Abstract
Cytogenetic analysis of chorionic villous sampling revealed a mosaic karyotype with gain of a rearranged X chromosome. Microarray and additional studies indicated that the rearranged X carried an inverted duplication, a deletion and a satellited Xqter. Gain of this rearranged X was confirmed by follow-up amniocentesis and postnatal cord blood sample. A full-term infant girl was delivered and showed normal physical findings at both birth and 21-month follow-up examinations. Late replication studies demonstrated that the rearranged X was inactivated in all abnormal cells analyzed. Skewed X-inactivation may suppress the potentially deleterious effects of genomic imbalance; however, gain of X chromosomes, particularly rearranged X chromosomes, often presents challenges for prenatal genetic counseling. The gradation of clinical phenotype severity generally correlates with the number of additional X chromosomes. However, the X chromosome regions responsible for the abnormal phenotypes are poorly understood. This case will further elucidate the phenotypic effects of X inactivation and X chromosome abnormalities.
Similar content being viewed by others
Background
Sex chromosome aneuploidies are considered the most common chromosomal abnormalities compatible with live births. A recent report suggested that the prevalence of sex chromosome trisomies is 0.19–5.36 per 1000 in European countries [1].
Sex chromosome abnormalities generally have less deleterious clinical effects as compare to autosomal aberrations. However, selective inactivation of the structurally rearranged X chromosome does not inevitably confer phenotypic normalcy on females. The presence of a complex rearranged third X chromosome often raises concern for the phenotypic significance of genomic dosage alteration and skewed X-inactivation. The wide phenotypic variation and lack of long term follow-up reports in current literature make definitive prognosis for genetic counseling more challenging and problematic. We report a normal female infant after the prenatal diagnosis of an extra satellited X chromosome with trisomy Xp and partial tetrasomy Xq.
Case presentation
A primiparous woman elected for chorionic villous sampling (CVS) with direct Fluorescence in situ hybridization (FISH) testing due to advanced maternal age. Family history was noncontributory. Cytogenetic analysis revealed a mosaic karyotype with gain of a rearranged X; 47,XX,add(X)(q?22)[11]/46,XX[9]. Both parental karyotypes were normal. The rearranged X chromosome was further characterized by array CGH, FISH, Ag-NOR, and X-late replication analysis. Following the diagnosis of an abnormal female karyotype, the pregnancy was monitored by serial ultrasound examinations and fetal echocardiograms, which revealed a normal female fetus. A healthy baby girl was delivered at 39 4/7 week’s gestation. Genetics evaluations at birth and 21 months of age determined that the baby presented with age-appropriate growth and development.
Results
Chromosome analysis
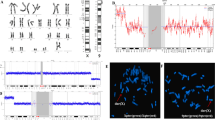
The chorionic villus cell karyotype was 47,XX,add(X)(q?22)[11]/46,XX[9] from four different initial cultures. Both of parental chromosomal studies were normal. Follow-up amniocentesis indicated full aneuploidy with an additional der(X) chromosome in all available 54 colonies; 47,XX,der(X)del(X)(q22.1)dup(X)(q22.1q11.1) (Fig. 1). Prenatal ultrasonography imaging including fetal echocardiography was unremarkable. Subsequent high resolution G-band study of postnatal cord blood showed 92 % of the analyzed cells with an extra satellited chromosome; 47,XX,der(X)del(X)(q22.1) dup(X)(q22.1q11.1) t(X;?)(q11.1;p11.2).
Cytogenomic microarray
Oligonucleotide array CGH of chorionic villus cells showed arr Xp22.33p11.1(2,327,474–57,973,187)x2 ~ 3, Xq11.1q22.1(61,977,455–101,490,234)x3 (Fig. 2), confirming mosaicism of a single copy gain on the Xp arm and possible 2 copies gain of the Xp11.1-q22.1.
Fluorescence in situ hybridization study
Direct FISH analysis of CVS cells revealed a female XXX signal pattern in 37 of 50 interphase nuclei scored; the remaining 13 nuclei showed a normal XX signal pattern. Subtelomere FISH revealed a negative hybridization of the long arm of the der(X) chromosome, indicating deletion of the Xqter region (Fig. 3a). A few cells with higher resolution analysis revealed an appearance of satellites located on the telomeric long arm of der(X). Ag-NOR stain was positive for acrocentric nucleolus organizer regions (Fig. 3b). Both of parental chromosomal studies were normal and showed no evidence of a satellited Xq.
X-inactivation assay
X chromosome late replication pattern of the villi cells using terminal 5- bromodeoxyuridine pulse [2] revealed delayed replication of der(X) and one of the normal X chromosomes (Fig. 4).
Short tandem repeat analysis
Polymorphic short tandem repeat markers, DXS6807, DXS6789, and DXS7133,were selected from ChrX-STR.org2.0 (www.chrx-str.org) to correspond with the genome region in the short arm of X chromosome, and the duplicated and deleted long arm of the der(X) chromosome, respectively. Results of the STR analysis on fetal and parental DNA are summarized as follows: marker DXS7133 on del(Xq) terminus showed one paternal and one maternal allele and indicated biparental inheritance of the normal two X chromosomes (Fig. 5a); marker DXS6807 on Xp demonstrated a 2:1 ratio of maternal allele versus paternal allele (Fig. 5b), indicating the der(X) is of maternal origin; marker DXS6789 detected three distinct alleles within the duplicated X region indicating two maternal X chromosomes of meiotic origin were involved in the rearrangement (Fig. 5c).
a,b,c: Short tandem repeat analysis. a: Marker DXS7133 showed biparental inheritance of the two normal X chromosomes and b: Marker DXS6807 on Xp demonstrated a 2:1 peak ratio corresponding to one maternal allele and one paternal allele and c: Marker DXS6789 (within the duplication region) indicated 2 identical maternal alleles with a 2:1 intensity ratio and one paternal allele
Discussion
The majority of cytogenetically detectable unbalanced rearrangements are associated with significant phenotypic abnormalities. Unbalanced rearrangements with additional sex chromosomes often present challenges for informed genetic counseling. We present a prenatally diagnosed supernumerary X chromosome with complex rearrangements of inverted duplication with deletion and gain of satellites in a phenotypically normal female infant.
Several mechanisms have been reported for the formation of inverted duplication with terminal deletion [3–7]. In this present case, an illegitimate crossing-over event took place between two maternal X homologous chromosomes at Xq22.1. The arising dicentric recombination was a segment of Xpter ->cen ->Xq22.1::Xq22.1 ->cen ->Xpter. The acentric Xq22.1 ->qter segment was lost in the process. At subsequent cell division, a breakage between two centromeres occurred at Xq11.1, resulting in an inverted duplication and an uncapped terminal deletion. A healing of the broken end with acrocentric satellite DNA would presumably follow (Fig. 6).
Proposed mechanism for the origin of the extra rearranged X chromosome. a: Interchromatid exchange in maternal meiosis, b: formation of a dicentric chromosome, loss of the acentric fragments, c: separation of two centromeres at anaphase resulting in breakage near one of the centromeres, d: inv dup(X) with del(X) and satellited Xq. Blue arrows indicate breakpoints
A satellited chromosome is a rare chromosomal anomaly and most reported de novo cases are associated with complex structural rearrangements [8, 9]. Scattered within the genome are repeat sequences with homology to the acrocentric repeat DNA. Homology between the repetitive sequences and spatial proximity in a common nucleolus may favor recombination. Various DNA end repair mechanisms may also play an important role in stabilizing terminal deletions with repetitive DNA-sequence elements [10–14].
In contrast to trisomy X with three structurally normal X chromosomes and random inactivation, the present karyotype has two copies of Xq22-qter, three copies of Xp, four copies of Xq11.1-q22 and skewed X inactivation. Genotype-phenotype correlations in individuals with extra copies of Xq and Xp are variable [15–26]. The presence of a normal phenotype and the identification of non-random X-inactivation of the abnormal X suggest that the inactivation minimizes the deleterious effect of structural and genomic imbalance. However, the presence of normal cells in other tissues is unknown.
There is no literature on the reproductive experience of individuals with the present chromosomal constitution. This karyotype might be associated with an increased risk of offspring with sex chromosome abnormality. Correlation with other clinical findings and characterization of the structural rearrangement at molecular level will assist in understanding the precise impact of rearranged supernumerary X and its role in phenotypic effects.
Conclusions
The clinical significance of gain of an X chromosome with a complex structural rearrangement containing four copies of the proximal Xq11.1-Xq22.1 region has not been previously described. Characterization of the structural rearrangement at molecular level further elucidated the phenotypic effects of X inactivation and X chromosome abnormalities. The identification of non-random X-inactivation of the abnormal X chromosome suggests that the inactivation minimizes the negative effects of the structural and genomic imbalances. The findings from this study demonstrated the importance of additional testing in the cases with unbalanced X chromosome rearrangements in prenatal genetic counseling.
Consent
Written informed consent for this case report was obtained from the patient.
A copy of the written consent is available for review by the Editor-in-Chief of this journal.
References
Boyd PA, Loane M, Garne E, Khoshnood B, Dolk H. EUROCAT working group: Sex chromosome trisomies in Europe: prevalence, prenatal detection and outcome of pregnancy. Eur J Hum Genet. 2011;19(2):231–4.
Barch M, Knutsen T, Spurbeck J. Lymphocyte culture initiation and staining procedures for late replication analysis. In: The AGT Cytogenetics Laboratory Manual. 3rd ed. Philadelphia: Lippincot-Raven; 1997. p. 302–9.
Rowe LR, Lee JY, Rector L, Kaminsky EB, Brothman AR, Martin CL, et al. U-type exchange is the most frequent mechanism for inverted duplication with terminal deletion rearrangements. J Med Genet. 2009;46:694–702.
Ciccone R, Mattina T, Giorda R, Bonaglia MC, Rocchi M, Pramparo T, et al. Inversion polymorphisms and non-contiguous terminal deletions: the cause and the (unpredicted) effect of our genome architecture. J Med Genet. 2006;43(5):e19.
Shimokawa O, Kurosawa K, Ida T, Harada N, Kondoh T, Miyake N, et al. Molecular characterization of inv dup del(8p): analysis of five cases. Am J Med Genet Part A. 2004;128A(2):133–7.
Jenderny J, Poetsch M, Hoeltzenbein M, Friedrich U, Jauch A. Detection of a concomitant distal deletion in an inverted duplication of chromosome 3. Is there an overall mechanism for the origin of such duplications/deficiencies? Eur J Hum Genet. 1998;6(5):439–44.
Genesio R, De Brasi D, Conti A, Borghese A, Di Micco P, Di Costanzo P, et al. Inverted duplication of 15q with terminal deletion in a multiple malformed newborn with intrauterine growth failure and lethal phenotype. Am J Med Genet Part A. 2004;128A(4):422–8.
Rujirabanjerd S, Suwannarat W, Sripo T, Dissaneevate P, Permsirivanich W, Limprasert P. De novo subtelomeric deletion of 15q associated with satellite translocation in a child with developmental delay and severe growth retardation. Am J Med Genet Part A. 2007;143(3):271–6.
Ki A, Rauen KA, Black LD, Kostiner DR, Sandberg PL, Pinkel D, et al. Ring 21 chromosome and a satellited 1p in the same patient: novel origin for an ectopic NOR. Am J Med Genet Part A. 2003;120A(3):365–9.
Ballif BC, Gajecka M, Shaffer LG. Monosomy 1p36 bkpts indicate repetitive DNA sequence elements may be involved in generating and/or stabilizing some terminal deletions. Chromosome Res. 2004;12(2):133–41.
Sarri C, Douzgou S, Gyftodimou Y, Tümer Z, Ravn K, Pasparaki A, et al. Complex distal 10q rearrangement in a girl with mild intellectual disability: follow up of the patient and review of the literature of non-acrocentric satellited chromosomes. Am J Med Genet A. 2011;155A(11):2841–54.
Lundblad V. DNA ends: maintenance of chromosome termini versus repair of double strand breaks. Mutat Res. 2000;451(1–2):227–40.
Bertuch A, Lundblad V. Telomeres and double-strand breaks: Trying to make ends meet. Trends Cell Biol. 1998;8:339–42.
Stahl A, Hartung M, Devictor M, Bergé-Lefranc JL. The association of the nucleolus and the short arm of acrocentric chromosomes with the XY pair in human spermatocytes: it’s possible role in facilitating sex-chromosome acrocentric translocations. Hum Genet. 1984;68(2):173–80.
Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet. 2011;12(6):429–42.
Lee JT. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell. 2000;103(1):17–27.
Schmidt M, Du Sart D, Kalitsis P, Leversha M, Dale S, Sheffield L, et al. Duplications of the X chromosome in males: evidence that most parts of the X chromosome can be active in two copies. Hum Genet. 1991;86(5):519–21.
Pramyothin P, Pithukpakorn M, Arakaki RF. A 47, XXY patient and Xq21.31 duplication with features of Prader-Willi syndrome: results of array-based comparative genomic hybridization. Endocr. 2010;37:379–82.
Calvano S, de Cillis GP, Croce AI, Perla G, Notarangelo A, Zelante L. A complex mosaicism 45, X/46, X, del(Xq)/46, X, idic(Xq) in a patient with secondary amenorrhea. Ann Genet. 2002;45(3):137–40.
Aughton DJ, AlSaadi AA, Johnson JA, Transue DJ, Trock GL. Dir dup(X) (q13-- > qter) in a girl with growth retardation, microcephaly, developmental delay, seizures, and minor anomalies. Am J Med Genet. 1993;46(2):159–64.
Thode A, Partington MW, Yip M-Y, Chapman C, Richardson VF, Turner G. A new syndrome with mental retardation, short stature and an Xq duplication. Am J Med Genet. 1988;30:239–50.
Van Dyke DL, Miller MJ, Weiss L. The origin of inverted tandem duplications, and phenotypic effects of tandem duplication of the X chromosome long arm. Am J Med Genet. 1983;15:441–50.
Vejerslev LO, Iiix M, Jespersen B. Inherited tandem duplication dup(X) (q131-q212) in a male proband. Clin Genet. 1985;27:276–81.
Yokoyama Y, Narahara K, Tsuji K, Moriwake T, Kanzaki S, Murakami M, et al. Growth hormone deficiency and empty sella syndrome in a boy with dup(X) (q13.3--q21.2). Am J Med Genet. 1992;42:660–4.
Bernstein R, Jenkins T, Dawson B, Wagner J, Dewald G, Koo GC, et al. Female phenotype and multiple abnormalities in sibs with a Y chromosome and partial X chromosome duplication: H--Y antigen and Xg blood group findings. J Med Genet. 1980;17(4):291–300.
Barnes IC, Curtis D, Duncan SL. A duplication/deficient X chromosome in a girl with mental retardation and dysmorphic features. J Med Genet. 1988;25(4):264–7.
Acknowledgements
This study was support by funds from the Department of Laboratory Medicine, University of California, San Francisco.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MY drafted and finalized the manuscript. ZQ carried out the STR, BrdU, and acrocentric. p-arm analyses. AK performed G-banding and Ag-NOR staining. JWY initiated the study and proved the final manuscript. All authors read and approved the final manuscript.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Yiu, M., Qi, Z., Ki, A. et al. Trisomy Xp and partial tetrasomy Xq resulting from gain of a rearranged X chromosome in a female fetus: pathogenic or not?. Mol Cytogenet 8, 53 (2015). https://doi.org/10.1186/s13039-015-0160-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-015-0160-5