Abstract
Background
Deletions involving the long arm of chromosome 18 have been associated with a highly variable phenotypic spectrum that is related to the extent of the deleted region. Duplications in chromosomal region 4p16.3 have also been shown to cause 4p16.3 microduplication syndrome. Most reported patients of trisomy 4p16.3 have more duplications, including the Wolf-Hirschhorn critical region (WHSCR). Here, we present a patient with speech delay and mental retardation caused by a deletion of 18q (18q22.1-qter) and terminal microduplication of 4p (4p16.3-pter) distal to WHSCR.
Case presentation
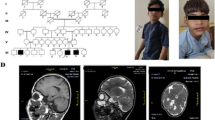
The patient was a 23-month-old boy with moderate growth retardation, severe speech delay, mental retardation, and dysmorphic features. Single nucleotide polymorphism (SNP) array analysis confirmed an 11.2-Mb terminal deletion at 18q22.1 and revealed a 1.8-Mb terminal duplication of 4p16.3. Our patient showed clinical overlap with these two syndromes, although his overall features were milder than what had been previously described. Some dosage-sensitive genes on the 18q terminal deleted region and 4p16.3 duplicated region of the present case may have contributed to his phenotype.
Conclusions
This is the first report of a patient with combined terminal deletion of 18q22.1 and duplication of 4p16.3. In this report, we provide clinical and molecular evidence supporting that the microduplication in 4p16.3, distal to WHSCR, is pathogenic. The coexistence of two chromosome aberrations complicates the clinical picture and creates a chimeric phenotype. This report provides further information on the genotype-phenotype correlation of 18q terminal deletion and 4p microduplication.
Similar content being viewed by others
Background
Deletions of the long arm of chromosome 18 (18q deletion syndrome, OMIM #601808) are common abnormalities involving chromosome 18 [1]. Individuals with a terminal 18q deletion display variable phenotypes, including short stature, microcephaly, characteristic dysmorphic facial feature, cleft lip/palate, delayed myelination, foot deformities, hypotonia, congenital aural atresia (CAA), mental retardation (MR), and genitourinary malformations [2,3,4,5]. Some dosage-sensitive genes and critical regions on 18q that contribute to the clinical features have been identified, thereby providing a foundation for establishing the genotype-phenotype correlation for 18q deletions [1, 6].
Terminal deletions of chromosome 4p cause Wolf-Hirschhorn syndrome (WHS, OMIM #194190). Duplications involving 4p16.3 have also been reported in several individuals, giving rise to a proposed 4p16.3 microduplication syndrome [7, 8]. Larger imbalanced rearrangements on chromosome 4p in the form of deletions and duplications involving the Wolf–Hirschhorn critical region (WHSCR) have defined clinical features, such as developmental delay, delayed psychomotor development, intellectual disability, and craniofacial and skeletal malformations [9,10,11,12]. However, the significance and clinical presentation of patients with microduplication distal to WHSCR are not well understood. To the best of our knowledge, only two patients have been reported to date [8, 13].
Here, we report another case of a 23-month-old boy with a 1.8-Mb duplication at 4p16.3 distal to the WHSCR combined with an 11.2-Mb terminal deletion at the 18q22.1, who presents with similar and different symptoms previously seen in trisomy 4p and 18q deletion syndrome. This case provides further information regarding the clinical features and genotype-phenotype correlation of 18q deletion and 4p16.3 microduplication.
Case presentation
The patient was a 23-month-old boy who was referred for cytogenetic studies because of speech delay and mental retardation. He was born at 38 weeks gestation following an unremarkable pregnancy by Caesarean section. His birth weight was 3.40 kg (<50th centile), and birth length was 52 cm (>75th centile). At birth, he had an umbilical hernia, which healed at 3 months of age.
The patient could sit at 8 months, and took his first steps at 18 months. At 23 months, his height was 90 cm (<75th centile), and his weight was 13 kg (<75th centile). He cannot speak meaningful words and walked with instability and large strides. Medical examination revealed developmental delay, sensory integration dysfunction, moderate MR, and reduced cognitive ability. Additional physical features included hypotonia, a moon face, midface hypoplasia, deep-set eyes, epicanthal folds, a wide nasal bridge, a flat nose, a protrusible mouth, short neck, and a longer fourth toe of the right foot. No significant defects such as cleft lip/palate, ears, heart, lung or genitourinary system were noted.
Materials and methods
Karyotyping
For chromosome analysis, metaphase chromosomes were obtained from peripheral blood lymphocytes after 72 h of incubation and were prepared for GTG banding according to standard protocols.
Fish
According to standard protocols, the cultured blood lymphocytes of the patient were harvested to obtain metaphase chromosomes. FISH analysis was performed with six different chromosome 18 BAC probes; RP11-7H17, 185,880 bp, 18q23 (chr18:77,115,373-77,301,252); RP11-55 N14, 173,736 bp, 18p11.31 (chr18:2,744,753-2,918,488); RP11-53 N15, 678 bp, 18q22.3 (chr18:71,881,306-71,881,983); RP11-90 L7, 165,866 bp, 18q11.2 (chr18:23,355,089-23,520,954); RP11-79A24, 141,894 bp, 18q22.1 (chr18:66,593,997-66,735,890); and RP11-90E1, 182,823 bp, and 18q12.3 (chr18:41,350,363-41,533,185). Images were captured using a fluorescent microscope (Leica DM5000B), and signals were analyzed using Leica CW 4000 software.
SNP array analysis
Infinium OmniZhongHua-8 SNP arrays (Illumina, San Diego, CA, USA) were utilized to analyze genome-wide copy number aberrations. DNA amplification, tagging, and hybridization were processed according to manufacturer’s protocols. Illumina’s iScan system was used to scan the beadchips. Image data was analyzed using Illumina’s GenomeStudio.
Results
The G-banding chromosomal analysis revealed a karyotype of 46, XY, del (18) (q22) (Figs. 1a and b). The karyotypes of his parents are normal. To investigate the deletion region of 18q, fluorescence in situ hybridization (FISH) analysis using chromosome 18 BAC probes was performed. Data showed hybridization signals in the region of 18q22.1 (chr18:66,593,997-66,735,890) (Fig. 1c) and no hybridization signal in the region of 18q23 (chr18:77,115,373-77,301,252) (Fig. 1d) and 18q22.3 (chr18:71,881,306-71,881,983) (Fig. 1e), indicating an 18q22 terminal deletion. To identify the size and position of the chromosomal aberrations, we performed SNP array analysis. The results showed a terminal deletion at 18q22.1 (Fig. 2a) and revealed the presence of a duplication at 4p16.3 (Fig. 2b). The terminal deletion 18q22.1 was approximately 11.2 Mb (66,794,478–78,015,180) in size and the 4p16.3 duplication was approximately 1.8 Mb (71,566–1,883,647) in size (UCSC genome browser, Human Assembly, GRCh37/hg19).
Identification of chromosome 18 terminal deletion. a GTG banded karyotype of the case showing an aberration at distal 18q indicated by an arrow. b G-banding of chromosome 18 showing the deletion of 18q. c BAC-probes RP11-79A24 (orange) (18q22.1) (chr18:66,593,997-66,735,890) and RP11-90E1 (green) (18q12.3) (chr18:41,350,363-41,533,185) showing no deletion at 18q22.1 (chr18:66,593,997-66,735,890). d BAC-probes (green) (18q22.3) (chr18:71,881,306-71,881,983) and RP11-90 L7 (orange) (18q11.2) (chr18:23,355,089-23,520,954) revealing the deletion of 18q22.3. e BAC-probes RP11-7H17 (orange) (18q23) (chr18:77,115,373-77,301,252) and RP11-55 N14 (green) (18p11.31) (chr18:2,744,753-2,918,488) revealing the deletion of 18q23
Discussion
Using conventional karyotyping, FISH, and SNP array analyses, we report a case involving a terminal deletion of 18q22.1 and duplication of 4p16.3. Patients with terminal deletions of the long arm of chromosome 18 or microduplications of the short arm of chromosome 4 display diverse phenotypes. The clinical phenotypes of our patient show similarities and differences from previously reported cases.
The 18q22 deletion syndrome is characterized by mental retardation and development delay, as well as a range of physical anomalies, including microcephaly, short stature, hearing loss, CAA, cleft palate with or without cleft lip (CL/CP), white matter abnormalities of the brain, hand and foot malformations. Several reports have explored the genotype-phenotype correlations of 18q22 deletion syndrome. Recently, Cody et al. [6] viewed more than 350 individuals with 18q22 deletion syndrome and classified 133 of a total of 196 confirmed genes on 18q as dosage-insensitive and 15 (8%) as dosage-sensitive leading to haploinsufficiency, whereas another 10 (5%) have effects that are conditionally haploinsufficient and dependent on another factor, such as genetic or environmental, to cause an abnormal phenotype. Our patient’s deletion region includes 5 of the 15 genes. Those genes are NETO1 (chr18:70,409,549-70,534,810), CYB5A (chr18:71,983,110-72,026,422), TSHZ1 (chr18:72,997,498-73,000,596), MBP (chr18:74,690,789-74,844,774), and NFATC1 (chr18:77,160,326-77,289,323).
Although the terminal deletion involving 18q was approximately 11.2 Mb in size and included five dosage-sensitive genes, our patient’s clinical features were relatively mild and distinct from those described in 18q- syndrome. Abnormal phenotypes in our case were only associated with NETO1 and MBP genes. In terms of NETO1 (chr18:70,409,549-70,534,810), patients who are hemizygous for this gene represent executive function difficulties [14]. Haploinsufficiency of this gene has been associated with impaired spatial learning and memory. The MBP gene lies within the 18q23 CNS dysmyelination critical region (chr18:72,980,819-75,485,284), and hemizygosity of the MBP gene can cause dysmyelination of the brain for people with distal 18q- [14, 15]. Our patient presents MR, walking instability, and large strides, which may be attributed to CNS dysmyelination. However, high-frequency sensorineural hearing loss, which has been associated with the MBP gene, was not observed in our patient. In addition, defects in the TSHZ1 gene would cause cleft soft palate and CAA [16], but our patient does not exhibit both symptoms, although the deletion region of this case included this gene. The other case with a larger deletion of 18q21.3-qter that we previously reported also did not present sensorineural hearing loss, CAA, and CL/CP [17]. Some cases presenting with a mild phenotype suggest that features associated with this deletion is highly heteregeneous. It seems that hemizygosity of some dosage-sensitive genes, such as TSHZ1 and MBP, may not lead to an abnormal phenotype. We infer that variable phenotypes may be attributed to potential gene-gene interactions and gene-environmental interactions.
Interestingly, our patient presents severe speech delay, although the deleted region in 18q does not include the dosage-sensitive gene SETBP1 (chr18:42,260,863-42,648,475), which is associated with severe delay in expressive speech with intact receptive language [6, 18, 19]. It may be due to a 1.8-Mb duplication involving 4p16.3 that was also observed in this case. Two critical regions within 4p16.3-WHSCR1 and WHSCR2 for WHS were identified [20, 21]. Most patients with trisomy 4p16.3 have more duplications, which include WHSCR1 and WHSCR2. In contrast, smaller duplications in 4p16.3 distal to WHSCR1 and WHSCR2 are rare. To the best of our knowledge, only two patients have been reported to have this defect [8, 13]. In addition, the duplication involving chromosome 4p16.3 in our patient does not involve WHSCR2 and the coding region of WHSCR1. His clinical features also overlap with the two previously reported cases, including developmental delay, dysmorphic facial features, and speech and cognitive delay. Clinical features of the three patients are discussed in detail and summarized in Table 1.
Some dosage-sensitive genes, such as LETM1, WHSC1, and WHSC2, can be responsible for the duplication 4p phenotype [22]. However, the phenotypic spectrum of patients without the involvement of the WHS critical region is not well understood [13]. The 1.8-Mb microduplication in the present case includes at least 34 RefSeq genes, and some of the genes in this region may have a role in the 4p trisomy phenotype, which include TACC3, FAM53A, FGFR3, LETM1, SLBP, and CRIPAK. TACC3 (transforming acidic coiled-coil containing protein 3) is involved in microtubule dynamic regulation during cell division. Microtubule dynamics are essential to mitotic spindle assembly for appropriate neurogenesis in the cerebral cortex [23, 24]. Peset and Vernos [25] showed that the overexpression of TACC3 cause defects in chromosome alignment and eventually mitotic arrest. Piekorz et al. [26] reported that TACC3 is a critical component of the centrosome/mitotic spindle apparatus, and its absence triggers p53-mediated apoptosis. Based on this evidence, we hypothesize that the overexpression of TACC3 influences early embryonic neural development by interfering with neuronal apoptosis and/or cell cycle. Another gene, FAM53A, is also known as a candidate gene for neurodevelopmental features because it is highly expressed in the early embryonic central nervous system, suggesting that it plays critical roles in neuronal development [8, 13]. In summary, neurodevelopmental delay, which we have described in this case study and in other patients, [13, 22] may be caused by an increased dosage of the TACC3 and FAM53A genes.
Another key gene of the 4p16.3 duplication may be LETM1. It encodes a leucine zipper EF hand-containing transmembrane protein 1 that is involved in mitochondrial morphology, protein transport, and mitochondrial K+/H+ exchange [27, 28]. It has been suggested that the overexpression of LETM1 can cause seizures [8, 13, 22, 29], but our patient did not exhibit this symptom. No duplication carriers of a three-generation family described by Schönewolf-Greulich’s group [12] experienced seizures either. Data from these patients with 4p16 duplications suggest variable penetrance of epilepsy [8, 13, 22, 29]. It is not clear how duplication of the FGFR3, CRIPAK, and SLBP genes could affect the phenotype expressed in our patient. Evaluation of additional patients with well-characterized 4p16.3 duplication and/or point mutations in this region will be useful to illuminate the role of individual genes in these clinical features.
In addition, severe speech delay is the outstanding clinical symptom of our patient, which has also been observed in the other two individuals. As of writing, the patient is nearly 4 years old. We followed the patient and found that he could not speak complete sentences. Hannes et al. [22] suggested that speech development may be impaired by the aberration involving the WHSCR, whereas the duplicated region of the three individuals does not include WHSCR. Although these evidences suggest there may be a candidate region or genes, such as TACC3, FAM53A, and LETM1, shared by all three patients associated with speech delay on 4p16.3 distal to WHSCR, additional experimental and clinical data are needed to support our hypothesis.
Conclusions
We describe a 23-month-old male with a combined terminal deletion of 18q22.1 and duplication of 4p16.3. Compared to other individuals with 18q22.1 deletions, our patient presents a relatively mild phenotype. The coexistence of two chromosomal rearrangements complicates the clinical symptoms and creates a chimeric disorder marked by characteristics of both chromosomal abnormalities. Additionally, our case report provides clinical and molecular evidence supporting the existence of a novel 4p16.3 microduplication syndrome. The TACC3 and LETM1 genes apparently play a key role in the etiology of the clinical phenotype of 4p16.3 microduplication.
Abbreviations
- FISH:
-
Fluorescence in situ hybridization
- SNP array:
-
Single nucleotide polymorphism array
- WHS:
-
Wolf–Hirschhorn syndrome
- WHSCR:
-
Wolf-Hirschhorn critical region
References
Feenstra I, Vissers LE, Orsel M, et al. Genotype-phenotype mapping of chromosome 18q deletions by high-resolution array CGH: an update of the phenotypic map. Am J Med Genet A. 2007;143(16):1858–67.
Cody JD, Heard PL, Crandall AC, et al. Narrowing critical regions and determining penetrance for selected 18q- phenotypes. Am J Med Genet A. 2009;149(7):1421–30.
Cody JD, Semrud-Clikeman M, Hardies LJ, et al. Growth hormone benefits children with 18q deletions. Am J Med Genet A. 2005;137(1):9–15.
Cody JD, Ghidoni PD, DuPont BR, et al. Congenital anomalies and anthropometry of 42 individuals with deletions of chromosome 18q. Am J Med Genet. 1999;85(5):455–62.
Kulikowski LD, Yoshimoto M, da Silva Bellucco FT, et al. Cytogenetic molecular delineation of a terminal 18q deletion suggesting neo-telomere formation. Eur J Med Genet. 2010;53(6):404–7.
Cody JD, Sebold C, Heard P, et al. Consequences of chromsome18q deletions. Am J Med Genet C Semin Med Genet. 2015;169(3):265–80.
Bi W, Cheung SW, Breman AM, et al. 4p16.3 microdeletions and microduplications detected by chromosomal microarray analysis: new insights into mechanisms and critical regions. Am J Med Genet A. 2016;170(10):2540–50.
Palumbo O, Palumbo P, Ferri E, et al. Report of a patient and further clinical and molecular characterization of interstitial 4p16.3 microduplication. Mol Cytogenet. 2015;8:15.
Dai Y, Yang J, Chen Y, et al. Microarray analysis of unbalanced translocation in wolf-Hirschhorn syndrome. Pediatr Int. 2013;55(3):368–70.
Partington MW, Fagan K, Soubjaki V, et al. Translocations involving 4p16.3 in three families: deletion causing the Pitt-Rogers-Danks syndrome and duplication resulting in a new overgrowth syndrome. J Med Genet. 1997;34(9):719–28.
Patel SV, Dagnew H, Parekh AJ, et al. Clinical manifestations of trisomy 4p syndrome. Eur J Pediatr. 1995;154(6):425–31.
Schonewolf-Greulich B, Ravn K, Hamborg-Petersen B, et al. Segregation of a 4p16.3 duplication with a characteristic appearance, macrocephaly, speech delay and mild intellectual disability in a 3-generation family. Am J Med Genet A. 2013;161(9):2358–62.
Cyr AB, Nimmakayalu M, Longmuir SQ, et al. A novel 4p16.3 microduplication distal to WHSC1 and WHSC2 characterized by oligonucleotide array with new phenotypic features. Am J Med Genet A. 2011;155(9):2224–8.
Miller G, Mowrey PN, Hopper KD, et al. Neurologic manifestation in 18q- syndrome. Am J Med Genet. 1990;37(1):128–32.
Gay CT, Hardies LJ, Rauch RA, et al. Magnetic resonance imaging demonstrates incomplete myelination in 18q- syndrome: evidence for myelin basic protein haploinsufficiency. Am J Med Genet. 1997;74(4):422–31.
Feenstra I, Vissers LE, Pennings RJ, et al. Disruption of teashirt zinc finger homeobox 1 is associated with congenital aural atresia in humans. Am J Hum Genet. 2011;89(6):813–9.
Liao YP, Liang YH, Dong HF, et al. 18q terminal deletion identified by high resolution chromosome and fluorescence in situ hybridization. Fudan Univ J Med Sci. 2010;37(4):479–82.
Filges I, Shimojima K, Okamoto N, et al. Reduced expression by SETBP1 haploinsufficiency causes developmental and expressive language delay indicating a phenotype distinct from Schinzel–Giedion syndrome. J Med Genet. 2011;48(2):117–22.
Marseglia G, Scordo MR, Pescucci C, et al. 372 kb microdeletion in 18q12.3 causing SETBP1 haploinsufficiency associated with mild mental retardation and expressive speech impairment. Eur J Med Genet. 2012;55(3):216–21.
Wright TJ, Ricke DO, Denison K, et al. A transcript map of the newly defined 165 kb wolf-Hirschhorn syndrome critical region. Hum Mol Genet. 1997;6(2):317–24.
Zollino M, Lecce R, Fischetto R, et al. Mapping the wolf-Hirschhorn syndrome phenotype outside the currently accepted WHS critical region and defining a new critical region, WHSCR-2. Am J Hum Genet. 2003;72(3):590–7.
Hannes F, Drozniewska M, Vermeesch JR, et al. Duplication of the wolf-Hirschhorn syndrome critical region causes neurodevelopmental delay. Eur J Med Genet. 2010;53(3):136–40.
Buchman JJ, Tsai LH. Spindle regulation in neural precursors of flies and mammals. Nat Rev Neurosci. 2007;8(2):89–100.
Yang YT, Wang CL, Van Aelst L. DOCK7 interacts with TACC3 to regulate interkinetic nuclear migration and cortical neurogenesis. Nat Neurosci. 2012;15(9):1201–10.
Peset I, Vernos I. The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol. 2008;18(8):379–88.
Piekorz RP, Hoffmeyer A, Duntsch CD, et al. The centrosomal protein TACC3 is essential for hematopoietic stem cell function and genetically interfaces with p53-regulated apoptosis. EMBO J. 2002;21(4):653–64.
Frazier AE, Kiu C, Stojanovski D, et al. Mitochondrial morphology and distribution in mammalian cells. Biol Chem. 2006;387(12):1551–8.
Nowikovsky K, Froschauer EM, Zsurka G, et al. The LETM1/YOL027 gene family encodes a factor of the mitochondrial K+ homeostasis with a potential role in the wolf-Hirschhorn syndrome. J Biol Chem. 2004;279(29):30307–15.
McQuibban AG, Joza N, Megighian A, et al. A Drosophila mutant of LETM1, a candidate gene for seizures in wolf-Hirschhorn syndrome. Hum Mol Genet. 2010;19(6):987–1000.
Acknowledgments
We thank the patient and his family for participating in this study.
Funding
The Natural Science Foundation of the Higher Education Institutions of Anhui Province, China (Nos. KJ2015A263 and KJ2016A473), and The National Undergraduate Innovative Training Program (201510367042) supported this study. These funding agencies had no role in this study.
Availability of data and materials
The datasets supporting the conclusions of this report are included within the article. More details are available upon request.
Author information
Authors and Affiliations
Contributions
LP designed the study. WJ and RH drafted the paper and interpreted the SNP-array data. WQ conducted conventional and molecular cytogenetic analysis. LM and DF performed SNP arrays. All of the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The patient’s parents have given their informed written consent for their son’s participation to this study.
Consent for publication
We obtained written informed consent from the subjects’ parents for publication of this case report and any accompanying images.
Competing interests
The authors have no competing interests to declare.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wang, C., Ren, H., Dong, H. et al. 18q22.1-qter deletion and 4p16.3 microduplication in a boy with speech delay and mental retardation: case report and review of the literature. Mol Cytogenet 11, 55 (2018). https://doi.org/10.1186/s13039-018-0404-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-018-0404-2