Abstract
Objective
This study aimed to report a fetus with maternal partial trisomy 9p and 14q and the phenotype detected in ultrasound.
Methods
The chromosome rearrangements in the fetus were characterized by G-banding and chromosome microarray analysis based on single nucleotide polymorphism (SNP) array of cultured amniocytes and compared with the parents’ karyotypes.
Results
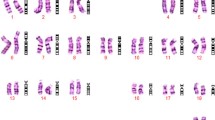
The fetal abnormal karyotype was 47,XY,+der(14)(9;14)(p23;q22). The SNP array revealed a duplicate 11.8-Mb 9p23-p24.3 fragment and a duplicate 29.6-Mb 14q11.2-q21.3 fragment. The peripheral blood karyotype of the mother was 46,XX,t(9;14)(p23;q22), while the father’s was normal at the level of 300~400 bands. A high-resolution karyotype analysis conformed the same abnormality of the mother at the level of 550~650 bands. These results indicated that the fetal chromosomal abnormality probably derived from the mother. The fetal nuchal translucency thickness was 3.5 mm, and the fetal heart was detected with around 1.0-mm ventricular defect by the ultrasound examination at 12-week gestation. The couple decided to terminate the pregnancy. They opted for in vitro fertilization and embryo transfer for the fourth pregnancy, which was successful.
Conclusions
The SNP array combined with cytogenetic analysis was particularly effective in identifying abnormal chromosomal rearrangements. These methods combined with the existing database information and fetal ultrasonography might provide a comprehensive and efficient way for the prenatal assessment of fetal situations. Preimplantation genetic diagnosis might effectively assist those women with an adverse pregnancy history in their next pregnancy.
Similar content being viewed by others
Introduction
Trisomy 9p is one of the most abnormal chromosomes in newborns. However, the case of partial 9p and 14q trisomy has been reported only once to date [1]. Chromosome trisomy is usually caused by the nondisjunction of homologous chromosomes during gamete formation, especially from the balanced translocation carriers in the parents. In most cases, the trisomic segments are transmitted from the mother or father carrying balanced translocation. However, genetic changes in the embryo often result in clinical phenotypic changes. The degree of phenotype is closely related to the extension of chromosome duplication or deletion segments. In other words, the phenotypes are connected with a small supernumerary marker chromosome (sSMC) [2]. Moreover, the degree of clinical symptoms is consistent with the important functional genes in the abnormal chromosome segments. The correlation studies between phenotype and genotype indicated that the region from 9p22 to 9p24 was the minimal critical extension to result in clinical syndromes [3, 4]. The derived duplication from 14q11.2 to 14q22.3 indicated severe physical and mental retardation defects [5]. The forkhead box protein G1 (FOXG1) gene encompassed on 14q11.2 to 14q12 could cause severe epilepsy and developmental delay and severe speech impairment [6, 7]. This study aimed to report a fetus inheriting maternal derivative chromosome 14 with partial 9p24.3p23 and 14q11.2q21.3 duplications and abnormal phenotype, which was detected by ultrasound examination.
Case presentation
A 28-year-old woman who had previously experienced two early spontaneous abortions was pregnant for the third time. The couple were not consanguineous and did not have any family hereditary diseases. The woman’s last menstruation was on January 24, 2017. The nuchal translucency thickness of the fetus was 3.5 mm, and his heart had an approximately 1.0-mm ventricular defect detected in ultrasound at 12-week gestation. Amniocentesis was performed at 18-week gestation with the consent of the parents because of the two previous spontaneous abortions and the fetal structural abnormality. The fetal abnormal karyotype by G-banding was 47,XY,+der(14)(9;14)(p23;q22) at the level of 300~400 bands (Fig. 1). The SNP array revealed a duplicate 11.8-Mb fragment and a duplicate 29.6-Mb fragment with the suspended amniotic cells (Figs. 4 and 5). The couple underwent karyotype analysis to further identify the source of fetal chromosomal abnormalities and the arrangement of the cytological changes.. The results showed the same chromosomal abnormalities in the mother (Fig. 2), but no abnormality in the father. A high-resolution karyotype analysis identified the same abnormal karyotype of the mother at the level of 550~650 bands once more (Fig. 3). Combined with the CMA results, this study concluded that the fetus had an extra derivative materal chromosome with partial 9p and 14p duplication. The couple decided to terminate the pregnancy at 24-week gestation after they were informed of the possible serious consequences. A 724 g fetus was delivered with low-set ears. They selected preimplantation genetic diagnosis (PGD) to assist the next pregnancy.
Cytogenetic and SNP array analyses
Amniocytes and peripheral blood lymphocytes of the couple were routinely collected, cultured, and harvested. G-banding was performed, followed by conventional cytogenetic analysis. Then 47,XY,+der(14)(9;14)(p23;q22) of the fetus and 46,XX,t(9;14)(p23;q22) of the mother were found according to the international system for human cytogenomic nomenclature (ISCN) 2016. Further, a high-resolution chromosome analysis of the mother’s peripheral blood was performed. The SNP array of suspended cultured amniocytes was conducted using the SNP array CytoScan 750 K probes (Affymetrix, CA, USA). The Chromosome Analysis Suite software (ChAS) was adopted for data analysis, and the results were analyzed using multiple databases, such as Online Mendelian Inheritance in Man (OMIM) and Genome.
Results
The fetal karyotype was 47,XY,+der(14)(9;14)(p23;q22)mat at the level of 300~400 bands (Fig. 1). The mother’s chromosome was the same as that of the fetus at the level of both 300~400 (Fig. 2) and 550~650 bands (Fig. 3). However, the karyotype of the father was normal. The fetus had a duplicate 11.8-Mb 9p24.3p23 fragment (arr[hg19] 9p24.3p23 (208 454–12 064 543) × 3, Fig. 4) containing 32 OMIM genes, including GLI-similar 3 (GLIS3) and SWI/SNF-related matrix associated, actin-dependent regulator of chromatin 2 (SMARCA2). The fetus also had a duplicate 29.6-Mb 14q11.2q21.3 fragment (arr[hg19] 14q11.2q21.3 (20 516 277–50 131 335) × 3, Fig. 5), containing 146 OMIM genes, including chromodomain helicase DNA-binding protein 8 (CHD8), suppressor of Ty 16 homolog (SUPT16H), forkhead box protein G1 (FOXG1) and protein kinase D1 (PRKD1).
Comparison with the literature
We compared the clinical phenotypes of the fetus with those previously reported cases with duplication of chromosome 9 and 14 (Tables 1 and 2). Table 1 gave an overview clinical abnormal performance of the patients with partial trisomy 9p at least overlapping with duplicated segment in our index fetus. At the same time, we listed clinical manifestations of the patients with partial trisomy 14q on the Table 2. There was mostly apparent consistency in the facial and limb anomalies and developmental delay and mental retardation in the patients with partial trisomy 9p and (or) 14q which might vary in degree listed in the tables above. Less common findings were congenital heart defects. A female infant born at 35 weeks gestation with duplicated 9p13p24.3 and 14p13q22 showed craniofacial anomalies and limbs abnormalities and a patent ductus arteriosus [1].
Follow-up outcomes
In early March 2018, the couple underwent one cycle of in vitro fertilization (IVF) and embryo transfer for the fourth pregnancy and selected the PGD pregnancy procedure in the People’s Hospital of Jiangsu Province. Subsequently, an amniocentesis chromosome examination was conducted at 18-week gestation, and the karyotype of the fetus was found to be normal. Fortunately, the mother succeeded in delivering a healthy baby girl on December 11, 2018.
Discussion
According to the principle of gamete distribution [33], the possibility of the living offspring inheriting an abnormal chromosome is 1/18 if either of a couple has a balanced translocation. The present study reported that fetal-derived chromosome 14 had partial 9p and 14q duplications. The chromosome analysis combined with the SNP array of cultured amniocyte results revealed that the fetal chromosomal abnormality probably derived from the mother. That was to say, the fetus not only inherited the normal chromosomes 9 and 14 of the parents’, but also had a derived abnormal chromosome 14 from the mother. Trisomy 9p was the fourth most frequent chromosome anomaly compatible with long-term survival in live-born infants [13, 14, 34], meanwhile trisomy 14q was not less than reported trisomy 9p in the literatures of 1970s [16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32, 35]. However, the case of partial 9p and 14q trisomy has been reported only once to date [1].
Patients with trisomy 9p are easily recognizable owing to their facial appearance. This results in complex rearrangements and the possibility that some of the duplicated genes will be dosage-sensitive, influencing the phenotype [15]. The pericentromeric region of chromosome 9 is rich in segmental duplication and low copy repeats that predispose it to nonallelic homologous recombination. With a high degree of sequence identity to sequences in 15p, 18p, and 21p, chromosome 9 is inclined to illegitimate intrachromosomal or interchromosomal recombination. The correlation studies between phenotype and genotype indicated that the region from 9p22 to 9p24 was the minimal critical extension to result in clinical syndromes [3, 4]. Patients with 9p trisomy display variable degrees of mental retardation and head and facial abnormal features, such as microcephaly with a large anterior fontanelle, micrognathia, a prominent or bulbous nose, malformed protruding ears, deep-set eyes, mild down slanting of the palpebral fissures, downturned corners of the mouth, congenital heart defects, mental retardation, and kidney and skeletal anomalies [13, 34]. A 3-year-old boy with de novo 9p24.2 to 9p23 was diagnosed with development lag and craniofacial anomalies [36]. Some studies reported that the partial duplication of 9p24.3p23 was related to microcephaly, autism, and other clinical phenotype-related diseases [4, 15, 37]. In the present study, the fetus with 9p24.3p23 contained 32 OMIM pathological genes, including GLIS3 and SMARCA2. The GLIS3 gene partially had the same chromosome segments as described in the aforementioned 3-year-old boy [36]. The fetus might be prone to neonatal diabetes complicated with congenital hypothyroidism, and have intrauterine developmental retardation during pregnancy and low-set ears and craniosynostosis after birth. SMARCA2 gene mutations are associated with Nicolaides–Baraitser syndrome of autosomal dominant inheritance, clinical manifestations of short stature, microcephalus, dysgnosia, epilepsy, and learning disabilities. The growth and structural abnormalities were observed through an ultrasound examination. Only low-set ears and abnormal nuchal translucency thickness and heart changes of the fetus occurred during the pregnancy, but some future symptoms such as epilepsy and learning disabilities could not be detected because of the termination of pregnancy.
Another duplication of 14q11.2q21.3 of the fetus was found with 146 OMIM genes, including CHD8, SUPT16H, FOXG1, and PRKD1 gene mutations closely correlated with the postnatal clinical phenotype. A 14-year-old male patient with a de novo 14q11.2 microduplication, a region significantly associated with quantitative trait loci for stature and a component of intelligence, was significantly characterized by short stature, mild mental retardation, and dysmorphic facial features [30]. A 445-kb 14q11.2 microduplication involving CHD8 and SUPT16H genes causes developmental delay, intellectual disability, autism spectrum disorders, and macrocephaly, which was found in an 8-year-old boy [31]. The clinical phenotype of 14q11.2 microduplication included postpartum slow growth, microcephalus, abnormal breathing patterns, gastroesophageal reflux, dysgnosia, and agenesis of the corpus callosum [5, 30]. The PRKD1 gene mutations are associated with autosomal dominant diseases, including congenital heart defects and ectodermal dysplasia [30, 31]. Furthermore, the thickness of the fetal nuchal translucency in the present case was 3.5 mm, and the heart had an approximately 1.0-mm ventricular defect detected during ultrasound examination at 12-week gestation, which might have been caused by the PRKD1 gene mutation.
In addition, based on the homozygosity or heterozygosity of polymorphic alleles inherited from the parent, uniparental disomy (UPD) can be classified into isodisomy and heterodisomy. Notably, balanced familial translocationincreases the risk of fetal UPD [38]. Human chromosome 14q32.2 carries a number of imprinted genes such as delta-like non-canonical Notch ligand 1 (DLK1), retrotransposon-like 1 (RTL1), and Deiodinase, iodothyronine, type III (DIO3). Both paternal UPD 14 and maternal UPD 14 can cause disorders. Paternal UPD14 has been reported to be associated with Kagami-Ogata syndrome, which is characterized of polyhydramnios, developmental delay, growth retardation, abdominal defects, thoracic dysplasia with respiratory failure, and facial abnormalities [39]. Maternal UPD 14 causes Temple syndrome with multiple serious phenotypes including prenatal and postnatal growth retardation, developmental delay, joint laxity, small hands and feet, muscular hypotonia, truncal obesity, precocious puberty, and short stature [40]. The SNP array analysis from the Allele difference and BAF showed no loss of heterozygosity(LOH)in this fetus. However, heterodisomy could not be excluded despite less phenotype of this fetus in ultrasound.
The pregnancy was terminated. The couple selected one cycle of IVF and embryo transfer. Also, they chose PGD for the fourth pregnancy in early March 2018 and accepted amniocentesis during middle gestation in the People’s Hospital of Jiangsu province. Fortunately, the mother succeeded in giving birth to a healthy baby girl on December 11, 2018.
In conclusion, the SNP array combined with cytogenetic analysis might help in identifying abnormal chromosomal rearrangements. These methods combined with the existing database information and fetal ultrasonography reports may provide a comprehensive and efficient way for prenatal assessment of fetal situations. PGD effectively assists women with an adverse pregnancy history for their next pregnancy.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Abbreviations
- ChAS:
-
Chromosome Analysis Suite
- CMA:
-
Chromosome microarray analysis
- DIO3 :
-
Deiodinase, iodothyronine, type III3
- DLK1 :
-
Delta-like non-canonical Notch ligand 1
- FOXG1 :
-
Forkhead box protein G1
- GLIS3 :
-
Genes including GLI-similar 3
- CHD8 :
-
Chromodomain helicase DNA-binding protein 8
- ISCN:
-
International system for human cytogenomic nomenclature
- LOH:
-
Loss of heterozygosity
- NT:
-
Nuchal translucency
- OMIM:
-
Online Mendelian Inheritance in Man
- PGD:
-
Plantation genetic diagnosis
- PRKD1 :
-
Protein kinase D1
- RTL1 :
-
Retrotransposon-like 1
- SMARCA2 :
-
SWI/SNF related matrix associated; actin dependent regulator of chromatin 2
- SNP:
-
Single nucleotide polymorphism
- sSMC:
-
Small supernumerary marker chromosome
- SUPT16H :
-
Suppressor of Ty 16 Homolog
- UPD:
-
Uniparental disomy
References
Angle B, Yen F, Cole CW. Case of partial trisomy 9p and partial trisomy 14q resulting from a maternal translocation: overlapping manifestations of characteristic phenotypes. Am J Med Genet. 1999;84(2):132–6.
Jafari-Ghahfarokhi H, Moradi-Chaleshtori M, Liehr T, Hashemzadeh-Chaleshtori M, Teimori H, Ghasemi-Dehkordi P. Small supernumerary marker chromosomes and their correlation with specific syndromes. Adv Biomed Res. 2015;4:140.
Fujimoto A, Lin MS, Schwartx S. Direct duplication of 9p22-p24 in a child with duplication 9p syndromel. Am J Med Geneet. 1998;77(4):268–71.
Haddad BR, Lin AE, Wyandt H, Milunsky A. Molecular cytogenetic characterization of the first familial case of partial 9p duplication (p22p24). J Med Genet. 1996;33(12):1045–7.
Wannenmacher B, Mitter D, KieβLing F, Liehr T, Weise A, Siekmeyer M, Kiess W. A 33-year-old male patient with paternal derived duplication of 14q11.2–14q22.1~22.3: clinical course, phenotypic and genotypic findings. J Pediatr Endocrinol Metab. 2016;2016(5):611–6.
Brunetti-Pierri N, Paciorkowski AR, Ciccone R, Della Mina E, Bonaglia MC, Borgatti R, Schaaf CP, Sutton VR, Xia Z, Jelluma N, Ruivenkamp C, Bertrand M, de Ravel TJ, Jayakar P, Belli S, Rocchetti K, Pantaleoni C, D'Arrigo S, Hughes J, Cheung SW, Zuffardi O, Stankiewicz P. Duplications of FOXG1 in 14q12 are associated with developmental epilepsy, mental retardation, and severe speech impairment. Eur J Hum Genet. 2011;19(1):102–7.
Allou L, Lambert L, Amsallem D, Bieth E, Edery P, Destrée A, Rivier F, Amor D, Thompson E, Nicholl J, Harbord M, Nemos C, Saunier A, Moustaïne A, Vigouroux A, Jonveaux P, Philippe C. 14q12 and severe Rett-like phenotypes: new clinical insights and physical mapping of FOXG1-regulatory elements. Eur J Hum Genet. 2012;20(12):1216–23.
Cuoco C, Gimelli G, Pasquali F, Poloni L, Zuffardi O, Alicata P, Battaglino G, Bernardi F, Cerone R, Cotellessa M, Ghidoni A, Motta S. Duplication of the short arm of chromosome 9. Analysis of five cases. Hum Genet. 1982;61:3–7.
Motegi T, Watanabe K, Nakamura N, Hasegawa T, Yanagawa Y. De novo tandeem duplication 9p(p12→p24) with normal GALT activity in red cells. J Med Genet. 1985;22:64–6.
Tsezou A, Kitsiou S, Galla A, Petersen MB, Karadima G, Syrrou M, Sahlèn S, Blennow E. Molecular cytogenetic characterization and origin of two de novo duplication 9p cases. Am J Med Genet. 2000;91(2):102–6.
Park IY, Kim Y, Lim J, Kim M, Han K, Sung IK, Shin JC. A case of duplication 9p syndrome with a small supernumerary marker derived from chromosome 9. Korean J Genet. 2006;28(4):421–4.
Phelan MC, Stevenson RE, Anderson EV. Recombinant chromosome 9 possibly derived from breakage and Reunion of sister chromatids within a paracentric inversion loop. Am J Med Genet. 1993;46:304–8.
Temtamy SA, Kamel AK, Ismail S, Helmy NA, Aglan MS, El Gammal M, El Ruby M, Mohamed AM. Phenotypic and cytogenetic Spectrum of 9p trisomy. Genet Couns. 2007;18(1):29–48.
Achkar WA, Wafa A, Moassass F, Liehr T. Partial trisomy 9p22 to 9p24.2 in combination with partial monosomy 9pter in a Syrian girl. Mol Cytogenet. 2010;3:18.
Guilherme RS, Meloni VA, Perez AB, Pilla AL, de Ramos MA, Dantas AG, Takeno SS, Kulikowski LD, Melaragno MI. Duplication 9p and their implication to phenotype. BMC Med Genet. 2014;15:142.
Coco R, Penchaszadeh VB. Partial trisomy 14q and familial translocation (2;14) (q12;q13). Ann Genet. 1977;20(1):41–4.
Simpson J, Zellweger H. Partial trisomy 14q -- and parental translocation of no. 14 chromosome. Report of a case and review of the literature. J Med Genet. 1977;14(2):124–7.
Laurent C, Dutrillaux B, Biemont MC, Genoud J, Bethenod M. T(14q- ; 21q+) Translocation in the father. Partial trisomy 14 and monosomy 21 in the daughter. Ann Génét. 1974;16(4):281–4.
Fryns JP, Cassiman JJ, Van den Berghe H. Tertiary partial 14 trisomy 47, XX, plus 14q minus. Humangenetik. 1974;24(1):71–7.
Fried K, Goldberg MD, Rosenblatt M. Proximal 14 trisomy 46,XX,?22+der(14)t(14;22)(q21;q11)mat. Teratology. 1980;21(3):309–12.
Raoul O, Rethoré MO, Dutriliaux B, Michon L, Lejeune J. Partial 14q trisomy. I. Partial 14q trisomy by maternal translocation t(10;14) (p15.2;q22). Ann Génét. 1975;18(1):35–9.
Turleau C, Grouchy J, Bocquentin F, Roubin M, Colin FC. Partial trisomy 14q II.--partial trisomy 14q due to a maternal t(12; 14) (q24.4; q21). Ann Génét. 1975;18(1):41–4.
Allderdice PW, Miller OJ, Miller DA, Breg WR, Gendel E, Zelson C. Familial translocation involving chromosomes 6, 14 and 20, identified by quinacrine fluorescence. Humangenetik. 1971;13(3):205–9.
Muldal S, Enoch BA, Ahmed A, Harris R. Partial trisomy 14q- and pseudoxanthoma elasticum. Clin Genet. 1973;4(6):480–9.
Fawcett WA , Mccord WK , Francke U. Trisomy 14q-. Birth Defects Orig Artic Ser, 1975, 11(5):223–228.
Yeatman GW, Riccardi VM. Partial trisomy of chromosome 14: (+14q−). Birth Defects Orig Artic Ser. 1976;12(5):119–24.
Reiss JA, Wyandt HE, Magenis RE, Lovrien EW, Hecht F. Mosaicism with translocation: autoradiographic and fluorescent studies of an inherited reciprocal translocation t(2q+;14q-). J Med Genet. 1972;9(3):280–6.
Lopez Pajares I, Delicado A, Cobos PV, Lledo G, Peralta A. Partial trisomy 14q. Hum Genet. 1979 Jan 25;46(2):243–7.
Short EM, Solitare GB, Breg WR. A case of partial 14 trisomy 47,XY,(14q-)+ and translocation t(9p+;14q-) in mother and brother. J Med Genet. 1972;9(3):367–73.
Monfort S, Blesa D, Roselló M, Orellana C, Oltra S, Cruz Cigudosa J, Martinez F. Duplication of 14q11.2 associates with short stature and mild mental retardation: a putative relation with quantitative trait loci. Am J Med Genet A. 2007;143A(4):382–4.
Smyk M, Poluha A, Jaszczuk I, Bartniik M, Bernaciak J, Nowakowska B. Novel 14q11.2 microduplication including the CHD8 and SUPT16H genes associated with developmental delay. Am J Med Genet A. 2016;170A(5):1325–9.
Ito M, Mutoh K, Okuno T, et al. De novo tandem duplication of the middle segment of the long arm of chromosome 14. Ann Génét. 1983;26(2):116–9.
Parker GA, Baker RR, Smith VG. The origin and evolution of gamete dimorphism and the male-female phenomenon. J Theor Biol. 1972;36(3):529–53.
Stagi S, Lapi E, Seminara S, Guarducci S, Pantaleo M, Giglio S, Chiarelli F, de Martion M. Long-term auxological and endocrinological evaluation of patients with 9p trisomy: a focus on the growth hormone-insulin-like growth factor-I axis. BMC Endocr Disord. 2014;14:3.
Tohyama J, Yamamoto T, Hosoki K, Nagasaki K, Akasaka N, Ohashi T, Kobayashi Y, Saitoh S. West syndrome associated with mosaic duplication of FOXG1 in a patient with maternal uniparental disomy of chromosome 14. Am J Med Genet A. 2011;155A(10):2584–8.
Martín-De Saro MD, Valdés-Miranda JM, Plaza-Benhumea L, Pérez-Cabrera A, Gonzalez-Huerta LM, Guevara-Yañez R, Cuevas-Covarrubias SA. Characterization of a complex chromosomal rearrangement involving a de novo duplication of 9p and 9q and a deletion of 9q. Cytogenet Genome Res. 2015;147(2–3):124–9.
Martínez-Jacobo L, Ortíz-López R, Rizo-Méndez A, García-Molina V, Santuario-Facio SK, Rivas F, Rojas-Martínez A. Clinical and molecular delineation of duplication 9p24.3q21.11 in a patient with psychotics behavior. Gene. 2015;560:124–7.
Heidemann S, Plendl H, Vater I, et al. Maternal uniparental disomy 15 in a fetus resulting from a balanced familial translocation t(2;15)(p11;q11.2). Prenat Diagn. 2010;30(2):183–5.
Ioannides Y, Lokulo-Sodipe K, Mackay DJ, et al. Temple syndrome: improving the recognition of an underdiagnosed chromosome 14 imprinting disorder: an analysis of 51 published cases. J Med Genet. 2014;51:495–501.
Bertini V, Antonella F, Bruno R, et al. Maternal Uniparental Disomy 14 (Temple syndrome) as a result of a Robertsonian translocation. Mol Syndromol. 2017;8:131–8.
Acknowledgements
We acknowledge and thank all participants for their cooperation and sample contributions.
Funding
The work was supported by the science and technology bureau project of Xuzhou (NO 18182).
Author information
Authors and Affiliations
Contributions
JBW and JFZ conceived of the study, designed the study, JS and YL collected the data. All authors analysed the data and were involved in writing the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by Xuzhou Central Hospital Ethics Committee. The approval number is XZXY-LJ-20161121-021.
Consent for publication
All authors consent for publication. The pregnant woman signed the consent form of BioMed central.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wu, J.B., Sha, J., Zhai, J.F. et al. Prenatal diagnosis of maternal partial trisomy 9p23p24.3 and 14q11.2q21.3 in a fetus: a case report. Mol Cytogenet 13, 6 (2020). https://doi.org/10.1186/s13039-020-0473-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-020-0473-x