Abstract
Background
Rasd1 is a member of the Ras family of monomeric G proteins that was first identified as a dexamethasone inducible gene in the pituitary corticotroph cell line AtT20. Using microarrays we previously identified increased Rasd1 mRNA expression in the rat supraoptic nucleus (SON) and paraventricular nucleus (PVN) of the hypothalamus in response to increased plasma osmolality provoked by fluid deprivation and salt loading. RASD1 has been shown to inhibit adenylyl cyclase activity in vitro resulting in the inhibition of the cAMP-PKA-CREB signaling pathway. Therefore, we tested the hypothesis that RASD1 may inhibit cAMP stimulated gene expression in the brain.
Results
We show that Rasd1 is expressed in vasopressin neurons of the PVN and SON, within which mRNA levels are induced by hyperosmotic cues. Dexamethasone treatment of AtT20 cells decreased forskolin stimulation of c-Fos, Nr4a1 and phosphorylated CREB expression, effects that were mimicked by overexpression of Rasd1, and inhibited by knockdown of Rasd1. These effects were dependent upon isoprenylation, as both farnesyltransferase inhibitor FTI-277 and CAAX box deletion prevented Rasd1 inhibition of cAMP-induced gene expression. Injection of lentiviral vector into rat SON expressing Rasd1 diminished, whereas CAAX mutant increased, cAMP inducible genes in response to osmotic stress.
Conclusions
We have identified two mechanisms of Rasd1 induction in the hypothalamus, one by elevated glucocorticoids in response to stress, and one in response to increased plasma osmolality resulting from osmotic stress. We propose that the abundance of RASD1 in vasopressin expressing neurons, based on its inhibitory actions on CREB phosphorylation, is an important mechanism for controlling the transcriptional responses to stressors in both the PVN and SON. These effects likely occur through modulation of cAMP-PKA-CREB signaling pathway in the brain.
Similar content being viewed by others
Background
The hypothalamo-neurohypophyseal system (HNS) is the source of the neuropeptide hormone arginine vasopressin (AVP). AVP is synthesised in magnocellular neurons (MCN) of the supraoptic nucleus (SON) and paraventricular nucleus (PVN) and is transported anterogradely to terminals in the posterior pituitary gland. A rise in plasma osmolality increases secretion of AVP into the blood stream where it promotes water reabsorption at the kidney [1].
While the SON contains a homogenous population of MCN, the PVN is divided into MCN and parvocelluar neurons (PCNs). The PCNs form part of the hypothalamo-pituitary-adrenal (HPA) axis that mediates the stress response. In response to restraint stress, AVP and corticotropin releasing hormone (CRH) are released from the PCN axon terminals in the median eminence into the portal vasculature [2–4] that supplies the anterior pituitary to stimulate the release of adrenocorticotropin hormone [5, 6], and, subsequently, glucocorticoids from the adrenal cortex. These secretory responses are accompanied by transcriptional increases in Avp and Crh in PCN by stress [7–9] and Avp in MCN of the hypothalamus by osmotic stress [10].
The signaling mechanisms governing transcriptional increases in Crh and Avp are believed to involve cAMP activation of the protein kinase A (PKA) pathway and the subsequent phosphorylation of cAMP response element binding protein (CREB) [11]. It is known that both hyperosmotic and restraint stress increase the abundance of phosphorylated CREB, a process that occurs within minutes of stimulation in MCN and PCNs [8, 9, 12, 13]. Stress induced transcriptional increases can be short-lived, particularly for Crh and Avp in PCNs, as the subsequent increase in circulating levels of glucocorticoid following stress, through its interactions with glucocorticoid receptors (GR) within these neurons [14], rapidly dampens this transcriptional response. This feedback by glucocorticoids has been reported to inhibit CREB phosphorylation in PCNs [12], through a proposed unknown intermediate intracellular signaling molecule regulating cAMP [7]. Less is known about inhibitory inputs controlling the transcriptional response to osmotic stress in MCN of the PVN and SON where phosphorylated CREB levels also increase [13]. MCN of the SON express GRs [15], and expression has been shown to increase during hypoosmotic stress [16], indicating that glucocorticoid negative feedback is a possible route for regulation, though studies suggest considerably lower levels of this receptor compared to PCNs of the PVN [14, 17]. Nonetheless, we reasoned that a glucocorticoid inducible gene might be important for regulating transcriptional feedback inhibition in both MCN and PCNs.
Our candidate was Rasd1 (dexamethasone inducible Ras protein 1, Dexras1), a member of the Ras family of monomeric G proteins that was first identified as a dexamethasone (DEX) inducible gene in the pituitary corticotroph cell line AtT20 [18]. A putative glucocorticoid response element identified by Kemppainen and colleagues [19] in the 3’ flanking region of the human Rasd1 gene was shown to confer rapid responsiveness to glucocorticoids by reporter assay. Indeed, the peripheral administration of DEX in rats and mice strongly and rapidly induces Rasd1 expression in several tissue types, including the brain [18, 20, 21]. Another important feature of Rasd1 is its role as an inhibitor of adenylyl cyclase (AC) activity [22–25]. An in vitro study by Graham et al. [22] showed that Rasd1 inhibits GαS- and forskolin (FSK)-induced increases in cAMP levels through ligand independent activation of the Gαi and Gβγ arms of the Gi signaling pathway. This results in inhibition of the cAMP-PKA-CREB signaling pathway and so forms a possible feedback mechanism for regulation of MCN and PCNs. There is also in vitro evidence from reporter assays that Rasd1 acts at cAMP response element (CRE) sites within target gene promoters, to directly inhibit transcription [26, 27]. Thus, Rasd1 has the capacity to influence transcriptional events either indirectly, by inhibiting AC activity at the cell membrane, or directly, through modulation of activity at the level of the promoter.
Using microarrays we identified increased Rasd1 expression in the rat and mouse SON in response to hyperosmotic stress (dehydration) [28, 29]. Rasd1 mRNA expression has previously been described in the mouse PVN and SON but its function in these brain nuclei is not known [30, 31]. Only in the suprachiasmatic nucleus (SCN) of the brain has Rasd1 expression been interrogated thoroughly [32]. Rasd1 cycles with a circadian rhythm that oscillates in antiphase with many of the cAMP inducible mRNAs in the SCN [31] including Avp. Interestingly, in the SCNs of Rasd1 knockout mice, Cheng et al. [26] reported higher cAMP levels alongside increased CREB phosphorylation compared to wild-type mice. We thus hypothesised that Rasd1 may control the transcription of cAMP inducible genes, and ultimately their downstream targets, Avp and Crh, in the hypothalamus. Using in vitro and in vivo approaches we demonstrate that Rasd1 negatively regulates the hypothalamic transcriptional response to stimulation by hyperosmotic stress in the rat.
Results
Rasd1 mRNA expression in the rat hypothalamus in response to different stressors
We first investigated the time course of Rasd1 mRNA expression in the PVN and SON of control, dehydrated (DH) and salt loaded (SL) rats using qPCR (Fig. 1a). Rasd1 mRNA expression was significantly higher in the PVN and SON of 1 day (day) DH and 1 day SL rats compared with control rats. Increasing the duration of hypertonic stress to 3 days DH and 7 days SL further increased the magnitude of this response in both PVN and the SON. When drinking water was returned after 1 day DH and 1 day SL, Rasd1 mRNA abundance was seen to quickly return to control levels (Fig. 1b).
Rasd1 mRNA expression in the rat PVN and SON in response of hyperosmotic stress (a–c). The expression of Rasd1 was examined in the PVN and SON in chronic and acute hyperosmotic conditions, and following water repletion. Relative mRNA expression of Rasd1 was investigated by qPCR in the PVN and SON of a control, DH (1 and 3 days), and SL (1 and 7 days) rats, b after water repletion for 1 day after 1 day DH and 1 day SL compared to controls, c after a single i.p injection of 1.5 ml/100 g body weight 1.5 M NaCl over a 4 h experimental period compared to controls. d, Rasd1 mRNA expression in PVN and SON after 30 min and 1 h restraint stress compared to control. All data was analysed by One-way ANOVA. Values are means + SEM of n = 5−6 animals per group. *p < 0.05, **p < 0.01, ***p < 0.001. RH, rehydration; Con, control
We then examined the acute response of Rasd1 transcripts to a single intraperitoneal (i.p) injection of hypertonic saline (HS) (Fig. 1c). The abundance of the Rasd1 mRNA increased rapidly after injection in both PVN and SON, and levels progressively increased until peaking at around 60 min, with levels remaining steadily elevated thereafter. As Rasd1 is known to be strongly induced by glucocorticoids [18], we also examined expression in PVN and SON resulting from stress induced by 30 min and 1 h restraint (Fig. 1d). An increase of Rasd1 mRNA was observed in PVN but interestingly not the SON.
Expression of RASD1 protein in the rat HNS
Immunofluorescent localisation studies were performed to determine the cell populations expressing RASD1 in the PVN and SON of control rats (Fig. 2). RASD1 was expressed MCN of the PVN and SON (Fig. 2a). In the PVN, RASD1 staining was predominantly found in the lateral magnocellular part with fewer RASD1 positive neurones in parvocellular regions. The MCN of the SON and PVN can be divided into two populations, expressing either AVP, or the closely related neuropeptide hormone oxytocin (OT), with only a small percentage (2–3%) of MCN express high, equivalent levels of both peptides [33]. Therefore, to examine the identity of neurons expressing RASD1 in the PVN and SON, co-immunostaining of RASD1 with AVP neurophysin-II or OT neurophysin-I was performed in euhydrated control rats (Fig. 2b–c). RASD1 expression was observed in AVP positive (Fig. 2b), but not OT expressing MCN of the PVN and SON (Fig. 2c). High magnification images using confocal microscopy are shown in Fig. 2d. Therefore, one can assign the potential functions of Rasd1 principally to vasopressinergic neurones in these two brain nuclei following hyperosmotic stress. The specificity of the RASD1 antibody was confirmed in N2a cells transfected with GFP-Rasd1 fusion construct (Fig. 2e).
Immunofluorescent localisation of RASD1 in the hypothalamus of the euhydrated rat. a, immunofluorescent localisation of RASD1 in MCN of the PVN and SON. b–c immunofluorescent colocalisation of RASD1 (green) with b AVP (red) and c OT (red) in MCN in the PVN and SON. d Higher magnification images showed the presence of Rasd1 in AVP, but not OT, positive cells. e, eGFP and Rasd1 fluorescence in N2a cells transfected with eGFP-Rasd1 fusion construct. Dapi staining indicates the nucleus. Scale bars a–c = 100 μm, d–e = 10 μm. OC, optic chiasm; 3 V, third ventricle
Effect of osmotic and restraint stress on RASD1 protein distribution
Immunofluorescence staining was performed to examine the expression of RASD1 after exposure to either osmotic or restraint stress (Fig. 3). The intensity of RASD1 staining in neuronal cell bodies of the PVN and SON appeared similar in DH but lower in SL compared to control rats (Fig. 3a). We then looked at the distribution of RASD1 staining in the PVN after restraint stress (Fig. 3b). The degree of RASD1 staining appeared higher in the dorsomedial parvocellular portions in response to restraint stress (Fig. 3b).
The effect of osmotic and restraint stress on RASD1 expression. a, immunofluorescent staining for RASD1 in the PVN and SON of control, 3 days DH and 7 days SL rat. High magnification confocal images are shown. b, immunofluorescent staining of RASD1 in the rat PVN in response 1 h restraint stress with 3 h recovery. Arrows indicate the dorsomedial parvocellular portion of PVN. Scale bars a–b = 100 μm, confocal images = 10 μm. OC, optic chiasm; 3 V, third ventricle; Con, control
Activation of Rasd1 transcription by glucocorticoids in the PVN and SON
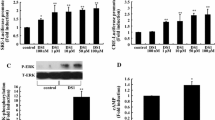
Peripheral treatment with DEX has been reported to rapidly and robustly increase the expression of Rasd1 mRNA in the brain [21]. To determine if Rasd1 expression was stimulated by glucocorticoids specifically in PVN and SON, we performed ex vivo and in vivo experiments with DEX (Fig. 4). Incubation of hypothalamic organotypic cultures for 4 h and 24 h in the presence of DEX significantly increased Rasd1 mRNA expression in both the PVN and SON compared to controls (Fig. 4a). Next, to investigate glucocorticoid actions on Rasd1 expression in vivo, rats were injected with DEX before injection of isotonic (IS) or HS. At 30 min after saline injection mRNA levels were examined in the PVN and SON (Fig. 4b). As expected, HS injection significantly increased Rasd1 mRNA expression in PVN and SON compared to the vehicle control. DEX injection increased Rasd1 mRNA expression in PVNs and SONs of rats administered IS, in agreement with our findings for organotypic cultures. However, there was no significant difference in Rasd1 mRNA in PVNs of rats treated with DEX and IS compared to vehicle and HS, whilst in the SON, HS treatment significantly increased Rasd1 expression compared to rats treated with DEX and IS. Moreover, DEX injection increased hypertonic induced Rasd1 mRNA expression in both PVN and SON compared to vehicle controls, perhaps suggesting activation of Rasd1 expression by separate molecular pathways.
DEX induced Rasd1 mRNA expression in the rat PVN and SON. a the addition of DEX to hypothalamic organotypic culture media 4 or 24 h before collection of samples increased the expression of Rasd1 in the PVN and SON compared to vehicle (DMSO) controls. b rats were injected with DEX (1 mg/kg) or vehicle (0.15 M NaCl) 2 h before i.p injection of IS (0.15 M NaCl) or HS (1.5 M NaCl; 1.5 ml/100 g body weight). Brains were collected 30 min after IS or HS injection. qPCR was performed to analyse the expression of Rasd1 c-Fos, Nr4a1, hnAvp and hnCrh in PVN and SON. c to block endogenous glucocorticoid synthesis rats were injected with MET or a combination of MET and DEX 4 h before i.p injection of IS or HS. Brains were collected after 30 min and qPCR analysis was performed on the cDNAs generated. a, independent-sample unpaired Student’s t tests; b–c, Two-way ANOVA. Values are means + SEM of n = 5−8 animals per group. *p < 0.05, **p < 0.01, ***p < 0.001. DEX, dexamethasone; MET, Metyrapone; VEH, vehicle
We next focused our attention on the expression of the Avp and Crh mRNAs, their respective precursor transcripts, heteronuclear Avp (hnAvp) and heteronuclear Crh (hnCrh), the assessment of which acts as a surrogate measure for transcription [34, 35]. We also looked at two established cAMP inducible transcription factors, c-Fos and nuclear receptor subfamily 4 group A member 1 (Nr4a1), the expression of which are known to be induced both by CREB phosphorylation and osmotic stress (reviewed by Yoshida, 2008 [11]). DEX injection reduced HS induced hnAvp, hnCrh, c-Fos and Nr4a1 expression in the PVN and c-Fos and Nr4a1 in the SON. Basal levels of gene expression of these genes were unchanged by DEX treatment, with the exception hnCrh expression which was lower in the PVN. We then asked if treatment with metyrapone (MET), which blocks corticosterone synthesis [36], could influence basal Rasd1 mRNA expression, or its response to HS in the PVN and SON (Fig. 4c). Basal Rasd1 mRNA expression was unchanged by MET treatment in the PVN and SON. However, hypertonic induced Rasd1 mRNA expression was diminished by pretreatment with MET, though still elevated above control measures, in the PVN and this effect was recovered by combined treatment with DEX. Rasd1 mRNA expression was unchanged by any of these treatments in the SON.
DEX and Rasd1 inhibit cAMP induced gene expression in AtT20 cells
The stimulatory actions of glucocorticoids on Rasd1 gene expression were first described in AtT20 cells [18]. We initially treated AtT20 cells with DEX concentrations ranging from 0.01–1000nM to establish a suitable concentration for our studies (Fig. 5a). Maximum stimulation of Rasd1 mRNA expression was observed at 100nM so we selected this dose for our in vitro studies. Treatment with DEX increased Rasd1 in a time dependent manner (Fig. 5b), with significance reached at 30 min, and maximum expression between 1–2 h, similar to the effect of acute hypertonic stimulation in vivo (Fig. 1c). We produced shRNAs to knockdown Rasd1 expression. Of the two Rasd1 shRNAs we tested, shRNA2 showed significant knockdown of Rasd1 mRNA (Fig. 5c). We then examined the effect of DEX treatment on Rasd1 mRNA expression in the Rasd1 knockdown cell line. The result showed that, from its significantly diminished baseline, Rasd1 mRNA increased in abundance, suggesting that the knockdown cell line is still able to respond to DEX, but the level of Rasd1 mRNA expression was significantly reduced compared to the control shRNA cells.
Rasd1 mediates actions of glucocorticoid on transcription factors c-Fos and Nr4a1 in AtT20 cells. a, Rasd1 mRNA expression in response to 1 h treatment with different concentrations of DEX. b changes in Rasd1 mRNA expression after treatment with 100nM DEX over a 4 h experimental period compared to time matched vehicle controls. c responses to Rasd1 knock down cell lines to 1 h treatment with 100nM DEX were examined by qPCR compared to a control non-targeting shRNA cell line. d the effect of pretreatment with DEX (2 h), 72 h of adenoviral mediated Rasd1 overexpression and Rasd1 knockdown on FSK (10 μM) induced c-Fos and Nr4a1 mRNA expression was examined compared to controls. e cells were infected with Rasd1 adenovirus for 4 h. Media was replaced and cells were grown in the presence of farensyltransferase inhibitor FTI-277 for 72 h and qPCR analysis of c-Fos and Nr4a1 expression was performed. f the effect of 72 h of adenoviral mediated dnRasd1 overexpression on FSK induced c-Fos and Nr4a1 mRNA expression was examined compared to controls. g responses of GR knockdown cell lines to treatment with 100nM DEX were examined by qPCR compared to a control non-targeting shRNA cell line. VEH, vehicle; NT, non-targeting; Con, control. Values are means + SEM of n = 3−4 per group. *p < 0.05, **p < 0.01, ***p < 0.001
We next asked if the DEX inhibition of c-Fos and Nr4a1 mRNAs seen in vivo was the result of increased Rasd1 expression. We used FSK to induce endogenous c-Fos and Nr4a1 expression over a time course of 4 h in AtT20 cells pretreated with DEX. As seen in our in vivo study, DEX treatment significantly inhibited c-Fos and Nr4a1 expression in AtT20 cells (Fig. 5d). To establish if Rasd1 could be mediating these DEX effects, we firstly overexpressed Rasd1 or eGFP in AtT20 cells using adenoviral vectors, and then treated the transfected cells with FSK to stimulate endogenous c-Fos and Nr4a1 expression. Increasing Rasd1 expression inhibited FSK stimulation of c-Fos and Nr4a1 with a remarkably similar time course and level of inhibition to that of DEX treatment. shRNA mediated knockdown of endogenous Rasd1 increased FSK-stimulated Nr4a1 expression, but not c-Fos expression compared to the control shRNA. DEX inhibited FSK-stimulated Nr4a1 but not c-Fos in both Rasd1 and non-targeting shRNA cell lines, but expression of Nr4a1 remained higher than the control cell line. This may be expected as DEX was still able to increase Rasd1 mRNA in the knockdown cell line (Fig. 5c). When we treated Rasd1 overexpressing cells with the farnesyltransferase inhibitor FTI-277, a highly potent CAAX peptidomimetic that inhibits Ras signaling [37], FSK-stimulated expression of c-Fos and Nr4a1 increased compared to control (Fig. 5e). Adenoviral mediated overexpression of a CAAX box deficient Rasd1 (dnRasd1) significantly increased c-Fos and Nr4a1 expression confirming the importance of this motif (Fig. 5f).
The GR is believed to be responsible for mediating the effects of DEX on Rasd1 mRNA expression. Therefore, we produced a GR knockdown AtT20 cell line and tested the responsiveness of Rasd1 expression to treatment with DEX (Fig. 5g). shRNA mediated silencing of GR expression abolished Rasd1 stimulation by DEX, confirming that Rasd1 activation is mediated by GR dependent signaling pathways in AtT20 cells. We speculated that Rasd1 provoked these changes in gene expression may result from altered CREB phosphorylation, which is a known regulator of c-Fos and Nr4a1 (Fig. 6). Indeed CREB phosphorylation was reduced by both DEX (Fig. 6a) and Rasd1 (Fig. 6b), but was increased by overexpression of dnRasd1 (Fig. 6c).
Rasd1 reduces the abundance of phosphorylated CREB in FSK stimulated AtT20 cells. Phosphorylated CREB expression in AtT20 cells (a) treated with FSK (5, 10, 20, 60 min) after 2 h pretreatment with DEX, b 72 h adenoviral overexpression of Rasd1 or c 72 h adenoviral expression of dnRasd1 by Western blot. Viral overexpression of Rasd1 was confirmed. GAPDH was used as the loading control. Con, control; VEH vehicle
Rasd1 expression controls the transcriptional response to hyperosmotic stress in vivo
We asked about the transcriptional effects of Rasd1 overexpression in the SON in vivo. We confirmed expression of the Rasd1 and dnRasd1 lentiviruses in N2a cells by immunoblot (Fig. 7a) and in MCN of the SON using the eGFP tag (Fig. 7b). Quantitative PCR confirmed the overexpression of Rasd1 in the SON (Fig. 7c). We then examined gene expression in SON injected with Rasd1 compared to eGFP controls. Basal levels of c-Fos, Nr4a1 and hnAvp were unaffected by overexpression of Rasd1 compared to eGFP controls. The expression of all of these genes was increased by HS injection in eGFP virus injected SONs. In Rasd1 injected SON, HS induced smaller rises in c-Fos and Nr4a1 expression compared to controls and inhibited HS induced hnAvp expression. We then directly assessed the importance of Rasd1 in the hypothalamic systems controlling homeostasis using the SL model in rats injected with dnRasd1 virus (Fig. 7d). Quantitative PCR confirmed higher expression of Rasd1 in SON injected with dnRasd1 compared to eGFP controls. The expression of dnRasd1 in the SON increased expression of c-Fos, Nr4a1 and hnAvp in SL rats compared to eGFP controls. However, the increase of c-Fos expression did not reach statistical significance.
Lentiviral mediated manipulation of Rasd1 expression in the rat SON alters c-Fos, Nr4a1 and hnAvp expression. a confirmation of Rasd1 protein expression 72 h after viral transduction in mouse neuronal cell line N2a. b lentiviral transduction of MCN in the SON 2 weeks after injection. c rats were injected into SON with lentiviral vectors expressing Rasd1 or eGFP. Two weeks after viral administration rats received a single i.p injection of either IS (0.15 M NaCl) or HS (1.5 M NaCl; 1.5 ml/100 g body weight). Brains were collected 30 min later. Relative expression of c-Fos, Nr4a1, hnAvp and Rasd1 was determined by qPCR. d two weeks after dnRasd1 injection rats were presented with 2% (w/v) NaCl solution in place of drinking water for 7 days. Relative expression of c-Fos, Nr4a1, hnAvp and Rasd1 in SON of dnRasd1 delivered animals compared to control eGFP control. c, Two-way ANOVA; d, independent-sample unpaired Student’s t tests. Scale bars = 100 μm. Con, control; MOI, multiplicity of infection. Values are means + SEM of n = 4−5 animals per group. *p < 0.05, **p < 0.01, ***p < 0.001
Discussion
Rasd1 is becoming somewhat of avant-garde member of the Ras family of GTPases by performing many non-conventional signaling functions. Our identification of Rasd1 in Avp neurons starts a new chapter for this small GTPase. Here we show that Rasd1 is rapidly induced by stress in the PVN, and by elevated plasma osmolality in the PVN and SON of the hypothalamus. We propose that the abundance of RASD1 in MCN and PCNs, based on its inhibitory actions on CREB phosphorylation, is an important mechanism for controlling the transcriptional responses to stressors in both the PVN and SON. In MCN we show, by virally mediated overexpression of Rasd1, that Rasd1 inhibits HS induced stimulation of cAMP inducible genes. When a CAAX box deficient mutant form of Rasd1 is expressed in the SON cAMP inducible genes were further increased by SL. These effects likely occur through modulation of cAMP-PKA-CREB signaling pathway.
Our interest in Rasd1 began following identification increased expression of this gene in the SON of the 3 days DH rat using microarrays [28]. Here, using qPCR, we confirmed increased Rasd1 expression in rat PVN and SON by DH, SL and HS injection. These protocols have all been shown to increase plasma corticosterone levels in the rat [38–41], suggesting that glucocorticoids may be responsible for increased Rasd1 mRNA expression in the brain. To test this notion, we performed restraint stress, where plasma corticosterone levels are increased [42, 43] in the absence of the osmotic cues presented by hyperosmotic stress. To our surprise, restraint stress rapidly increased Rasd1 expression, but only in the PVN, suggesting differing mechanisms of Rasd1 activation in the PVN and SON by restraint stress and osmotic stress. We show here in AtT20 cells that the GR is important for Rasd1 induction by DEX similar to previous reports for other cell lines [44]. We thus proposed that the location of GRs in the hypothalamus likely underlies these differing responses to restraint stress. Indeed, in support of this concept, PCNs of the PVN are known to express comparatively higher levels of the GR than MCN of the SON [14–17]. These data thus imply different mechanisms of Rasd1 activation in PCNs and MCN.
The observed expression of Rasd1 exclusively in AVP positive neurons suggested a role in the regulation of these neurons. Such regulation may occur at multiple levels: as we observed Rasd1 expression in neuronal cell body, dendrites and axons of control rats. The presence of Rasd1 in axons is novel, but this property is not unique to this particular Ras protein. In the mouse hippocampus another member of the Ras family, H-Ras, a potent activator of ERK signaling, has previously been identified in axon terminals [45]. Transgenic mice overexpressing a constitutively active mutant of H-Ras, which strongly localises to axon terminals in the hippocampus, resulted in several presynaptic changes including a higher density of docked neurotransmitter vesicles in glutamatergic terminals [45]. The granular cytoplasmic pattern of RASD1 immunoreactivity in the neuronal cell body suggests that RASD1 may be associated with vesicular structures in the cytoplasm. The presence of RASD1 in cell processes suggests that RASD1 has secretory role in AVP neurons. The idea that Rasd1 could influence hormone secretion is not new. A number of in vitro studies have reported inhibitory actions of Rasd1 on hormone secretion but the precise mechanisms mediating these inhibitory effects remains to be established [21, 46–48]. It is established that high plasma osmolality results in the secretion and depletion of AVP in MCN cell bodies, even though mRNA expression of Avp is up-regulated in the PVN and SON [49, 50]. This is also true for RASD1 in SL rats. What is known is that RASD1 regulates a number of cell signaling processes [51, 52], in particular it has inhibitory actions on the cAMP-PKA-CREB signaling pathway [21–25]. Therefore, any changes in cellular abundance of RASD1, as we observe here in the MCN cell bodies by hyperosmotic stress and the increase in PCN by restraint stress, would be expected to influence the cAMP signalling pathway.
The expression of Rasd1 in the brain has been shown to be under hormonal control [20]. We show here that DEX increases Rasd1 expression in the PVN and SON of the hypothalamus. These findings differed from our observations in restraint stress where Rasd1 increased only in the PVN. One possibility for these differences is the greater potency of DEX in its glucocorticoid effects than endogenous corticosterone. Our data showed that pretreatment with DEX potentiated HS induced increases in Rasd1 expression in PVN and SON, consistent with our hypothesis of two separate mechanisms activating Rasd1 transcription in these nuclei. This was confirmed by treatment with MET, which decreased Rasd1 expression only in the PVN, a response that was effectively rescued by DEX. These findings are consistent with the presence of GRs in MCN of the SON a well as PCN of the PVN [15, 16]. These data thus imply two separate mechanisms of Rasd1 activation in the PVN and SON, one sensitive to glucocorticoids, and one mediated by a currently undefined mechanism in response to elevated plasma osmolality.
The negative feedback of glucocorticoids, in our case using DEX, on hnAvp and hnCrh transcription in PVN has been known for many years [7, 53–55]. Withdrawal of glucocorticoids by adrenalectomy results in increased Crh and Avp mRNA expression in the PVN [56, 57], which can be reversed by glucocorticoid replacement [7]. In adrenalectomised rats, by replacing corticosterone, Kovacs et al. demonstrated that glucocorticoids selectively targets Avp transcription in PCNs and not MCN of the PVN [7]. In PCNs of the rat PVN, these transcriptional events are thought to be mediated by stress induced increases in CREB phosphorylation, which results in increased Crh and Avp transcription [8, 9, 12]. Inhibition CREB phosphorylation is thought to be one of the targets for the negative feedback of glucocorticoids on Avp and Crh transcription in PCN of the PVN [7, 12]. Furthermore, it is known that cAMP increases in the SON in response to hyperosmotic stress [58–60], through activation of the PKA pathway and subsequent CREB phosphorylation [13], and this is thought to stimulate Avp transcription.
In recent years many in vivo actions of DEX have been attributed to molecular signaling through Rasd1 [44, 46, 47] suggesting that DEX actions in the hypothalamus were the result of increased Rasd1 expression. Our in vivo observations of altered c-Fos and Nr4a1 expression by DEX treatment in the PVN and SON, where Rasd1 expression is elevated, prompted us to compare DEX treatment with Rasd1 overexpression on gene expression in mouse corticotroph AtT20 cells. Both c-Fos and Nr4a1 are well established cAMP inducible genes that are induced by phosphorylated CREB [61], and respond robustly to increases in neuronal activity arising from osmotic stress and restraint stress [62, 63]. There is also evidence that DEX inhibits c-Fos and Nr4a1 expression [53, 55, 64], so we speculated that these effects could be mediated by Rasd1 via modulation of cAMP-PKA-CREB signaling pathway. We show by overexpression and knockdown, that Rasd1 can reproduce DEX effects on c-Fos and Nr4a1 in AtT20 cells, suggesting that glucocorticoids primarily act through Rasd1 to inhibit the expression of these transcription factors.
RASD1 has also been shown to inhibit cAMP inducible genes through interactions at the promoter. RASD1 interacts with NonO, a member of RNA-Recognition motif gene family, at cAMP response element (CRE) sites within target genes, including Nr4a1, inhibiting their transcription [27]. Furthermore, in cortical neuron cultures, transfection of a CRE driven luciferase construct also demonstrated the inhibitory actions of RASD1 on FSK induced CRE mediated transcription [26]. The transcriptional changes in the present study were isoprenylation dependent as determined using the farnesyltransferase inhibitor FTI-277 and the CAAX deficient dnRasd1 suggesting membrane translocation is necessary. RASD1 undergoes posttranslational modifications by farnesylation of its CAAX box [25] similar to other members of the Ras family [65]. Isoprenylation of this CAAX consensus site is required for translocation of Ras proteins to the cell membrane [65, 66], stimulation of protein-protein interactions, and can affect protein stability [67]. Isoprenylation has been shown to reduce the stability of Ras proteins [68] and is thought to be necessary for rapid protein turnover of GTPase RhoB [69]. The decreased stability of farnesylated proteins may provide an explanation for the lower levels RASD1 observed in the PVN and SON during SL, which may be the result of increased turnover of RASD1 protein. These data imply that the observed actions of Rasd1 in AtT20 cells occur through regulation of G proteins at the plasma membrane.
The expression of over 100 non-chemosensory G protein coupled receptors has been described in the PVN and SON [70], many of which could be subject to regulation by RASD1. Indeed, it has been proposed that RASD1 may act to antagonise G protein coupled receptor signaling by altering the pool of heterotrimeric G-proteins available for receptor coupling [71]. Interestingly, Rasd1 has been reported to selectively activate transient receptor potential channel 4 (TRPC4) in INS-I cells [72], and altered expression of these channels has been described in vasopressinergic MCNs of the PVN and SON in response to hyperosmotic stress [28, 73]. The activation of TRPC4 in neurosecretory cells is known to trigger a robust secretory response [74], suggesting that Rasd1 may affect the secretion of vasopressin. Furthermore, Rasd1 has been shown to modulate N-type calcium channels in HEK293 cells [75]. Thus, Rasd1 has the capacity to modulate calcium influx into cells and may influence calcium dependent events such as vasopressin secretion from MCNs. Therefore, in hypertonic stress altered RASD1 expression may perhaps modulate the excitability of MCNs by altering channel activity, though this remains to be determined.
Rasd1 overexpression has been shown to inhibit GαS and FSK mediated activation of AC through activation of Gαi and Gβγ [22]. Interestingly, SL in rats increases Gαi expression in MCN of the PVN and SON [60] consistent with the idea of inhibitory inputs on cAMP production in osmotic stress. In addition, Rasd1 has been seen to abolish dopamine D2L receptor mediated potentiation of AC2 activity by blocking protein kinase C and Gβγ activity [25], and to block dopamine D2L receptor mediated heterologous sensitization of AC1 [24]. Both AC1 and AC2 have been reported in the PVN and SON [76], so could be targets of Rasd1 actions in these brain nuclei as well as other brain regions that express Rasd1.
To validate our in vitro findings in the physiological context of the whole organisms, we used lentiviral vectors to overexpress Rasd1 in the rat SON. Analogous to our findings in AtT20 cells, overexpression of Rasd1 in the SON reduced HS induced increases in c-Fos and Nr4a1, and completely blocked induced hnAvp expression. Therefore, Rasd1 alone could mimic the actions of DEX on these target genes in the SON, suggesting that DEX effects in PVN and SON are mediated through altered Rasd1 expression. We suggest that this is due to inhibition of CREB phosphorylation. In a recent in vivo study, injection of a recombinant adeno-associated virus expressing ACREB, a dominant negative inhibitor of endogenous CREB, into the SON resulted in decreased c-Fos and Nr4a1 mRNA expression [77]. Furthermore in the SCNs of Rasd1 knockout mice, Cheng et al. [26] reported higher cAMP levels, alongside increased c-Fos and CREB phosphorylation, compared to wild-type mice. These changes in Rasd1 knockout mice are consistent with removal of the inhibitory influences of Rasd1 on cAMP dependent signaling pathways. This is consistent with our findings with dnRasd1 overexpression in the SON of the SL rat where expression levels of cAMP inducible genes were increased. We would predict a similar mode of action for Rasd1 overexpression in PCNs of the PVN based on our findings presented here. Unfortunately, our viruses are not able to selectively discriminate between MCN and PCNs of the PVN to extend these findings to PCNs of the PVN.
RASD1 has been shown to exist in ternary complex with carboxy-terminal PDZ ligand CAPON and neuronal nitric oxide synthase (nNOS) [30]. This enables activation of Rasd1 by s-nitrosylation on single cysteine (Cys11) residue at the N-terminus of this protein [78]. In cortical neuronal cultures, Rasd1 is activated by nitric oxide donors as well as N-methyl-D-aspartate receptor stimulated nitric oxide synthesis [78]. The AVP expressing MCN of the SON express nNOS, RASD1, CAPON [30] and nNos is also subjected to regulation by DH and SL [79, 80]. We do not know whether RASD1 complexes with nNOS and CAPON regulate any signaling events in the SON, or indeed if s-nitrosylation of RASD1 is important. However, we do know that AtT20 cells do not express nNos [81], so nNos is not a necessary component for Rasd1 mediated inhibition of cAMP pathways.
Conclusion
In summary, our results showed that Rasd1 is expressed in vasopressinergic neurons in the PVN and SON of the hypothalamus. We identified two mechanisms of Rasd1 induction in the hypothalamus, one by elevated glucocorticoids in response to stress, and one in response to increased plasma osmolality resulting from osmotic stress. The ability to influence c-Fos, Nr4a1, and hnAVP in vivo by lentiviral gene transfer greatly strengthened our hypothesis that Rasd1 was inhibiting cAMP-PKA-CREB signaling pathway in the hypothalamus (Fig. 8). We propose that the abundance of RASD1 in MCN, based on these inhibitory actions, is important for controlling the transcriptional response to osmotic stress in the PVN and SON. In PCNs of the PVN, Rasd1 has all the necessary credentials to facilitate the rapid glucocorticoid negative feedback of the HPA axis on Crh and Avp expression. Studies have long proposed that an intermediate signalling molecule activated by the GR may be necessary for glucocorticoid negative feedback on CREB phosphorylation and our data points towards this being Rasd1.
Proposed mechanism of action of Rasd1 in cAMP-dependent gene regulation. Rasd1 mRNA expression is upregulated by glucocorticoid (dexamethasone) and increased plasma osmolality (hypertonic stress). In the cytoplasm RASD1 undergoes posttranslational processing by isoprenylation of its CAAX box motif enabling trafficking to the cell membrane. RASD1 activates Gαi which inhibits the cAMP dependent pathway by inhibiting adenylyl cyclase. This results in the inhibition of the cAMP-PKA-CREB signalling pathway
Methods
Animals
Male Sprague–Dawley rats weighing 250–300 g were used in this study, except for the restraint stress experiment (performed in Brazil), where male Wistar rats weighing 250–300 g were used. Rats were maintained under a 14:10 light dark cycle (lights on at 0500) with food and water ad libitum for at least 1 week prior to experimentation. Animal experiments were performed between 9 am–2 pm. All experiments in the UK were performed under a Home Office UK licence held under, and in strict accordance with, the provision of the UK Animals (Scientific Procedures) Act (1986); they had also been approved by the University of Bristol Animal Welfare and Ethical Review Board. The experiments in Brazil were conducted according to the “Guide for the Care and Use of Laboratory Animals” (NIH Publication No. 85–23, revised 1996), and experimental protocols were approved by the local Ethics Committee on Animal Use in the School of Medicine of Ribeirão Preto, University of São Paulo.
Hypertonic stress experiment
We used two protocols a chronic and an acute hypertonic stress protocol. To induce chronic hyperosmotic stress, water was removed (DH) for 1 or 3 days or replaced by 2% (w/v) NaCl in drinking water for 1 or 7 days SL. The control group had access to food and water ad libitum. In some instances water was returned after DH and SL for 24 h (rehydration). The acute responses were assessed (10 min, 30 min, 1, 2, or 4 h) after a single i.p injection of 1.5 ml/100 g body weight of 1.5 M NaCl solution HS. After injection, HS rats were placed back in their home cages and water was removed. The reference group (time 0) had access to food and water ad libitum. In acute experiments with time-point comparisons the control groups received a single i.p injection of IS solution (1.5 ml/100 g body weight of 0.15 M NaCl solution).
Restraint stress experiment
Rats were randomly allocated to 3 groups (control, 0.5 h and 1 h restraint). The control group stayed in their home cages and had access food and water ad libitum throughout the experimental period. For RNA analysis, animals were placed in clear plexiglass tubes with access to air for either 0.5 h or 1 h duration, before being immediately killed. For immunofluorescence protein analysis, the rats were killed 3 h after removal from the tube to allow time for de novo protein synthesis to take place.
Glucocorticoid studies
To investigate actions of glucocorticoid on gene expression in the hypothalamus, rats were injected intraperitoneally with 0.5 ml 0.15 M NaCl (vehicle) or 0.5 ml of 1 mg/kg body weight DEX (Sigma D2915) 2 h before IS or HS injection. To inhibit the synthesis of endogenous glucocorticoid, rats were injected subcutaneously with 0.5 ml sesame oil (vehicle), 150 mg/kg body weight MET (Sigma 856525), which inhibits endogenous corticosterone synthesis by inhibiting steroid 11-β hydroxylase activity, or 150 mg/kg MET and 0.2 mg/kg body weight DEX 4 h before IS or HS injection. After injection of HS or IS rats were placed back in their home cages, and water, but not food, was removed for the remaining 30 min of the experiment. For RNA analyses, rats were killed by striking of the cranium, followed by decapitation, using a guillotine (Harvard Apparatus). Brains were removed and frozen on dry ice before being stored at −80 °C.
RNA extraction and cDNA synthesis
Frozen brains were sliced into 60 μm coronal sections in a cryostat. Sections were mounted on glass slides and stained with 0.1% (w/v) toludine blue then visualised on a light microscope until brain nuclei were visible, then SON and PVN samples were collected using a 1 mm micro punch (Fine Scientific Tools). The optic chiasm (SON), or neurons lateral to the third ventricle (PVN), were used as a reference. SON and PVN samples were then dispensed into 1.5 ml tubes and kept on dry ice within the cryostat. Total RNA was extracted from punched samples by combining Qiazol Reagent with Qiagens RNeasy kit protocols (Qiagen). The punched samples were removed from dry ice and rapidly resuspended, by vortexing, in 1 ml Qiazol reagent. Following Qiazol phase separation with chloroform, 350 μl of the upper aqueous phase was removed, mixed with 350 μl 70% (v/v) ethanol and applied to RNeasy columns. The remaining steps were performed as recommended by the manufacturer. For cDNA synthesis 200 ng (tissue) or 500 ng (cells) of total RNA was reverse transcribed using the Quantitect reverse transcription kit (Qiagen).
Real-time quantitative PCR analysis
Primers for Rasd1 (Rat/Mouse, 5’-CCCTCAGCGTTGTGCCTACT-3’ and 5’-AAAGAGCGCACGGAACATCT-3’), c-Fos (Rat, 5’-AGCATGGGCTCCCCTGTCA-3’ and 5'-GAGACCAGAGTGGGCTGCA-3'; Mouse, 5’-TCCCCAAACTTCGACCATGA-3’ and 5’-GGCTGGGGAATGGTAGTAGG-3’), Nr4a1 (Rat, 5'-CTGCGACTGGGTCCTGGGTC-3' and 5'-TGTCAGGTGGTCACGCGGTC-3'; Mouse, 5’-AAAGTTGGGGGAGTGTGCTA-3’ and 5’-GAATACAGGGCATCTCCAGC-3’), rat hnCrh (5'-GGGCGAATAGCTTAAACCTG-3' and 5'-CAGGTGACCCTTCCTTGGAGA-3'), mouse GR (5’-TGTCACTGCTGGAGGTGATT-3’ and 5'-ATCACTTGACGCCCACCTAA-3'), rat hnAvp (5'-GAGGCAAGAGGGCCACATC-3' and 5'-CTCTCCTAGCCCATGACCCTT-3'), rat mature Avp (5'-TGCCTGCTACTTCCAGAACTGC-3' and 5'-AGGGGAGACACTGTCTCAGCTC-3'), eGFP (5’-ATCATGGCCGACAAGCAGAAGAAC-3’ and 5’-GTACAGCTCGTCCATGCCGAGAGT-3’), rat Rpl19 (5'-GCGTCTGCAGCCATGAGTA-3' and 5'-TGGCATTGGCGATTTCGTTG-3') and Gapdh (Rat, 5’-ATGATTCTACCCACGGCAAG-3’ and 5’-CTGGAAGATGGTGATGGGTT-3’; Mouse, 5’-CAACTCCCACTCTTCCACCT-3’ and 5’-CTTGCTCAGTGTCCTTGCTG-3’) were synthesised by Eurofins MWG Operon. The qPCRs were carried out in duplicate using SYBR green (Roche) on an ABI StepOnePlus Sequence Detection System (ABI, Warrington, UK). For relative quantification of gene expression the 2-ΔΔCT method was employed [82]. The internal control gene used for these analyses were the housekeeping gene Rpl19 and Gapdh. To analyse Rasd1 and GR gene knockdown PCRs for gel electrophoresis were performed using TaqDNA polymerase (New England Biolabs).
Immunofluorescence
Rats were deeply anesthetised with sodium pentobarbitone (100 mg/kg i.p.) and transcardially perfused with 0.1 M phosphate buffered saline (PBS, pH 7.4) followed by 4% (w/v) paraformaldehyde (PFA) in PBS. Brains were removed and post-fixed overnight in 4% (w/v) PFA followed by 30% (w/v) sucrose prepared in PBS. Tissues were sectioned to 40 μm on a cryostat, washed in PBS and blocked for 30 min in 5% (v/v) horse serum in PBS containing 0.25% (v/v) Triton X-100 (PBST). Sections were incubated with 1:500 dilution of rabbit anti-RASD1 antibody (Abcam, ab78459), 1:100 mouse anti-OT (neurophysin-I, PS38, [83]) or 1:100 mouse anti-AVP (neurophysin-II, PS41) prepared in 1% (v/v) horse serum in PBST at 4 °C overnight. The Rasd1 antibody has previously been shown to detect Rasd1 in mouse brain extracts by Western blot (data sheet), mouse kidney by immunohistochemistry (data sheet), and rat pancreas by immunofluorescence [47]. The sections were washed three times in PBS for 5 min and incubated with 1:500 dilution of anti-rabbit IgG-biotinylated secondary antibody in PBST for 1 h at room temperature. Sections were washed three times for 5 min with PBS and incubated for 1 h with secondary antibodies conjugated with fluorophore Alexa Fluor 488 streptavidin-conjugated and Alexa Fluor 594 donkey anti-mouse (Invitrogen). Sections were mounted and sealed with VectorShields mounting media (Vector Laboratories).
The GFP-Rasd1 fusion construct was generated by overlap extension PCR. Primers for GFP (5’-CGCGGATCCATGGTGAGCAAGGGCGAGGA-3’ and 5’-CTTGTACAGCTCGTCCATGCCGA-3’) and Rasd1 (5’-TCGGCATGGACGAGCTGTACAAGAAACTGGCCGCGATGAA-3’ and 5’-ACGCGTCGACCTAACTGATGACACAGCGCT-3’) were used for PCRs using Phusion High-FidelityDNAPolymerase (New England BioLabs). The final PCR product was digested with BamH1 and SalI and cloned into the corresponding sites of pcDNA3.1. N2a cells were grown on coverslips in 12-well tissue culture plates and transfected with GFP-Rasd1 fusion construct using Lipofectamine LTX (Life Technologies). At 48 h after transfections cells were fixed with 4% (w/v) PFA in PBS for 10 min and washed three times with PBS for 5 min. Cells were then incubated with 0.3% (v/v) Triton X-100 in PBS for 10 min for permeabilisation followed by 5% (v/v) horse serum prepared in PBS with 0.03% (v/v) Triton X-100 for 30 min for blocking. Cells were incubated with 1:500 dilution of rabbit anti-RASD1 antibody (Abcam, ab78459) prepared in 1% (v/v) horse serum in PBS-0.03 T at 4 °C overnight. After three washes, cells were incubated with Alexa Fluor 594 donkey anti-rabbit IgG (1:500; Invitrogen) for 1 h at room temperature, followed by three washes with PBS-0.03 T. Coverslips were mounted onto glass slides using VectorShields hard mounting media with DAPI. Images were captured on a confocal microscope (Leica).
Protein extraction and immunoblotting
Cells were washed twice with cold PBS pH 7.4 (Gibco; 10010–015), and harvested by scraping into RIPA buffer, consisting of PBS containing 1% (v/v) IGEPAL CA-630 (Sigma I3021), 0.5% (w/v) sodium deoxycholate, and 0.1% (w/v) sodium dodecyl sulfate, 1 mM PMSF, protease inhibitor (Sigma: P8340) and phosphatase inhibitor (Roche: 04906845001). The lysate was incubated on ice for 15 min with vortexing every 5 min, followed by centrifugation at 10,000 × g for 10 min. Supernatants were collected and kept at −80 °C. Protein concentrations were determined using the Bradford assay (Bio-Rad).
For immunoblotting, proteins (50 μg) were separated by SDS-PAGE and transferred to 0.45 μm PVDF membranes (Millipore). The membranes were blocked with 3% (w/v) ECL Prime blocking agent (GE Healthcare) in Tris-buffered saline-Tween 20 (0.1% (v/v) TBS-T) for 1 h at room temperature, followed by incubation with primary antibody diluted in 3% (w/v) ECL Prime blocking agent in TBST at 4 °C overnight. This was followed by incubation with appropriate secondary antibody conjugated with horseradish peroxidase (HRP) at room temperature for 1 h. Membranes were washed with TBST. Signal was visualized using high sensitivity WESTAR EtaC or WESTAR SuperNova extreme sensitivity HRP Detection Substrate (Cyanagen). Primary antibodies used: a rabbit polyclonal anti-RASD1 (1:1000; Abcam, ab78459), goat polyclonal anti-phosphoCREB (1:2500; Santa Cruz, sc-7978), and a mouse polyclonal anti-GAPDH (1:20,000; Santa Cruz, sc-32233) antibody. Immunoblots were stripped in Restore Western blot stripping buffer (ThermoScientific) and re-probed to assess multiple proteins in the same blot.
Organotypic studies
Organotypic cultures were prepared as described previously [84]. Sprague–Dawley pups (P5–P7) were purchased from Harlan Laboratories (UK). Pups were decapitated using scissors, and brains were removed and incubated in cold-Hank’s solution for 5 min, and then dissected using the optic chiasm as a landmark to produce a hypothalamic block. Slices (400 μm) were cut on a Mcllwain Tissue Chopper and placed onto hydrated Millipore Millicell-CM filter inserts in 6 well tissue culture plates containing 1.1 ml of culture medium. After 10 day, culture medium was replaced with serum free medium. The cultures were incubated at 35 °C in 5% (v/v) CO2 enriched air and medium was replaced every 2 days. Experimental treatments were performed after 4 days in serum free medium. Slice cultures were incubated with 0.01% (v/v) DMSO (vehicle) or 100nM DEX prepared in serum free medium for 4 h or 24 h. The inserts were frozen on dry ice in 6-well tissue culture plates, SON and PVN were punched (1 mm diameter micropunch) from 3 slices in a cryostat and RNA was extracted as described above.
Cells and treatments
Mouse pituitary cell line AtT20/D16v-F2 (Sigma; 94050406), Human Embryonic Kidney cells HEK293T/17 (ATTC CRL-11268), and Mouse Neuroblastoma N2a cells (ATTC CCL-131) were cultured in DMEM (Sigma; D6546) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Gibco), 2 mM L-glutamine and 100 unit/ml of penicillin-streptomycin. Cells were incubated at 37 °C in a humidified incubator with 5% (v/v) CO2. For chemical treatments, cells were seeded onto tissue culture plates to 60–70% confluence. After 24–72 h chemical treatments were performed at the time points indicated in the figure legend; 10 μM FSK (Sigma: F6886) and/or 100nM DEX (Sigma: D1756), or 10 μM FTI-277 (Sigma). Stock solutions of FSK (10 mM) and DEX (100 μM) were prepared in DMSO while FTI-277 (2.5 mM) was prepared in water.
To produce knockdown cell lines, AtT20 cells were transduced with a lentivirus containing shRNAs targeting mouse Rasd1 or GR. The shRNA sequences (Rasd1 shRNA1-GCCGTTTCGAGGATGCTTCAA, shRNA2-GCTCAAACAGCAGATCCTAGA; GR- GGAGATACAATCTTATCAAGC) were obtained from the RNAi consortium shRNA library. Sense and antisense oligonucleotides for shRNAs were synthesised (European MWG Operon) and cloned into lentiviral transfer vector pLKO.1 puro according to manufacturer’s guidelines (pLKO.1 puro was a gift from Bob Weinberg, Addgene plasmid 8453) [85]. A non-targeting shRNA sequence (ATCATGTTAGGCGTACGGACT) was used as a control. Virus particles were produced as previously described [86]. Twenty four hours after transduction, culture media was replaced with fresh media containing puromycin (2 μg/ml, Life Technologies). The cells were cultured in presence of puromycin for two weeks before use in experiments. The level of knockdown was confirmed by qPCR.
Adenoviral vector synthesis
cDNA clones encoding mouse Rasd1 and a dnRasd1 were the kind gift of Professor Richard Dorin, University of New Mexico. cDNAs were excised from pcDNA3.1 with restriction enzymes KpnI and XhoI and ligated into compatible restriction sites of adenoviral vector pacAd5.CMV (Cell Biolabs). Adenoviral vector pacAd5.CMV.eGFP was used as a control. The adenoviruses were generated by co-transfection of viral shuttle and backbone (pacAd5 9.2-100) vectors in HEK293T cells by calcium phosphate method in accordance with manufacturer’s guidelines (Cell Biolabs). Adenoviruses were purified by two rounds of CsCl ultracentrifugation and desalted using Slide-A-Lyzer dialysis cassettes (Thermoscientific). The purified viruses were aliquoted and stored at −80 °C. The virus titers were determined in triplicate by standard plaque assay. A multiplicity of infection of 10 was used for cell experiments.
Lentiviral vector gene transfer into SON
We used lentiviral vectors to study Rasd1 overexpression in the rat brain due to their higher infectivity of neurons compared to adenoviruses [87]. The cDNA clones of Rasd1 and dnRasd1 were excised from plasmid pcDNA3.1 and expressed in lentiviral vector pRRL.SIN.CPPT.CMV.IRES.eGFP.WPRE (modified from Addgene plasmid 12252). A lentiviral vector expressing eGFP (pRRL.SIN.CPPT.CMV.eGFP.WPRE) was used as a control. Viruses were generated as described previously [86]. Titers were calculated in transduction units per ml (TU/ml) with all viruses in the present study having a titer of > 5 x 109 TU/ml.
Stereotaxic injections of lentiviral vectors into the SON were performed as previously described [84]. Lentiviral vector tropism was assessed by visualising GFP expression in perfused tissue. To investigate gene expression following overexpression of Rasd1 or dnRasd1 in the SON, rats received bilateral lentiviral injections, where Rasd1 or dnRasd1 virus was administered to one nuclei and eGFP into the other and viral expression was allowed to proceed for two weeks. Two weeks after Rasd1 virus administration rats received a single i.p injection of either IS (0.15 M NaCl) or HS (1.5 M NaCl; 1.5 ml/100 g body weight) and were killed 30 min later. Two weeks after dnRasd1 virus administration rats were SL for 7 days.
Statistical analysis
Statistical differences between two experimental groups were evaluated using independent-sample unpaired Student’s t tests. One-way ANOVA with Tukey’s post hoc test were used to determine the difference between more than two samples with only a single influencing factor. Two-way ANOVA with Bonferonni post hoc test was used to determine interactions between two independent variables on the dependent variable. p < 0.05 was considered significant.
References
Antunes-Rodrigues J, de Castro M, Elias LL, Valenca MM, McCann SM. Neuroendocrine control of body fluid metabolism. Physiol Rev. 2004;84(1):169–208.
Papadimitriou A, Priftis KN. Regulation of the hypothalamic-pituitary-adrenal axis. Neuroimmunomodulation. 2009;16(5):265–71.
Swanson LW, Sawchenko PE, Rivier J, Vale WW. Organization of ovine corticotropin-releasing factor immunoreactive cells and fibers in the rat brain: an immunohistochemical study. Neuroendocrinology. 1983;36(3):165–86.
Vandesande F, Dierickx K, De Mey J. The origin of the vasopressinergic and oxytocinergic fibres of the external region of the median eminence of the rat hypophysis. Cell Tissue Res. 1977;180(4):443–52.
Antoni FA. Vasopressinergic control of pituitary adrenocorticotropin secretion comes of age. Front Neuroendocrinol. 1993;14(2):76–122.
DeBold CR, DeCherney GS, Jackson RV, Sheldon WR, Alexander AN, Island DP, et al. Effect of synthetic ovine corticotropin-releasing factor: prolonged duration of action and biphasic response of plasma adrenocorticotropin and cortisol. J Clin Endocrinol Metab. 1983;57(2):294–8.
Kovacs KJ, Foldes A, Sawchenko PE. Glucocorticoid negative feedback selectively targets vasopressin transcription in parvocellular neurosecretory neurons. J Neurosci. 2000;20(10):3843–52.
Kovacs KJ, Sawchenko PE. Regulation of stress-induced transcriptional changes in the hypothalamic neurosecretory neurons. J Mol Neurosci. 1996;7(2):125–33.
Kovacs KJ, Sawchenko PE. Sequence of stress-induced alterations in indices of synaptic and transcriptional activation in parvocellular neurosecretory neurons. J Neurosci. 1996;16(1):262–73.
Murphy D, Carter D. Vasopressin gene expression in the rodent hypothalamus: transcriptional and posttranscriptional responses to physiological stimulation. Molecular endocrinology (Baltimore. Md. 1990;4(7):1051–9.
Yoshida M. Gene regulation system of vasopressin and corticotropin-releasing hormone. Gene regulation and systems biology. 2008;2:71–88.
Legradi G, Holzer D, Kapcala LP, Lechan RM. Glucocorticoids inhibit stress-induced phosphorylation of CREB in corticotropin-releasing hormone neurons of the hypothalamic paraventricular nucleus. Neuroendocrinology. 1997;66(2):86–97.
Shiromani PJ, Magner M, Winston S, Charness ME. Time course of phosphorylated CREB and Fos-like immunoreactivity in the hypothalamic supraoptic nucleus after salt loading. Brain Res. 1995;29(1):163–71.
Ceccatelli S, Cintra A, Hokfelt T, Fuxe K, Wikstrom AC, Gustafsson JA. Coexistence of glucocorticoid receptor-like immunoreactivity with neuropeptides in the hypothalamic paraventricular nucleus. Exp Brain Res. 1989;78(1):33–42.
Kiss JZ, Van Eekelen JA, Reul JM, Westphal HM, De Kloet ER. Glucocorticoid receptor in magnocellular neurosecretory cells. Endocrinology. 1988;122(2):444–9.
Berghorn KA, Knapp LT, Hoffman GE, Sherman TG. Induction of glucocorticoid receptor expression in hypothalamic magnocellular vasopressin neurons during chronic hypoosmolality. Endocrinology. 1995;136(2):804–7.
Han F, Ozawa H, Matsuda K, Nishi M, Kawata M. Colocalization of mineralocorticoid receptor and glucocorticoid receptor in the hippocampus and hypothalamus. Neurosci Res. 2005;51(4):371–81.
Kemppainen RJ, Behrend EN. Dexamethasone rapidly induces a novel ras superfamily member-related gene in AtT-20 cells. J Biol Chem. 1998;273(6):3129–31.
Kemppainen RJ, Cox E, Behrend EN, Brogan MD, Ammons JM. Identification of a glucocorticoid response element in the 3'-flanking region of the human Dexras1 gene. Biochim Biophys Acta. 2003;1627(2–3):85–9.
Brogan MD, Behrend EN, Kemppainen RJ. Regulation of Dexras1 expression by endogenous steroids. Neuroendocrinology. 2001;74(4):244–50.
Graham TE, Key TA, Kilpatrick K, Dorin RI. Dexras1/AGS-1, a steroid hormone-induced guanosine triphosphate-binding protein, inhibits 3',5'-cyclic adenosine monophosphate-stimulated secretion in AtT-20 corticotroph cells. Endocrinology. 2001;142(6):2631–40.
Graham TE, Qiao Z, Dorin RI. Dexras1 inhibits adenylyl cyclase. Biochem Biophys Res Commun. 2004;316(2):307–12.
Harrison LM, He Y. Rhes and AGS1/Dexras1 affect signaling by dopamine D1 receptors through adenylyl cyclase. J Neurosci Res. 2011;89(6):874–82.
Nguyen CH, Watts VJ. Dexras1 blocks receptor-mediated heterologous sensitization of adenylyl cyclase 1. Biochem Biophys Res Commun. 2005;332(3):913–20.
Nguyen CH, Watts VJ. Dexamethasone-induced Ras protein 1 negatively regulates protein kinase C delta: implications for adenylyl cyclase 2 signaling. Mol Pharmacol. 2006;69(5):1763–71.
Cheng HY, Dziema H, Papp J, Mathur DP, Koletar M, Ralph MR, et al. The molecular gatekeeper Dexras1 sculpts the photic responsiveness of the mammalian circadian clock. J Neurosci. 2006;26(50):12984–95.
Ong SA, Tan JJ, Tew WL, Chen KS. Rasd1 modulates the coactivator function of NonO in the cyclic AMP pathway. PLoS One. 2011;6(9):e24401.
Hindmarch C, Yao S, Beighton G, Paton J, Murphy D. A comprehensive description of the transcriptome of the hypothalamoneurohypophyseal system in euhydrated and dehydrated rats. Proc Natl Acad Sci U S A. 2006;103(5):1609–14.
Stewart L, Hindmarch CC, Qiu J, Tung YC, Yeo GS, Murphy D. Hypothalamic transcriptome plasticity in two rodent species reveals divergent differential gene expression but conserved pathways. J Neuroendocrinol. 2011;23(2):177–85.
Fang M, Jaffrey SR, Sawa A, Ye K, Luo X, Snyder SH. Dexras1: a G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron. 2000;28(1):183–93.
Takahashi H, Umeda N, Tsutsumi Y, Fukumura R, Ohkaze H, Sujino M, et al. Mouse dexamethasone-induced RAS protein 1 gene is expressed in a circadian rhythmic manner in the suprachiasmatic nucleus. Brain Res. 2003;110(1):1–6.
Cheng HY, Obrietan K. Dexras1: shaping the responsiveness of the circadian clock. Semin Cell Dev Biol. 2006;17(3):345–51.
Mezey E, Kiss JZ. Coexpression of vasopressin and oxytocin in hypothalamic supraoptic neurons of lactating rats. Endocrinology. 1991;129(4):1814–20.
Ma XM, Levy A, Lightman SL. Rapid changes in heteronuclear RNA for corticotrophin-releasing hormone and arginine vasopressin in response to acute stress. J Endocrinol. 1997;152(1):81–9.
Ponzio TA, Yue C, Gainer H. An intron-based real-time PCR method for measuring vasopressin gene transcription. J Neurosci Methods. 2007;164(1):149–54.
de Quervain DJ, Roozendaal B, McGaugh JL. Stress and glucocorticoids impair retrieval of long-term spatial memory. Nature. 1998;394(6695):787–90.
Lerner EC, Qian Y, Blaskovich MA, Fossum RD, Vogt A, Sun J, et al. Ras CAAX peptidomimetic FTI-277 selectively blocks oncogenic Ras signaling by inducing cytoplasmic accumulation of inactive Ras-Raf complexes. J Biol Chem. 1995;270(45):26802–6.
Aguilera G, Lightman SL, Kiss A. Regulation of the hypothalamic-pituitary-adrenal axis during water deprivation. Endocrinology. 1993;132(1):241–8.
Dohanics J, Kovacs KJ, Folly G, Makara GB. Long-term salt loading impairs pituitary responsiveness to ACTH secretagogues and stress in rats. Peptides. 1990;11(1):59–63.
Dohanics J, Verbalis JG. Naloxone disinhibits magnocellular responses to osmotic and volemic stimuli in chronically hypoosmolar rats. J Neuroendocrinol. 1995;7(1):57–62.
Kiss A, Aguilera G. Regulation of the hypothalamic pituitary adrenal axis during chronic stress: responses to repeated intraperitoneal hypertonic saline injection. Brain Res. 1993;630(1–2):262–70.
Dallman MF, Akana SF, Scribner KA, Bradbury MJ, Walker CD, Strack AM, et al. Stress, feedback and facilitation in the hypothalamo-pituitary-adrenal axis. J Neuroendocrinol. 1992;4(5):517–26.
Pitman DL, Ottenweller JE, Natelson BH. Plasma corticosterone levels during repeated presentation of two intensities of restraint stress: chronic stress and habituation. Physiol Behav. 1988;43(1):47–55.
Cha JY, Kim HJ, Yu JH, Xu J, Kim D, Paul BD, et al. Dexras1 mediates glucocorticoid-associated adipogenesis and diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110(51):20575–80.
Kushner SA, Elgersma Y, Murphy GG, Jaarsma D, van Woerden GM, Hojjati MR, et al. Modulation of presynaptic plasticity and learning by the H-ras/extracellular signal-regulated kinase/synapsin I signaling pathway. J Neurosci. 2005;25(42):9721–34.
Compton SL, Kemppainen RJ, Behrend EN. Prenylated Rab acceptor domain family member 1 is involved in stimulated ACTH secretion and inhibition. Cell Signal. 2009;21(12):1901–9.
Lellis-Santos C, Sakamoto LH, Bromati CR, Nogueira TC, Leite AR, Yamanaka TS, et al. The regulation of Rasd1 expression by glucocorticoids and prolactin controls peripartum maternal insulin secretion. Endocrinology. 2012;153(8):3668–78.
McGrath MF, Ogawa T, de Bold AJ. Ras dexamethasone-induced protein 1 is a modulator of hormone secretion in the volume overloaded heart. Am J Physiol Heart Circ Physiol. 2012;302(9):H1826–37.
Meister B, Cortes R, Villar MJ, Schalling M, Hokfelt T. Peptides and transmitter enzymes in hypothalamic magnocellular neurons after administration of hyperosmotic stimuli: comparison between messenger RNA and peptide/protein levels. Cell Tissue Res. 1990;260(2):279–97.
Reaux-Le Goazigo A, Morinville A, Burlet A, Llorens-Cortes C, Beaudet A. Dehydration-induced cross-regulation of apelin and vasopressin immunoreactivity levels in magnocellular hypothalamic neurons. Endocrinology. 2004;145(9):4392–400.
Cismowski MJ, Ma C, Ribas C, Xie X, Spruyt M, Lizano JS, et al. Activation of heterotrimeric G-protein signaling by a ras-related protein. Implications for signal integration. J Biol Chem. 2000;275(31):23421–4.
Graham TE, Prossnitz ER, Dorin RI. Dexras1/AGS-1 inhibits signal transduction from the Gi-coupled formyl peptide receptor to Erk-1/2 MAP kinases. J Biol Chem. 2002;277(13):10876–82.
Imaki T, Xiao-Quan W, Shibasaki T, Yamada K, Harada S, Chikada N, et al. Stress-induced activation of neuronal activity and corticotropin-releasing factor gene expression in the paraventricular nucleus is modulated by glucocorticoids in rats. J Clin Invest. 1995;96(1):231–8.
Kovacs KJ, Mezey E. Dexamethasone inhibits corticotropin-releasing factor gene expression in the rat paraventricular nucleus. Neuroendocrinology. 1987;46(4):365–8.
Lauand F, Ruginsk SG, Rodrigues HL, Reis WL, de Castro M, Elias LL, et al. Glucocorticoid modulation of atrial natriuretic peptide, oxytocin, vasopressin and Fos expression in response to osmotic, angiotensinergic and cholinergic stimulation. Neuroscience. 2007;147(1):247–57.
Kiss JZ, Mezey E, Skirboll L. Corticotropin-releasing factor-immunoreactive neurons of the paraventricular nucleus become vasopressin positive after adrenalectomy. Proc Natl Acad Sci U S A. 1984;81(6):1854–8.
Suemaru S, Hashimoto K, Ogasa T, Takao T, Ota Z, Hirakawa M, et al. Effects of hyperosmotic stimulation and adrenalectomy on vasopressin mRNA levels in the paraventricular and supraoptic nuclei of the hypothalamus: in situ hybridization histochemical analysis using a synthetic oligonucleotide probe. Acta Med Okayama. 1990;44(5):233–41.
Carter DA, Murphy D. Cyclic nucleotide dynamics in the rat hypothalamus during osmotic stimulation: in vivo and in vitro studies. Brain Res. 1989;487(2):350–6.
Ceding P, Schilling K, Schmale H. Vasopressin Expression in Cultured Neurons is Stimulated by Cyclic AMP. J Neuroendocrinol. 1990;2(6):859–65.
Young 3rd WS, Shepard EA, Burch RM. Plasma hyperosmolality increases G protein and 3',5'-cyclic adenosine monophosphate synthesis in the paraventricular and supraoptic nuclei. Molecular endocrinology (Baltimore, Md). 1987;1(12):884–8.
Fass DM, Butler JE, Goodman RH. Deacetylase activity is required for cAMP activation of a subset of CREB target genes. J Biol Chem. 2003;278(44):43014–9.
Chan RK, Brown ER, Ericsson A, Kovacs KJ, Sawchenko PE. A comparison of two immediate-early genes, c-fos and NGFI-B, as markers for functional activation in stress-related neuroendocrine circuitry. J Neurosci. 1993;13(12):5126–38.
Kawasaki M, Yamaguchi K, Saito J, Ozaki Y, Mera T, Hashimoto H, et al. Expression of immediate early genes and vasopressin heteronuclear RNA in the paraventricular and supraoptic nuclei of rats after acute osmotic stimulus. J Neuroendocrinol. 2005;17(4):227–37.
Martin LJ, Tremblay JJ. Glucocorticoids antagonize cAMP-induced Star transcription in Leydig cells through the orphan nuclear receptor NR4A1. J Mol Endocrinol. 2008;41(3):165–75.
Kato K, Cox AD, Hisaka MM, Graham SM, Buss JE, Der CJ. Isoprenoid addition to Ras protein is the critical modification for its membrane association and transforming activity. Proc Natl Acad Sci U S A. 1992;89(14):6403–7.
Clarke S. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu Rev Biochem. 1992;61:355–86.
Basso AD, Kirschmeier P, Bishop WR. Lipid posttranslational modifications. Farnesyl transferase inhibitors. J Lipid Res. 2006;47(1):15–31.
Khan OM, Ibrahim MX, Jonsson IM, Karlsson C, Liu M, Sjogren AK, et al. Geranylgeranyltransferase type I (GGTase-I) deficiency hyperactivates macrophages and induces erosive arthritis in mice. J Clin Invest. 2011;121(2):628–39.
Stamatakis K, Cernuda-Morollon E, Hernandez-Perera O, Perez-Sala D. Isoprenylation of RhoB is necessary for its degradation. A novel determinant in the complex regulation of RhoB expression by the mevalonate pathway. J Biol Chem. 2002;277(51):49389–96.
Hazell GG, Hindmarch CC, Pope GR, Roper JA, Lightman SL, Murphy D, et al. G protein-coupled receptors in the hypothalamic paraventricular and supraoptic nuclei--serpentine gateways to neuroendocrine homeostasis. Front Neuroendocrinol. 2012;33(1):45–66.
Blumer JB, Cismowski MJ, Sato M, Lanier SM. AGS proteins: receptor-independent activators of G-protein signaling. Trends Pharmacol Sci. 2005;26(9):470–6.
Wie J, Kim J, Ha K, Zhang YH, Jeon JH, So I. Dexamethasone activates transient receptor potential canonical 4 (TRPC4) channels via Rasd1 small GTPase pathway. Pflugers Arch. 2015;467(10):2081–91.
Nedungadi TP, Cunningham JT. Differential regulation of TRPC4 in the vasopressin magnocellular system by water deprivation and hepatic cirrhosis in the rat. American journal of physiology. 2014;306(5):R304–14.
Obukhov AG, Nowycky MC. TRPC4 can be activated by G-protein-coupled receptors and provides sufficient Ca(2+) to trigger exocytosis in neuroendocrine cells. J Biol Chem. 2002;277(18):16172–8.
Thapliyal A, Bannister RA, Hanks C, Adams BA. The monomeric G proteins AGS1 and Rhes selectively influence Galphai-dependent signaling to modulate N-type (CaV2.2) calcium channels. Am J Physiol Cell Physiol. 2008;295(5):C1417–26.
Mons N, Cooper DM. Adenylyl cyclase mRNA expression does not reflect the predominant Ca2+/calmodulin-stimulated activity in the hypothalamus. J Neuroendocrinol. 1994;6(6):665–71.
Lubelski D, Ponzio TA, Gainer H. Effects of A-CREB, a dominant negative inhibitor of CREB, on the expression of c-fos and other immediate early genes in the rat SON during hyperosmotic stimulation in vivo. Brain Res. 2012;1429:18–28.
Jaffrey SR, Fang M, Snyder SH. Nitrosopeptide mapping: a novel methodology reveals s-nitrosylation of dexras1 on a single cysteine residue. Chem Biol. 2002;9(12):1329–35.
Kadowaki K, Kishimoto J, Leng G, Emson PC. Up-regulation of nitric oxide synthase (NOS) gene expression together with NOS activity in the rat hypothalamo-hypophysial system after chronic salt loading: evidence of a neuromodulatory role of nitric oxide in arginine vasopressin and oxytocin secretion. Endocrinology. 1994;134(3):1011–7.
Ueta Y, Levy A, Chowdrey HS, Lightman SL. Water deprivation in the rat induces nitric oxide synthase (NOS) gene expression in the hypothalamic paraventricular and supraoptic nuclei. Neurosci Res. 1995;23(3):317–9.
Soriano FX, Martel MA, Papadia S, Vaslin A, Baxter P, Rickman C, et al. Specific targeting of pro-death NMDA receptor signals with differing reliance on the NR2B PDZ ligand. J Neurosci. 2008;28(42):10696–710.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods (San Diego, Calif). 2001;25(4):402–8.
Ben-Barak Y, Russell JT, Whitnall MH, Ozato K, Gainer H. Neurophysin in the hypothalamo-neurohypophysial system. I. Production and characterization of monoclonal antibodies. J Neurosci. 1985;5(1):81–97.
Greenwood MP, Greenwood M, Paton JF, Murphy D. Control of Polyamine Biosynthesis by Antizyme Inhibitor 1 Is Important for Transcriptional Regulation of Arginine Vasopressin in the Male Rat Hypothalamus. Endocrinology. 2015;156(8):2905–17.
Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA (New York, NY). 2003;9(4):493–501.
Greenwood M, Bordieri L, Greenwood MP, Rosso Melo M, Colombari DS, Colombari E, et al. Transcription factor CREB3L1 regulates vasopressin gene expression in the rat hypothalamus. J Neurosci. 2014;34(11):3810–20.
Duale H, Kasparov S, Paton JF, Teschemacher AG. Differences in transductional tropism of adenoviral and lentiviral vectors in the rat brainstem. Exp Physiol. 2005;90(1):71–8.
Acknowledgements
We would like to thank Prof. Harold Gainer (NINDS, NIH, USA) for providing us with antibodies recognising AVP neurophysin-II and OT neurophysin-I. We gratefully acknowledge the support of the MRC (G0700954, DM), BBSRC (BB/G006156/1, MPG, JP, DM; BB/J015415/1, MG, JP, DM), University of Malaya (HIR award H-20001-E0000865, ASM, DM), and Sao Paulo Research Foundation FAPESP (2011/52108-4, JAR; 2013/09799-1, JAR).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare no competing financial or non-financial interests.
Authors’ contributions
MPG established knockdown cell lines, carried out animal studies, participated in organotypic culture, performed the statistical analysis and drafted the manuscript. MG carried out cell culture, organotypic culture and molecular studies, participated in the design of the study and helped to draft the manuscript. ASM participated in animal experiment and participated in design of the study. JAR, JFRP and DM conceived of the study, participated in its design and coordination and helped to draft the manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Greenwood, M.P., Greenwood, M., Mecawi, A.S. et al. Rasd1, a small G protein with a big role in the hypothalamic response to neuronal activation. Mol Brain 9, 1 (2016). https://doi.org/10.1186/s13041-015-0182-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13041-015-0182-2