one of the symptoms found in post-traumatic stress disorder (PTSD) patients. The functional abnormality of the hippocampus is known to be implicated in the development of such pathology. Peroxiredoxin 6 (PRDX6) belongs to the peroxiredoxin family. This antioxidant enzyme is expressed throughout the brain, including the hippocampus. Recent evidence reveals that PRDX6 plays an important role in redox regulation and the modulation of several signaling molecules involved in fear regulation. Thus, we hypothesized that PRDX6 plays a role in the regulation of fear memory. We subjected a systemic Prdx6 knockout (Prdx6−/−) mice to trace fear conditioning and observed enhanced fear response after training. Intraventricular injection of lentivirus-carried mouse Prdx6 into the 3rd ventricle reduced the enhanced fear response in these knockout mice. Proteomic analysis followed by validation of western blot analysis revealed that several proteins in the MAPK pathway, such as NTRK2, AKT, and phospho-ERK1/2, cPLA2 were significantly upregulated in the hippocampus of Prdx6−/− mice during the retrieval stage of contextual fear memory. The distribution of PRDX6 found in the astrocytes was also observed throughout the hippocampus. This study identifies PRDX6 as a participant in the regulation of fear response. It suggests that PRDX6 and related molecules may have important implications for understanding fear-dysregulation associated disorders like PTSD.
Similar content being viewed by others
Introduction
Fear acquisition and expression to threatening stimuli are innate responses to avoid dangers or predators to ensure safety and survival [1, 2]. Several pieces of evidence suggest that brain regions, including the amygdala, medial prefrontal cortex, and hippocampus, are required for an appropriate level of fear response [3,4,5]. Dysregulation of these brain regions leads to an excessive fear response in post-traumatic stress disorder (PTSD) [6]. The underlying molecular mechanism is still unclear. Peroxiredoxin 6 (PRDX6) is a multifunctional enzyme belonging to the peroxiredoxin superfamily [7]. Among the peroxiredoxin superfamily, PRDX6 is the only member that displays multiple functions, including the glutathione peroxidase (GPx), acidic calcium-independent phospholipase A2 (aiPLA2), and lysophosphatidylcholine acyltransferase (LPCAT) activities [7, 8]. These activities determine their roles in various organs under different physiological and pathobiological conditions [9, 10]. Although PRDX6 is expressed in various brain regions associated with fear regulation, including the hippocampus [11, 12] and expressed in all cell types with high expression level in the astrocytes [11, 13, 14], its function regarding cognition, particularly fear memory regulation has not yet been identified. Previous findings confirmed the association between enhanced fear memory and decreased overall enzymatic activity of GPx in the hippocampus, suggesting that GPx-PRDX6 may be involved in the regulation of fear response [15]. Besides, activation of PLA2 is required to acquire and retrieve emotional memory [16], indicating that aiPLA2-PRDX6 may also have a similar function. All the evidence mentioned above led us to hypothesize that PRDX6 may play an important role in regulating fear memory.
Trace fear conditioning (TFC) is a behavioral paradigm widely used to study associative fear memory [17]. The molecular mechanisms underlying fear memory processes are commonly approached with a fear conditioning paradigm, which shares similar mechanisms across species [18, 19]. This task causes fear memory formation by triggering a series of molecular and cellular changes to strengthen synaptic plasticity in emotion-related brain regions, including the hippocampus and the amygdala [20]. Tyrosine kinase receptor B (TrkB) and its downstream targets such as extracellular signal-regulated protein kinases 1 and 2 (ERK1/2) and protein kinase B (AKT) [21] are involved in the mediation of synaptic plasticity for fear memory formation. Interestingly, PRDX6 can modulate both ERK1/2 and AKT [22] expression, supporting our hypothesis that PRDX6 may participate in the neurobiological process of fear memory.
We performed behavioral, cellular, and molecular studies in the Prdx6 knockout (Prdx6−/−) mice in the present study. We first identified the function of PRDX6 by employing Prdx6−/− mice to trace fear conditioning (TFC) and found that this knockout strain exhibited enhanced contextual fear memory. We further confirmed with a gain-of-function study by injecting lentivirus-carrying mouse PRDX6 (mPRDX6) into the lateral ventricle of Prdx6−/− mice, which mitigated their enhanced contextual fear memory. We also investigated their general behaviors using open field, three-chambers tests, marble burying, and elevated plus-maze. Proteomic and immunoblotting analyses were also performed in this study to understand the molecular mechanism better.
Materials and methods
Animals
All experiments on animals were approved by the Institutional Animal Care and Use Committee of Tzu Chi University, Taiwan (approval #104099), and complied with the Taiwan Ministry of Science and Technology guidelines for animals' ethical treatment. Twelve- to 14-week-old wild-type (C57BL/6J) and Prdx6−/− mice were originally generated by Wang X. and colleagues and provided by Dr. Shun-Ping Huang at Tzu Chi University, Taiwan [23]. All mice were maintained in the Laboratory Animal Center of Tzu Chi University and were housed with ad libitum access to food and water under a constant 12-h light/dark cycle. Heterozygous knockout mice with one male and two females were crossed to reproduce Prdx6−/− mice and their wild-type littermates. Genotyping (Additional file 1: Fig. S1a) was conducted to confirm the absence of the Prdx6 gene in knock-out mice before every behavioral test. After the completion of trace fear conditioning, qRT-PCR (Additional file 1: Fig. S1b) and immunoblotting (Additional file 1: Fig. S1c) were carried out to visualize mRNA and protein level of PRDX6 in Prdx6−/− mice. Moreover, we also recorded the morphology and bodyweight of the Prdx6−/− mice. We found that both morphology (Additional file 1: Fig. S1d) and body weight (Additional file 1: Fig. S1e) (t19 = − 1.426, p = 0.170) of the Prdx6−/− mice appeared to be normal.
Behavioral tests
Trace fear conditioning (TFC)
Trace fear conditioning was modified from the protocol used in our previous study [17]. The conditioned chamber (17 cm (W) × 17 cm (L) × 25 cm (H)) illuminated with a white 30-lx light under the top-view camera was used in this study. After three days of habituation, mice were placed into the chamber for 2 min as a baseline and were then trained with three pairs of tone (CS) and electric foot shock (US) with an inter-trial interval of 1 min. One pair of CS-US consisted of a 20 s of tone (6000 Hz, 85 dB) followed by 1 s electric foot shock (2 mA) with a 10 s training interval. The mice were maintained in the conditioned chamber for a total of 9 min. To test their contextual fear memory retention, the mice were re-exposed to a conditioned chamber for 6 min without giving any tone and footshock after 24 h of the training session. One hour later, the mice were tested with cue fear memory by exposing them to 6 min of tone only after 1 min of habituation in an unconditioned context. The freezing behavior, defined as no movement except breathing, was analyzed using tracking software (EthoVision XT 15, Noldus Information Technology). The freezing time was converted to freezing percentage using the following formula:
Open field test
An open chamber (50 cm (W) × 50 cm (L) × 50 cm (H)) was used to test the locomotor function and anxiety-like behavior of the mice under the light-on condition [17]. The camera hung on top recorded the animals' locomotor activity within 10 min of the test. Their locomotor activity (distance traveled and moving speed) and time spent in the center and outer area were measured and analyzed by the tracking software (EthoVision XT 15, Noldus Information Technology).
Three-chambers test
This task was composed of three trials with 10 min of exploration time for each. The intertrial interval was 20 min. During the habituation trial, the experimental mice were placed into the middle compartment. Mice freely explored all three compartments that contained empty cups at the end of the left and right compartments. For the second trial, a sex- and age-matched stranger mouse (S1) was kept inside the cup in the right compartment. The experimental mice were then allowed to explore all compartments. For the third trial, another stranger mouse (S2) was placed in the cup located in the left compartment. The experimental mice were again placed in the middle compartment and allowed to explore the chamber. The time spent interacting with the empty cups or stranger mice was analyzed by tracking software (EthoVision XT 15, Noldus Information Technology). We followed the protocol described in a previous study [24].
Marble burying test
The protocol was described in a previous study [25]. Briefly, the cage (30 cm × 27 cm × 26 cm) was filled with 5 cm autoclaved bedding containing 20 marbles arranged centrally 4 by 5 and was kept in a soundproof box with 10 lx. Mice were placed and then filmed for 30 min. The number of unburied marbles was counted after 25 min.
Elevated-plus maze test
The elevated-plus maze is used to assess the anxiety-related behavior in rodents [26]. The apparatus consists of a "plus"-shaped maze at 60 cm height above ground with two oppositely positioned closed arms and two oppositely positioned open arms and a center region. The experiment was conducted during day time under the same light intensity (~ 130 lx) as provided in the animal housing room. The mice were placed in the center region facing one of the closed arms and allowed to explore the maze freely for 10 min. We used a video camera and tracking system (EthoVision XT 15, Noldus Information Technology) to record and analyze their anxiety-like behavior, respectively.
Lentiviral vector preparation
Total RNA was isolated from the mouse hippocampus and converted to cDNA using oligo (dT) 18 primers. The cDNA was then amplified using a specific forward primer (5′-CTA GCTAGC ATG CCC GGA GGG TTG CTT C-3′ containing a NheI site) and reverse primer (5′-GC GAA TTC TTA AGG CTG GGG TGT ATA ACG-3′containing an EcoRI site) [52]. Full-length mouse Prdx6 cDNA was purified by a PrestoTM Mini Plasmid Kit (catalog #PHD300, Geneaid Biotech Ltd., Taiwan). pLAS3wPpuro vectors containing EGFP and Prdx6 were designed for the production of lentiviral vectors. HEK293T cell lines were used to produce lentiviruses containing either EGFP or Prdx6 gene. Harvested lentivirus was concentrated using PEG-it (™) virus precipitation solution (System Biosciences, CA) and processed for titration.
Stereotaxic surgery and intracerebroventricular injections of lentivirus containing mouse PRDX6
The procedures for stereotaxic injection were performed according to our previous study with slight modification [27]. The mice were anesthetized by intraperitoneal (IP) injection of ketamine/xylazine mixture (0.45 ml/25 g of body weight) and then fixed on the stereotaxic frame (Stoelting, US). The lentivirus containing either EGFP or mouse PRDX6 was dissolved in sterile 1× phosphate-buffered saline (PBS) to obtain the final titer of 7 × 105 in 2 µl volume. The lentiviral vectors were then unilaterally injected into the right lateral third ventricle with the following brain coordinates: anterior–posterior (AP), − 0.5 mm; medial–lateral (ML), 1 mm (from the bregma): and DV, 2.33 mm (from the skull surface). A 10-µl Hamilton syringe with a 26 G needle was placed on the microinfusion pump (KD Scientific Inc. MA, USA) and connected via polyethylene—28 mm I.D. tubing to the internal cannula. We injected the lentiviruses with a flow rate of 0.5 µl/min over 4 min. The cannulas were placed for another 5 min to allow diffusion before removing them. Following surgery, mice were given pain killers (meloxicam, Achefree, Taiwan) and allowed to recover for 4 weeks before the behavioral tests.
Liquid chromatography–tandem mass spectrometry (LC/MS–MS)
After completing a contextual test, protein samples were collected from the whole hippocampi of Prdx6+/+ and Prdx6−/− mice. Protein samples from 3 mice were pooled together for each group and measured the protein concentration using Lowry assay [28]. For in-solution digestion, 5 µg of protein were used for each group of mice. The samples were treated with 10 mM ammonium bicarbonate and the disulfide bonds were reduced with 5 mM dithiothreitol (DTT) in 10 mM ammonium bicarbonate at 60 °C for 1 h. Samples were subsequently alkylated with 15 mM Iodoacetamide (IAA) in 10 mM ammonium bicarbonate for 45 min in the dark at room temperature. Protein digestion was done by incubating the samples with 50 ng/µl of sequencing grade trypsin (1:20 trypsin:protein) (Promega, Germany) o/n at 37 °C. Before the injection into the LC–MS/MS, the samples were protonated with 0.1% formic acid.
The tryptic peptides from the digested samples were injected into an Ultimate3000 Nano/Capillary LC System (Thermo Scientific, UK) coupled to a Hybrid quadrupole Q-Tof impact II™ (Bruker Daltonics) equipped with a Nano-captive spray ion source. The peptides were enriched on a µ-Precolumn 300 µm i.d. × 5 mm C18 Pepmap 100, 5 µm, 100 A (Thermo Scientific, UK), separated on a 75 μm I.D. × 15 cm and packed with Acclaim PepMap RSLC C18, two μm, 100 Å, nanoViper (Thermo Scientific, UK). Solvent A and B containing 0.1% formic acid in water and 0.1% formic acid in 80% acetonitrile were supplied on the analytical column. A gradient of 5–55% solvent B was used to elute the peptides at a constant flow rate of 0.30 μl/min for 30 min. Electrospray ionization was carried out at 1.6 kV using the CaptiveSpray. Mass spectra (MS) and MS/MS spectra were obtained in the positive-ion mode over the range (m/z) 150–2200 (Compass 1.9 software, Bruker Daltonics). We performed the LC–MS analysis of each sample in triplicate.
Bioinformatics and data analysis
The MS data were quantified with MaxQuant 1.6.6.0 using Andromeda search engine to correlate MS/MS spectra to the Uniprot Mus Musculus database [29]. Using MaxQuant's standard settings, label-free quantitation was performed. We used trypsin as a digesting enzyme, carbamidomethylation of cysteine as a fixed modification, and the oxidation of methionine and acetylation of the protein N-terminus as variable modifications. We set two miss cleavages as the maximum and a 0.6 Dalton as the main search's mass tolerance. At least one unique peptide with a minimum of 7 amino acids was used for further analysis [30, 31].
The log2 fold change > 1.2 was a cut off for differential expression proteins (DEPs) [31, 32]. The list of differential expression proteins (DEPs) was then inputted to Venn diagrams [33]. The list of up-and down-regulated proteins was then inputted in Panther software for protein classification [34]. Enrichr software was used to analyze enrichment terms from gene ontology (GO) biological processes (https://amp.pharm.mssm.edu/Enrichr/). The functional interaction networks between DEPs and memory-associated molecules were analyzed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database version 11 (http://string-db.org/cgi/input.pl). The MultiExperiment Viewer (MeV) software [35] was used to produce a heatmap for up-and down-regulated proteins extracted from the GO term "protein phosphorylation."
Detection of oxidative stress levels in the hippocampus
To measure reactive oxygen species (ROS) levels in the hippocampus, mice were sacrificed, and the brains were isolated 20 min after the contextual test. The procedure was conducted according to a previous study with minor modifications [36]. Briefly, the fixed brains were sectioned by cryostat with 20 µm thickness. Hippocampal sections were then immersed in 1 μmol/l dihydroethidium (DHE) in PBS solution at room temperature for 5 min. The stained sections were washed with 1× PBS three times and cover-slipped. DHE is oxidized by superoxide anion to form ethidium binding to DNA in the nucleus and emits red fluorescence. The images were viewed and taken under a fluorescent microscope (Nikon model #ECLIPSE Ni-E, Japan) with an excitation/emission wavelength of 380/420 nm.
Immunofluorescence staining
For immunohistochemistry, mice were anesthetized and transcardially perfused using 0.9% saline and 4% paraformaldehyde. Brains were exercised immediately and postfixed with 4% PFA for another 2 days. After that, the brains were washed with 1× PBS three times and then stored in 30% sucrose at 4 °C. After the dehydration period, the brains were embedded in an optimal cutting compound (Sakura Finetek USA, Inc., USA) and stored at − 80 °C until sectioning. Cryopreserved brains were sectioned at 20 µm using cryostat. Brain sections were washed with a washing buffer (1× PBS containing 0.3% Triton X-100) and treated with a permeating buffer (1% Triton X-100 and 2% Tween 20 in 1× PBS) for 30 min. Sections were further blocked with a blocking buffer containing 1% normal goat serum, 0.25% Triton X-100 dissolved in 1× PBS for 1 h. Subsequently, samples were double-stained with polyclonal rabbit anti-GFAP (1:200, Abcam, UK) and monoclonal mouse anti-PRDX6 (1:150, Bethyl laboratories, Inc, USA). The samples were then washed with washing buffer and incubated in secondary antibody: Alexa Fluor 546 anti-mouse and Alexa Fluor 488 anti-rabbit IgG (1:200, ThermoFisher Scientific, USA) for 1 h, followed by washes with PBS, and counterstained with DAPI (1:10,000) for 5 min. The images were obtained by either fluorescent microscope (Nikon model# ECLIPSE Ni-E, Japan) or confocal microscope (Nikon model#C2+, Japan).
Western blot analysis
The mice were sacrificed immediately after the completion of acute immobilization stress. Under trace fear conditioning, hippocampal proteins were extracted at 3 h after training and 20 min after the contextual test. After decapitation, the whole hippocampi were isolated and homogenized in ice-cold RIPA lysis buffer 1× (Millipore, USA) containing protease and phosphatase inhibitors. The protein samples were kept on ice for 30 min before centrifuging at 13,000 rpm for 15 min at 4 °C. The supernatants were collected for further experiments. For non-reducing SDS-PAGE, protein (30 or 45 μg) samples were boiled at 95 °C in 1× sample buffer without reducing agent for 10 min, and samples were cooled for 5 min. Similar to non-reducing conditions, adding reducing agents into protein samples were included to study total PRDX6 and other proteins of interest under reducing condition. The samples were loaded and run on 8% or 10% SDS-PAGE at 80 V in stacking gel and 120 V in resolving gel. The separated proteins were then transferred to a PVDF membrane (0.2 and 0.4 μm pore size) at 30 V overnight. The blots were incubated with anti-NTRK2 (1:1000; Abcam, UK), anti-cPLA2 (1:1000; Santa Cruz, USA), anti-pERK1/2 (1:1000; Cell Signaling, Danvers, MA), anti-total ERK1/2 (1:1000; Cell Signaling, Danvers, MA), anti-PSD95 (1:2000; ThermoFisher, USA), anti-PRDX6 (1:2000; Abcam, UK) or anti-β-actin antibody (1:10,000; Sigma-Aldrich) in TBST containing 0.1% BSA (ThermoFisher, USA) overnight at 4 °C room on a shaker. The next day, the blots were incubated with horseradish peroxidase-conjugated secondary antibody goat anti-mouse IgG (Cell signaling, Danvers, MA) for cPLA2, PSD95, PRDX6, and β-actin and goat anti-rabbit (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for total NTRK2 or TrkB, pERK1/2 and ERK1/2 with the dilution of 1: 10,000 in blocking buffer for 1 h at room temperature. A list of antibodies used in this study was provided in Additional file 2: Table S5. After three washes for 5-min in the TBST buffer, the membranes were developed using ECL (Western lightning® Plus ECL, PerkinElmer Inc, MA, USA) and detected under the UVP Biospectrum 810 imaging system. The band intensities were quantified using ImageJ 1.52a (National Institutes of Health, USA).
Statistical analysis
Based on previous studies [37,38,39], we decided to use a sample size from 3 to 20 per group with enough power to see a statistically significant difference. Statistical analysis was performed using SPSS (version 25, IBM Corporation), and the graphs were made using GraphPad Prism version 8. After assessing the normality using the Shapiro–Wilk test, Student's t-tests were conducted compared to two independent groups with a normal distribution. In contrast, data that is not normally distributed were assessed by Mann–Withney U-test. One-way ANOVA followed by Bonferroni's post hoc analysis was used for multiple comparisons. For learning ability of TFC and social interaction of three-chamber test, the results were analyzed as mixed-design repeated-measures ANOVA with trials as within-subjects factor and genotypes as a between-subjects factor. The significant interaction was then followed up with the Bonferroni-corrected t-test when a significant F-value was determined. All data are presented as mean ± SEM, with statistically significant at p < 0.05. Sample sizes are indicated in figure legends.
Results
Prdx6 −/− mice exhibited enhanced fear learning and memory
To identify the function of PRDX6 in fear response, Prdx6−/− mice underwent trace fear conditioning (TFC) according to the protocol schemed in Fig. 1a. During the first three days, mice were placed in the conditioning chamber and acclimatized to the context for 15 min per day. On day 4, TFC was applied, followed by a contextual test 24 h later. Using mixed design repeated ANOVA, there was no significant effect of the interaction between the genotypes and trials on freezing percentage (F(2.254,58.606) = 1.042, p = 0.366, Fig. 1b) during TFC. The two genotypes exhibited normal learning during the training session, indicated by an increased freezing percentage from baseline to trial 3 as shown by the main effect of trials (F(2.254,58.606) = 125.868, p = 0.000, Fig. 1b). There was a significant effect of genotypes on freezing percentage during TFC (F(1,26) = 6.638, p = 0.016, Fig. 1b). Bonferroni-corrected t-test revealed significant difference between the two genotypes at trials 2 (t26 = − 2.580, p = 0.016, Fig. 1b). These results suggested that deficiency of PRDX6 leads to fast acquisition of fear memory. No significant difference in total freezing percentage during TFC training (t26 = − 1.302, p = 0.204, Fig. 1c) between the Prdx6+/+ and Prdx6−/− mice. Interestingly, the Prdx6−/− mice exhibited a significantly higher freezing response to conditioned context (t26 = − 2.985, p = 0.006, Fig. 1d) and cue (t26 = − 2.956, p = 0.007, Fig. 1e) than the Prdx6+/+ mice suggesting the impact of the Prdx6 gene on the regulation of contextual and cued fear memories.
Loss of the Prdx6 gene caused fast learning and enhanced fear memories. a General procedure for trace fear conditioning: Habituation of mice in the chamber was performed for 3 consecutive days. The next day, mice were conditioned with three tone and shock pairs. Contextual fear memories were tested 24 h later, followed by a tone test to evaluate cue fear memory (n = 14/group). b The learning curve for baseline and three trials of TFC indicated both groups of mice learnt normally though significant differences appeared in trial 2. c Total freezing percentage of Prdx6+/+ or Prdx6−/− mice during the training session. d Total freezing percentage during the contextual test of mice. e Total freezing percentage during the tone test of mice. All data represent the mean ± the SEM. *p < 0.05. TFC trace fear conditioning
Lentivirus containing mouse PRDX6 (LV-mPRDX6) attenuated contextual fear memory of Prdx6 −/− mice
To further confirm the role of PRDX6 in the expression of fear memory, the gain-of-function study was conducted by intracerebroventricularly injecting LV-mPRDX6 into the lateral ventricle near the hippocampal region of Prdx6−/− mice. The mice were then subjected to TFC 4 weeks after the injection (Fig. 2a). Figure 2b and c illustrate the site of injection and lentiviral construct, respectively. There was no effect of group on learning ability as shown in Fig. 2d (F(1,16) = 0.551, p = 0.469; Prdx6−/− mice with LV-EGFP vs LV-mPRDX6). Both groups displayed normal learning during training sessions indicating an increased freezing percentage from baseline to trial 3 as shown by the main effect of trials (F(3,48) = 26.691, p = 0.000, Fig. 2d). During the training session, the total freezing percentage was similar between the two groups (t16 = − 0.654, p = 0.522, Fig. 2e), indicating that the injection of LV-mPRDX6 did not affect the learning ability of the Prdx6−/− mice. Importantly, lentiviral injection of mPRDX6 successfully reduced enhanced contextual fear response of the Prdx6−/− mice (t16 = 2.698, p = 0.016, Fig. 2f). However, re-expression of mPRDX6 failed to rescue cue fear memory (t16 = − 0.700, p = 0.494, Fig. 2g). Fluorescent images demonstrated the expression of mPRDX6 (Fig. 2h) and EGFP (Additional file 1: Fig. S2a, b) in three hippocampal regions, including the CA1, CA3, and DG after the completion of the tone test. We also detected the expression of mPRDX6 in the amygdala and prefrontal cortex (Fig. 2h and Additional file 1: Fig. S3a, b). These results suggest that hippocampal PRDX6 is involved in regulating fear expression, at least for contextual fear memory.
Intraventricular injection of mouse PRDX6 (mPRDX6) lentiviruses attenuated enhanced contextual fear memory of Prdx6−/− mice. a The procedure for the overexpression study. Mice were injected with lentivirus and housed for 2 weeks before performing trace fear conditioning. b Representative image of cannula tip position (red) in the right lateral ventricle. c The schematic lentivirus construct pLAS3w. Ppuro contains either EGFP or mPRDX6. d The learning curve of baseline and after each tone-shock pair (n = 10/group). e Total freezing percentage of Prdx6+/+ and Prdx6−/− mice during the training session. f Total freezing percentage during the contextual test of mice. g Total freezing percentage of the mice during the tone test. h mPRDX6 expression across the different subregions in the hippocampus of Prdx6−/− mice. All data represent the mean ± the SEM. *p < 0.05. TFC trace fear conditioning, LV-mPRDX6 lentivirus containing mouse PRDX6
Deletion of the Prdx6 gene caused hyperlocomotion without affecting social exploration and recognition
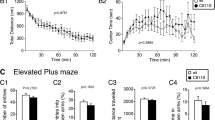
The heatmap during 10 min of exploration in the open field chamber was presented in Fig. 3a. The total distance traveled (t31 = − 2.191, p = 0.036, Fig. 3b) and moving speed (t31 = − 2.197, p = 0.036, Fig. 3c) of the Prdx6−/− mice were significantly higher than those of the Prdx6+/+ mice. An open field test indicated that Prdx6−/− mice exhibited hyperlocomotion compared with Prdx6+/+ mice; hence higher freezing response to context did not result from reduced locomotion. We then assess object exploration, sociability and social novelty behaviors of the Prdx6−/− mice using a three-chamber apparatus [40]. For the novel object exploration test (trial 1), both genotypes demonstrated a significant preference for exploring empty cups, and no significant genotype effect was observed (side: F(1.604,52.935) = 46.642, p = 0.000; genotype: F(1,33) = 0.003, p = 0.958; genotype × side: F(1.604,52.935) = 0.794, p = 0.432, Fig. 3d). The stranger mouse 1 (S1) was placed in the right compartment within an inverted wire cup for the sociability test. Both genotypes demonstrated a significant preference for exploring stranger mouse 1 and no significant genotype effect was observed (side: F(2,66) = 28.869, p = 0.000; genotype: F(1,33) = 0.232, p = 0.633; genotype × side: F(2,66) = 0.118, p = 0.889, Fig. 3e). In the social novelty preference test, the interaction duration with the novel mouse (S2) appeared to be normal since the Prdx6−/− mice spent similar time with the novel mouse compared to wild-type group (F(1,20) = 0.000; p = 0.991, Fig. 3f). Both genotypes stayed with the novel mouse longer than the familiar mouse (S1) (F(1.495,29.895) = 11.089; p = 0.001, Fig. 3f), representing the normal response of social novelty. In each test, no significant difference in locomotor activity was recorded, measured by equal distance traveled (t38 = − 1.056, p = 0.297, Fig. 3g) and moving speed (t38 = 0.340, p = 0.736, Fig. 3h) between the two genotypes.
Increased locomotor function, but normal anxiety-like behavior, exploration, sociability, and social novelty in Prdx6−/− mice. a Heatmaps during 10 min of exploration in an open field chamber. b Quantification data of distance traveled for 10 min (n = 15–18/group, Student’s t-test). c The mean moving speed (cm/s) of the mice introduced in the open field test. d Time spent on each side of the chamber containing empty wire cups (novel object) (n = 17/group). e Time spent on each side of the chamber containing a stranger mouse 1 (S1) or empty wire cup. f Time spent on each side of the chamber containing familiar mouse 1 (S1) or novel mouse (S2). g The mean distance traveled during three trials of the task. h The mean moving speed during three trials of a task. i The mean percentage of center zone time (n = 15–18/group, Student's t-test). j The mean percentage of outer zone time. k Percent time spent in open arms (n = 18–20/group). l Percent time spent in close arms. m Percent marbles buried in the marble-burying test. All data represent the mean ± the SEM. *p < 0.05
Normal anxiety-like behavior and hypervigilance in Prdx6 −/− mice
We next investigated anxiety response and hypervigilance in Prdx6−/− mice using an open field, elevated plus-maze, and marble burying tests, respectively. We observed equal time spent in the center (t31 = − 0.493, p = 0.632, Fig. 3i) and outer zone (t31 = 0.235, p = 0.816, Fig. 3j) in an open-field chamber between the two genotypes. The Prdx6−/− mice showed similar results in elevated plus-maze as controls indicated by equal time spent in open arms (U = 171, Z = − 0.263, p = 0.792, Fig. 3k) and close arms (t36 = − 0.180, p = 0.858, Fig. 3l) indicating normal anxiety-like behavior in Prdx6−/− mice. Performing the marble burying test, we observed no significant difference in the percentage of buried marbles between the two genotypes (t20 = 0.378, p = 0.709, Fig. 3m). This result demonstrated that deletion of the Prdx6 gene did not cause hypervigilance.
Proteomic analysis for total hippocampal proteins extracted during the contextual memory retrieval stage
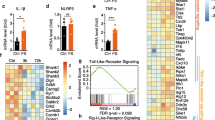
To understand what hippocampal proteins are involved in the retrieval process of contextual memory, we conducted a proteomic analysis for total hippocampal proteins collected during the retrieval stage of TFC (Fig. 4a). Liquid chromatography–tandem mass spectrometry (LC/MS–MS) provided a total of 937 proteins that differentially expressed in the hippocampus of Prdx6−/− and Prdx6+/+ mice. The top 20 up- and down-regulated differential expression proteins (DEPs) was provided in Additional file 2: Table S1. All proteins on the list from both genotypes were plotted in Venn diagrams based on their expressions (Fig. 4b). There were 11 proteins specifically expressed in Prdx6+/+ mice, 7 proteins expressed only in Prdx6−/− mice, and 919 proteins expressed in both genotypes. Using Panther software, the differential expression proteins (DEPs) were classified into three gene ontologies (GO): molecular function, biological process, and cellular component. According to molecular functions, the most overrepresented groups were catalytic activity (40.50% up- and 25.23% down-regulated proteins, Fig. 4c) and binding (35.10% up- and 47.85% down-regulated proteins). In the GO biological process, the main biological processes of DEPs were cellular processes (27.13% up- and 25.93% down-regulated proteins, Fig. 4d) and metabolic processes (16.32% up- and 3.61% down-regulated proteins). The analysis of cellular components indicated that cell (the plasma membrane and any external encapsulating structures; 23.78% up- and 23.83% down-regulated proteins, Fig. 4e) and cell part (any constituent part of a cell; 23.78% up- and 23.83% down-regulated proteins) were the main cellular components of DEPs.
Three functional classifications of the proteins in the hippocampus of mice re-exposed to conditioned chambers. a The schematic diagram of trace fear conditioning and protein collection (n = 3/group). b Venn diagram defining the difference in protein expressions between Prdx6−/− and Prdx6+/+ mice. The 125 upregulated proteins and 130 down-regulated proteins affected by the contextual test were classified into three functional classifications: c molecular function, d biological process, and e cellular component
PRDX6 regulates fear memory retrieval via the MAPK signaling pathway
Oxidative stress is known to be involved in the modulation of fear memory [41]. To measure the oxidative status in the hippocampus of Prdx6−/− mice during memory retrieval, we performed dihydroethidium (DHE) staining. The brains were collected 20 min after the contextual test (Additional file 1: Fig. S4a). Additional file 1: Figure S4b shows the ethidium fluorescence of DHE. Quantitative analysis showed no significant difference in the DHE-positive density was found between genotypes in both CA1 (t4 = − 0.508, p = 0.638, Additional file 1: Fig. S4c) and CA3 (t4 = − 0.060, p = 0.0.955, Additional file 1: Fig. S4d) subregions of the hippocampus. The results demonstrated that PRDX6 might not regulate fear response by controlling cellular oxidation. Moreover, it led us to question whether PRDX6 directly modulates the cellular signaling cascade to control fear memory expression.
To delineate the molecular pathways responsible for the enhanced fear response of the Prdx6−/− mice, Enrichr software was then conducted to identify the enriched biological process of the DEPs (cut-off 1.2 fold change) in Prdx6−/− mice. The protein phosphorylation (GO: 0006468) (p = 0.0056) was one of the significant enrichment terms from the GO biological process of DEPs in Prdx6−/− mice (Additional file 2: Table S2; the top 25 enrichment terms from GO biological process). We next extracted 15 DEPs from the GO term "protein phosphorylation" (GO: 0006468) (Additional file 2: Table S3). The 8 proteins that were upregulated and 7 proteins that were down-regulated in Prdx6−/− mice were input to STRING software to obtain the networks of protein–protein interaction involved in memory processes (Fig. 5a, b). The significant nodes of DEPs were identified according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (Additional file 2: Table S4). The DEPs of Prdx6−/− mice were strongly associated with the neurotrophin signaling pathway (false discovery rate or FDR = 0.00012), Ras signaling pathway (FDR = 0.00073), and MAPK signaling pathway (FDR = 0.0011). These pathways are well known to fear memory consolidation and retrieval [42, 43]. To study the molecular changes that participate in regulating contextual fear memory retrieval of Prdx6−/− mice, we extracted DEPs from the MAPK signaling pathway, including AKT2, CHUK, NTRK2, and RPS6KA1. We created new networks, including MAPK1, MAPK3, BDNF, cPLA2, and PRDX6, using STRING software (Fig. 5c). During retrieval (Fig. 6a), western blot analysis was performed to confirm the expression of the key proteins from the network, including NTRK2, AKT, ERK1/2, and cPLA2, during retrieval of contextual memory. Significant upregulation of NTRK2 (or TrkB) (t6 = − 2.798, p = 0.031, Fig. 6b), AKT (t6 = − 4.242, p = 0.005, Fig. 6c), ERK1/2 phosphorylation (t5 = − 5.336, p = 0.003, Fig. 6d) and cPLA2 (t6 = − 2.761, p = 0.033, Fig. 6e) were recorded in the hippocampus of Prdx6−/− mice after a contextual test. Postsynaptic density protein 95 (PSD95), a postsynaptic marker, was also detected and no significant difference was recorded (t6 = − 1.843, p = 0.115, Fig. 6f) in Prdx6−/− mice. These results demonstrated the correlation of the MAPK pathway with PRDX6 to regulate fear memory retrieval.
Proteomic analysis revealed differential expression proteins (DEPs) MAPK and Ras signaling pathways in the hippocampus of Prdx6−/− mice during retrieval of contextual fear memory. a Functional protein–protein interaction networks of 15 proteins related to GO term "protein phosphorylation" (GO:0006468). The significant nodes were labeled in red for Neurotrophin signaling pathway (FDR 0.00012), blue for the Ras signaling pathway (FDR 0.00073), and green for the MAPK signaling pathway (FRD 0.0011). b Heatmap of 15 proteins in GO term "protein phosphorylation" (GO:0006468) with 8 upregulated proteins and 7 down-regulated proteins in Prdx6−/− mice. c STRING showed a predicted functional protein–protein interaction network of proteins in the KEGG pathway termed "MAP kinase signaling pathway" with memory-associated proteins and PRDX6. The significant nodes were labeled in red for the MAPK signaling pathway (FDR 1.25e−12) and green for the Ras signaling pathway (FDR 3.33e−11)
Activation of MAPK signaling pathways in the hippocampus of Prdx6−/− mice during retrieval of contextual fear memory. a Hippocampal protein samples were collected 20 min after the contextual test for the retrieval process to validate proteins in the MAPK signaling pathway (mmu04010), including TrkB (NTRK2), AKT, pERK1/2, and cPLA2. (b–f; upper panels) Immunoblots of TrkB, AKT, pERK1/2, tERK1/2, cPLA2, PSD95, and β-actin expression in the hippocampus during memory retrieval. (b–f; lower panels) Quantification data for the expression levels of TrkB, AKT, phosphorylated ERK1/2, cPLA2, and PSD95 in the hippocampi of mice (n = 3–5/group). All data represent the mean ± the SEM. *p < 0.05. TrkB tyrosine receptor kinase B, AKT or PKB Protein kinase B, PSD95 postsynaptic protein density 95
Co-localization of PRDX6 with the astrocytic marker, GFAP, in the hippocampus
Since we focused on identifying the function of PRDX6 in the regulation of fear memory, we then confirmed the distribution of PRDX6 in three brain regions primarily involved in fear memory formation—the hippocampus [13], amygdala [11], and prefrontal cortex [44]. Previous studies report that PRDX6 is highly expressed in the astrocytes [11] under various conditions but not known in TFC. We thus performed double staining with anti-PRDX6 and anti-GFAP (astrocyte marker) antibodies to examine whether PRX6 is also expressed in astrocytes after TFC. Our results demonstrated that PRDX6 is expressed in the hippocampal astrocytes within the CA1, CA2, CA3, and DG (Fig. 7a, b). We also recorded the expression of PRDX6 in the amygdala (Additional file 1: Fig. S5a, b) and prefrontal cortex (Additional file 1: Fig. S6a, b).
PRDX6 protein is highly expressed in the astrocytes throughout the hippocampus. a Sagittal section of the brain showing colocalization of PRDX6 (red) with GFAP (green), scale bar 1000 μm. b Confocal images of PRDX6-GFAP colocalization in the astrocytes of CA1, CA2, CA3, and DG subregions of the hippocampus, scale bar 20 μm
Discussion
The present study reports that the loss of the Prdx6 gene in the brain led to enhanced trace fear memory to context. The intracerebroventricular injection (i.c.v) of LV-mPRDX6 could reverse the enhanced contextual fear response, a hippocampal-dependent memory, of Prdx6−/− mice. We confirmed that the observed effect was attributable to PRDX6. Proteomic and western blot analysis revealed that mitogen-activated protein kinase (MAPK) signaling pathways were highly activated in the hippocampi of Prdx6−/− mice during the expression of contextual fear memory. These results suggest that PRDX6 plays a critical role in the regulation of fear memory expression.
In humans, the feeling of intense fear has been defined using the Diagnostic and Statistical Manual of Mental Disorders, fifth edition (DSM-V) as one primary symptom of PTSD [45, 46]. Three brain regions, the hippocampus, amygdala, and prefrontal cortex, are important for fear memory formation [6]. Here we demonstrated that PRDX6 is expressed in the astrocytes of the amygdala (Additional file 1: Fig. S5), prefrontal cortex (Additional file 1: Fig. S6), and hippocampus (Fig. 7a, b) after TFC. The activation of hippocampal astrocytes plays a crucial role in synaptic plasticity and contextual fear memory [47, 48]. It is known that PRDX6 can modulate astrocyte activation [49, 50]. Whether PRDX6 may regulate the activation of astrocytes during the synaptic process and memory formation requires further study to verify.
In the present study, we used systemic Prdx6 knockout strain, which lacks PRDX6 in the whole brain [23], for trace fear conditioning (TFC). Since intracerebroventricular injection of mouse PRDX6 lentivirus reduced contextual fear memory, we thus focused on identifying the function of PRDX6 in the required brain region—the hippocampus [51, 52]. We found that PRDX6 is colocalized with an astrocytic marker, EGFP, within the hippocampus. Although high expression level of PRDX6 in astrocytes was confirmed, its expression in other cell types was not examined in the present study. Given that PRDX6 expression in different cell types would affect animal behavior, designing a construct containing a neuron or oligodendrocyte specific promotor may help identify related molecular and cellular mechanisms regarding PRDX6′s function in memory formation.
Our results also demonstrate that the Prdx6−/− mice displayed hyperlocomotion activity. This phenotype confirms that enhanced freezing behavior exhibited in Prdx6−/− mice was attributable to the lack of PRDX6, not reduced locomotor activity. Anxiety-like, motivation, and exploration behaviors may also affect response to fear conditioning [53, 54]. These behaviors are normal in Prdx6−/− mice, indicating loss of PRDX6 does not cause these phenotypes. This series of behavior tests suggest that the ablation of Prdx6 is specifically responsible for the enhanced fear memory.
The excessive fear expression to TFC exhibited by Prdx6−/− mice was also observed in activating transcription factor 3 (Atf3) deficient mice [17]. ATF3 is a leucine zipper-containing (bZIP) transcription factor-induced upon stress [55]. Using a computer-based search program (Alggen Promo software, version 8.3), we found that the promoter region of the Prdx6 gene contains binding sites for activating transcription factor 3 (ATF3). Moreover, proteomic analysis (Additional file 2: Table S1) reveals that expression of gelsolin (GSN), an actin-severing protein essential for synaptic plasticity [56], is reduced in the hippocampus of Prdx6−/− mice. This phenomenon is also recorded in the Atf3−/− mice after TFC [17]. Besides, in rats subjected to predator-scent-stress (PSS), a PTSD-like model, gelsolin (Gsn) expression levels were also downregulated [57]. We thus speculate that ATF3, PRDX6, and GSN may participate in the same or related pathways for the regulation of fear memory. Further experiments are necessary to verify their relationship.
Inhibition of the memory retrieval process is proved to attenuate excessive fear response [42]. Stress and stress hormone, glucocorticoid (GC) may positively or negatively affect fear response involving stress coping mechanisms [58, 59]. PRDX6 can be regulated by dexamethasone, a glucocorticoid analog suggesting a possible role of PRDX6 in stress coping mechanisms, including fear response [60]. Previous studies have shown the physiological and pathological role of reactive oxygen species (ROS) in fear response [61], and the hippocampal pyramidal neurons of the CA1 and CA3 subregions related to fear memory retrieval are more vulnerable to oxidative stress [51, 62, 63]. During contextual memory retrieval, ROS level in the hippocampal CA1 and CA3 regions of the Prdx6−/− mice remained similar as wild-type mice (Additional file 1: Fig. S4), indicating that PRDX6 did not regulate the expression of contextual fear memory through modulating ROS level in the hippocampus.
The proteomic and western blot analysis revealed that upregulation of several proteins (Additional file 2: Table S1) involved in the MAPK signaling pathway in the hippocampus of Prdx6−/− mice during the retrieval stage of contextual fear memory. It is known that ongoing protein synthesis is required for maintaining GluA1-AMPA receptors at the synapses for cue memory retrieval [64]. Tropomyosin receptor kinase B (TrkB) and its downstream molecules (AKT and ERK1/2) participate in the regulation of production and trafficking of GluA1-AMPA receptors [65]. A previous study revealed immediate upregulation of total TrkB after a probe test [66]. Another study showed upregulation of total AKT in the basolateral amygdala (BLA) 15 min after reexposure to conditioned context [67]. We also found immediate upregulation of these proteins in the hippocampi of Prdx6−/− mice. Further studies are necessary to verify whether upregulation of these proteins results from local protein synthesis during contextual fear memory retrieval. One previous research has shown that PRDX6 participates in the modulation of ERK1/2 activity in the lung [68]. Another study revealed that inhibition of ERK1/2 before a memory test blocks contextual fear memory retrieval [42]. These studies suggest that hyperphosphorylation of ERK1/2 is associated with enhanced contextual fear memory in the Prdx6−/− mice. Among differential expression proteins listed on Additional file 2: Table S1, total TrkB is highly expressed in the hippocampus of Prdx6−/− mice. This neurotrophin receptor is encoded by the neurotrophic receptor tyrosine kinase 2 (Ntrk2) gene and plays an important role in neuronal plasticity and fear memory [69]. Piazza and colleagues reported that the mice administered with stress hormone GC exhibited enhanced fear response via the activation TrkB/MAPK pathway [58]. Thus, hyperactivation of ERK1/2 in the absence of PRDX6 may be correlated with increased TrkB level.
Interestingly, we also observed upregulation of cytosolic phospholipase A2 (cPLA2), a downstream target of ERK1/2 [70], in the Prdx6−/− mice, which may be the compensation effect for the functional loss of aiPLA2-PRDX6 [71]. This increased cPLA2 level may promote contextual fear memory retrieval in Prdx6−/− mice, since blocking cPLA2 activity before memory test suppresses memory retrieval [72]. TrkB signaling can also activate phosphoinositide 3-kinase (PI3K) and protein kinase B (AKT) [73, 74]. Blocking of PI3K reduced activation of ERK1/2 and AKT, in turn, impaired fear memory retrieval [73]. These pieces of data suggest that activation of TrkB signaling and its downstream molecules-ERK1/2, cPLA2, and AKT in the hippocampus may help enhance retrieval of fear memory in Prdx6−/− mice. Other brain regions may also be responsible for the Prdx6−/− mice's enhanced contextual fear response, particularly the amygdala, but the protein changes were not examined in the present study. A further experiment is worth pursuing the amygdala’s significance in regarding this phenotype of the Prdx6−/− mice.
In conclusion, this study is the first to report PRDX6′s function in negative regulation of contextual fear memory along with hyperactivation of the MAPK pathway in the hippocampus during the retrieval stage of contextual memory. The results obtained from this study reveal the physiological role of PRDX6 in memory formation and help better understand the mechanism underlying homeostatic fear regulation. It also suggests that PRDX6 may be a potential drug target for treating fear-dysregulated disorders like PTSD.
Data and materials availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Abbreviations
- aiPLA2:
-
Acidic calcium-independent phospholipase A2
- ATF3:
-
Activating transcription factor 3
- CA1:
-
Cornu ammonis 1
- CA3:
-
Cornu ammonis 3
- cPLA2:
-
Cytosolic phospholipase A2
- DEPs:
-
Differential expression proteins
- DG:
-
Dentate gyrus
- DHE:
-
Dihydroethidium
- DSM-V:
-
Diagnostic and Statistical Manual of Mental Disorders, fifth edition
- ERK1/2:
-
Extracellular signal-regulated protein kinases 1 and 2
- FDR:
-
False discovery rate
- GC:
-
Glucocorticoid
- GO:
-
Gene ontology
- GPx:
-
Glutathione peroxidase
- LC/MS–MS:
-
Liquid chromatography–tandem mass spectrometry
- LPCAT:
-
Lysophosphatidylcholine acyltransferase
- LV-mPRDX6:
-
Lentivirus-carrying mouse PRDX6
- MAPK:
-
Mitogen-activated protein kinase
- NTRK2:
-
Neurotrophic receptor tyrosine kinase 2
- PI3K:
-
Phosphoinositide 3-kinase
- PRDX6:
-
Peroxiredoxin 6
- PTSD:
-
Post-traumatic stress disorder
- ROS:
-
Reactive oxygen species
References
Jhang J, Lee H, Kang MS, Lee HS, Park H, Han JH. Anterior cingulate cortex and its input to the basolateral amygdala control innate fear response. Nat Commun. 2018;9(1):2744.
Raio CM, Phelps EA. The influence of acute stress on the regulation of conditioned fear. Neurobiol Stress. 2015;1:134–46.
Stevenson CW. Role of amygdala-prefrontal cortex circuitry in regulating the expression of contextual fear memory. Neurobiol Learn Mem. 2011;96(2):315–23.
Etkin A, Wager TD. Functional neuroimaging of anxiety: a meta-analysis of emotional processing in PTSD, social anxiety disorder, and specific phobia. Am J Psychiatry. 2007;164(10):1476–88.
Jovanovic T, Norrholm SD, Blanding NQ, Phifer JE, Weiss T, Davis M, et al. Fear potentiation is associated with hypothalamic-pituitary-adrenal axis function in PTSD. Psychoneuroendocrinology. 2010;35(6):846–57.
Mahan AL, Ressler KJ. Fear conditioning, synaptic plasticity and the amygdala: implications for posttraumatic stress disorder. Trends Neurosci. 2012;35(1):24–35.
Fisher AB. Peroxiredoxin 6: a bifunctional enzyme with glutathione peroxidase and phospholipase A(2) activities. Antioxid Redox Signal. 2011;15(3):831–44.
Arevalo JA, Vazquez-Medina JP. the role of peroxiredoxin 6 in cell signaling. Antioxidants (Basel). 2018;7(12):172.
Asuni AA, Guridi M, Sanchez S, Sadowski MJ. Antioxidant peroxiredoxin 6 protein rescues toxicity due to oxidative stress and cellular hypoxia in vitro, and attenuates prion-related pathology in vivo. Neurochem Int. 2015;90:152–65.
Shanshan Y, Beibei J, Li T, Minna G, Shipeng L, Li P, et al. Phospholipase A2 of peroxiredoxin 6 plays a critical role in cerebral ischemia/reperfusion inflammatory injury. Front Cell Neurosci. 2017;11:99.
Power JH, Asad S, Chataway TK, Chegini F, Manavis J, Temlett JA, et al. Peroxiredoxin 6 in human brain: molecular forms, cellular distribution and association with Alzheimer’s disease pathology. Acta Neuropathol. 2008;115(6):611–22.
Figueiredo HF, Dolgas CM, Herman JP. Stress activation of cortex and hippocampus is modulated by sex and stage of estrus. Endocrinology. 2002;143(7):2534–40.
Asuni AA, Gray B, Bailey J, Skipp P, Perry VH, O’Connor V. Analysis of the hippocampal proteome in ME7 prion disease reveals a predominant astrocytic signature and highlights the brain-restricted production of clusterin in chronic neurodegeneration. J Biol Chem. 2014;289(7):4532–45.
Tan L, Zhao Y, Jiang B, Yang B, Zhang H. Knockdown of PRDX6 in microglia reduces neuron viability after OGD/R injury. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2016;32(8):1014–20.
Hassan W, Gomes VdC, Pinton S, Batista Teixeira da Rocha J, Landeira-Fernandez J. Association between oxidative stress and contextual fear conditioning in Carioca high- and low-conditioned freezing rats. Brain Res. 2013;1512:60–7.
Schaeffer EL, Zorron PuL, Gagliotti DA, Gattaz WF. Conditioning training and retrieval increase phospholipase A(2) activity in the cerebral cortex of rats. J Neural Transm (Vienna). 2009;116(1):41–50.
Pai CS, Sharma PK, Huang HT, Loganathan S, Lin H, Hsu YL, et al. The Activating Transcription Factor 3 (Atf3) homozygous knockout mice exhibit enhanced conditioned fear and down regulation of hippocampal GELSOLIN. Front Mol Neurosci. 2018;11:37.
Briscione MA, Jovanovic T, Norrholm SD. Conditioned fear associated phenotypes as robust, translational indices of trauma-, stressor-, and anxiety-related behaviors. Front Psychiatry. 2014;5:88.
Haaker J, Maren S, Andreatta M, Merz CJ, Richter J, Richter SH, et al. Making translation work: Harmonizing cross-species methodology in the behavioural neuroscience of Pavlovian fear conditioning. Neurosci Biobehav Rev. 2019;107:329–45.
Dai JX, Han HL, Tian M, Cao J, Xiu JB, Song NN, et al. Enhanced contextual fear memory in central serotonin-deficient mice. Proc Natl Acad Sci USA. 2008;105(33):11981–6.
Yoshii A, Constantine-Paton M. Postsynaptic BDNF-TrkB signaling in synapse maturation, plasticity, and disease. Dev Neurobiol. 2010;70(5):304–22.
He Y, Xu W, Xiao Y, Pan L, Chen G, Tang Y, et al. Overexpression of peroxiredoxin 6 (PRDX6) promotes the aggressive phenotypes of esophageal squamous cell carcinoma. J Cancer. 2018;9(21):3939–49.
Wang X, Phelan SA, Forsman-Semb K, Taylor EF, Petros C, Brown A, et al. Mice with targeted mutation of peroxiredoxin 6 develop normally but are susceptible to oxidative stress. J Biol Chem. 2003;278(27):25179–90.
Vyas Y, Lee K, Jung Y, Montgomery JM. Influence of maternal zinc supplementation on the development of autism-associated behavioural and synaptic deficits in offspring Shank3-knockout mice. Mol Brain. 2020;13(1):110.
Lebow M, Neufeld-Cohen A, Kuperman Y, Tsoory M, Gil S, Chen A. Susceptibility to PTSD-like behavior is mediated by corticotropin-releasing factor receptor type 2 levels in the bed nucleus of the stria terminalis. J Neurosci. 2012;32(20):6906–16.
Katano T, Takao K, Abe M, Yamazaki M, Watanabe M, Miyakawa T, et al. Distribution of Caskin1 protein and phenotypic characterization of its knockout mice using a comprehensive behavioral test battery. Mol Brain. 2018;11(1):63.
Huang CH, Chiang YW, Liang KC, Thompson RF, Liu IY. Extracellular signal-regulated kinase 1/2 (ERK1/2) activated in the hippocampal CA1 neurons is critical for retrieval of auditory trace fear memory. Brain Res. 2010;1326:143–51.
Sukkaew A, Suksatu A, Roytrakul S, Smith DR, Ubol S. Proteomic analysis of CHIKV-infected human fibroblast-like synoviocytes: identification of host factors potentially associated with CHIKV replication and cellular pathogenesis. Microbiol Immunol. 2020;64(6):445–57.
Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13(9):731–40.
Losuwannarak N, Maiuthed A, Kitkumthorn N, Leelahavanichkul A, Roytrakul S, Chanvorachote P. Gigantol targets cancer stem cells and destabilizes tumors via the suppression of the PI3K/AKT and JAK/STAT pathways in ectopic lung cancer xenografts. Cancers (Basel). 2019;11(12):2032.
Forester CM, Zhao Q, Phillips NJ, Urisman A, Chalkley RJ, Oses-Prieto JA, et al. Revealing nascent proteomics in signaling pathways and cell differentiation. Proc Natl Acad Sci USA. 2018;115(10):2353–8.
Dybas JM, O’Leary CE, Ding H, Spruce LA, Seeholzer SH, Oliver PM. Integrative proteomics reveals an increase in non-degradative ubiquitylation in activated CD4(+) T cells. Nat Immunol. 2019;20(6):747–55.
Bardou P, Mariette J, Escudie F, Djemiel C, Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinform. 2014;15:293.
Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47(D1):D419–26.
Howe EA, Sinha R, Schlauch D, Quackenbush J. RNA-Seq analysis in MeV. Bioinformatics. 2011;27(22):3209–10.
Seo JS, Park JY, Choi J, Kim TK, Shin JH, Lee JK, et al. NADPH oxidase mediates depressive behavior induced by chronic stress in mice. J Neurosci. 2012;32(28):9690–9.
Yau SY, Bettio L, Vetrici M, Truesdell A, Chiu C, Chiu J, et al. Chronic minocycline treatment improves hippocampal neuronal structure, NMDA receptor function, and memory processing in Fmr1 knockout mice. Neurobiol Dis. 2018;113:11–22.
Han RW, Liu ZP, Lin HR, Tian AW, Xiao YF, Wei J, et al. Role of lateral amygdala calstabin2 in regulation of fear memory. Mol Brain. 2020;13(1):35.
Kikuchi M, Takase K, Hayakawa M, Hayakawa H, Tominaga SI, Ohmori T. Altered behavior in mice overexpressing soluble ST2. Mol Brain. 2020;13(1):74.
Zhang JB, Chen L, Lv ZM, Niu XY, Shao CC, Zhang C, et al. Oxytocin is implicated in social memory deficits induced by early sensory deprivation in mice. Mol Brain. 2016;9(1):98.
Miller MW, Wolf EJ, Logue MW, Baldwin CT. The retinoid-related orphan receptor alpha (RORA) gene and fear-related psychopathology. J Affect Disord. 2013;151(2):702–8.
Besnard A, Caboche J, Laroche S. Recall and reconsolidation of contextual fear memory: differential control by ERK and Zif268 expression dosage. PLoS ONE. 2013;8(8):e72006.
Mazzucchelli C, Brambilla R. Ras-related and MAPK signalling in neuronal plasticity and memory formation. Cell Mol Life Sci. 2000;57(4):604–11.
Hamezah HS, Durani LW, Yanagisawa D, Ibrahim NF, Aizat WM, Bellier JP, et al. Proteome profiling in the hippocampus, medial prefrontal cortex, and striatum of aging rat. Exp Gerontol. 2018;111:53–64.
Regier DA, Kuhl EA, Kupfer DJ. The DSM-5: classification and criteria changes. World Psychiatry. 2013;12(2):92–8.
Cahill SP, Pontoski K. Post-traumatic stress disorder and acute stress disorder I: their nature and assessment considerations. Psychiatry (Edgmont). 2005;2(4):14–25.
Ota Y, Zanetti AT, Hallock RM. The role of astrocytes in the regulation of synaptic plasticity and memory formation. Neural Plast. 2013;2013:185463.
Ben Menachem-Zidon O, Avital A, Ben-Menahem Y, Goshen I, Kreisel T, Shmueli EM, et al. Astrocytes support hippocampal-dependent memory and long-term potentiation via interleukin-1 signaling. Brain Behav Immun. 2011;25(5):1008–16.
Pankiewicz JE, Diaz JR, Marta-Ariza M, Lizinczyk AM, Franco LA, Sadowski MJ. Peroxiredoxin 6 mediates protective function of astrocytes in Abeta proteostasis. Mol Neurodegener. 2020;15(1):50.
Yun HM, Choi DY, Oh KW, Hong JT. PRDX6 exacerbates dopaminergic neurodegeneration in a MPTP mouse model of Parkinson’s disease. Mol Neurobiol. 2015;52(1):422–31.
Jimenez JC, Berry JE, Lim SC, Ong SK, Kheirbek MA, Hen R. Contextual fear memory retrieval by correlated ensembles of ventral CA1 neurons. Nat Commun. 2020;11(1):3492.
Raineki C, Holman PJ, Debiec J, Bugg M, Beasley A, Sullivan RM. Functional emergence of the hippocampus in context fear learning in infant rats. Hippocampus. 2010;20(9):1037–46.
Maren S, Fanselow MS. Appetitive motivational states differ in their ability to augment aversive fear conditioning in rats (Rattus norvegicus). J Exp Psychol Anim Behav Process. 1998;24(3):369–73.
McHugh TJ, Tonegawa S. Spatial exploration is required for the formation of contextual fear memory. Behav Neurosci. 2007;121(2):335–9.
Green TA, Alibhai IN, Unterberg S, Neve RL, Ghose S, Tamminga CA, et al. Induction of activating transcription factors (ATFs) ATF2, ATF3, and ATF4 in the nucleus accumbens and their regulation of emotional behavior. J Neurosci. 2008;28(9):2025–32.
Hlushchenko I, Koskinen M, Hotulainen P. Dendritic spine actin dynamics in neuronal maturation and synaptic plasticity. Cytoskeleton (Hoboken). 2016;73(9):435–41.
Daskalakis NP, Cohen H, Cai G, Buxbaum JD, Yehuda R. Expression profiling associates blood and brain glucocorticoid receptor signaling with trauma-related individual differences in both sexes. Proc Natl Acad Sci USA. 2014;111(37):13529–34.
Revest JM, Kaouane N, Mondin M, Le Roux A, Rouge-Pont F, Vallee M, et al. The enhancement of stress-related memory by glucocorticoids depends on synapsin-Ia/Ib. Mol Psychiatry. 2010;15(12):1125, 40–51.
Schutsky K, Ouyang M, Castelino CB, Zhang L, Thomas SA. Stress and glucocorticoids impair memory retrieval via beta2-adrenergic, Gi/o-coupled suppression of cAMP signaling. J Neurosci. 2011;31(40):14172–81.
Chowdhury I, Fisher AB, Christofidou-Solomidou M, Gao L, Tao JQ, Sorokina EM, et al. Keratinocyte growth factor and glucocorticoid induction of human peroxiredoxin 6 gene expression occur by independent mechanisms that are synergistic. Antioxid Redox Signal. 2014;20(3):391–402.
Massaad CA, Klann E. Reactive oxygen species in the regulation of synaptic plasticity and memory. Antioxid Redox Signal. 2011;14(10):2013–54.
Rajasethupathy P, Sankaran S, Marshel JH, Kim CK, Ferenczi E, Lee SY, et al. Projections from neocortex mediate top-down control of memory retrieval. Nature. 2015;526(7575):653–9.
Medvedeva YV, Ji SG, Yin HZ, Weiss JH. Differential vulnerability of CA1 versus CA3 pyramidal neurons after ischemia: possible relationship to sources of Zn accumulation and its entry into and prolonged effects on mitochondria. J Neurosci. 2017;37(3):726–37.
Lopez J, Gamache K, Schneider R, Nader K. Memory retrieval requires ongoing protein synthesis and NMDA receptor activity-mediated AMPA receptor trafficking. J Neurosci. 2015;35(6):2465–75.
Fortin DA, Srivastava T, Dwarakanath D, Pierre P, Nygaard S, Derkach VA, et al. Brain-derived neurotrophic factor activation of CaM-kinase kinase via transient receptor potential canonical channels induces the translation and synaptic incorporation of GluA1-containing calcium-permeable AMPA receptors. J Neurosci. 2012;32(24):8127–37.
Boondam Y, Songvut P, Tantisira MH, Tapechum S, Tilokskulchai K, Pakaprot N. Inverted U-shaped response of a standardized extract of Centella asiatica (ECa 233) on memory enhancement. Sci Rep. 2019;9(1):8404.
Slouzkey I, Maroun M. PI3-kinase cascade has a differential role in acquisition and extinction of conditioned fear memory in juvenile and adult rats. Learn Mem. 2016;23(12):723–31.
Yun HM, Park KR, Lee HP, Lee DH, Jo M, Shin DH, et al. PRDX6 promotes lung tumor progression via its GPx and iPLA2 activities. Free Radic Biol Med. 2014;69:367–76.
Yamada K, Nabeshima T. Brain-derived neurotrophic factor/TrkB signaling in memory processes. J Pharmacol Sci. 2003;91(4):267–70.
Andersen JM, Myhre O, Fonnum F. Discussion of the role of the extracellular signal-regulated kinase-phospholipase A2 pathway in production of reactive oxygen species in Alzheimer’s disease. Neurochem Res. 2003;28(2):319–26.
Blanchard H, Taha AY, Cheon Y, Kim HW, Turk J, Rapoport SI. iPLA2beta knockout mouse, a genetic model for progressive human motor disorders, develops age-related neuropathology. Neurochem Res. 2014;39(8):1522–32.
Fujita S, Ikegaya Y, Nishiyama N, Matsuki N. Ca2+-independent phospholipase A2 inhibitor impairs spatial memory of mice. Jpn J Pharmacol. 2000;83(3):277–8.
Chen X, Garelick MG, Wang H, Lil V, Athos J, Storm DR. PI3 kinase signaling is required for retrieval and extinction of contextual memory. Nat Neurosci. 2005;8(7):925–31.
Nakai T, Nagai T, Tanaka M, Itoh N, Asai N, Enomoto A, et al. Girdin phosphorylation is crucial for synaptic plasticity and memory: a potential role in the interaction of BDNF/TrkB/Akt signaling with NMDA receptor. J Neurosci. 2014;34(45):14995–5008.
Acknowledgements
The authors highly appreciated everyone for your contributions to this study. We are also thankful for the service from the Core Research Laboratory, Tzu Chi University.
Funding
This work was supported by the Ministry of Science and Technology (MOST), Taiwan (MOST-107-2410-H320-DOI-MY3), and Tzu Chi University/Tzu Chi Foundation (TCMF-SP-108-04).
Author information
Authors and Affiliations
Contributions
SP: conceptualized and performed experiments, analyzed data, and wrote the manuscript. TP, PS: performed experiments and wrote parts of the manuscript. CHY, SR: analyzed data and advised experimental design. HKC: performed experiments. SPH: provided essential mouse strain and advised experimental designs. CCC, NP, SC, SNT: advised experimental designs. IYL: supervised the research, analyzed the data, and prepared manuscripts. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All animal experiments were done following the Taiwan Ministry of Science and Technology guidelines for animals' ethical treatment. Experiments were approved by the Institutional Animal Care and Use Committee of Tzu Chi University, Taiwan (Approval #104099).
Consent for publication
Not applicable.
Competing interests
The authors report no biomedical financial interests or potential conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1–S6.
The characteristics of Prdx6−/− mice (Fig. S1). Expression of EGFP in the hippocampus of Prdx6−/− mice (Fig. S2). Immunostaining of mPRDX6 expressed in the amygdala and prefrontal cortex (Fig. S3). The level of reactive oxygen species (ROS) measured by superoxide-sensitive DHE staining in hippocampal CA1 and CA3 regions of Prdx6+/+ and Prdx6−/− mice (Fig. S4). Co-localization of PRDX6 with GFAP in the amygdala (Fig. S5) and prefrontal cortex (Fig. S6).
Additional file 2: Table S1–S5.
This file contains the list of differential expression proteins (DEPs) in the hippocampus of Prdx6+/+ and Prdx6−/− mice after a contextual test (Table S1). First 25th enrich terms of GO biological process of differential expression proteins (DEPs) (Table S2). Up- and down-regulated enrich proteins differentially expressed relative to GO biological process termed MAPK signaling pathway during contextual memory retrieval in Prdx6−/− mice (Table S3). List of KEGG pathways for GO biological process termed "protein phosphorylation" (GO:0006468) of up-and down-regulated proteins (Table S4). And the details of antibodies and vectors used in this study (Table S5).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Phasuk, S., Pairojana, T., Suresh, P. et al. Enhanced contextual fear memory in peroxiredoxin 6 knockout mice is associated with hyperactivation of MAPK signaling pathway. Mol Brain 14, 42 (2021). https://doi.org/10.1186/s13041-021-00754-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13041-021-00754-1