Abstract
Background
Copy number variants (CNVs) are an important source of normal and pathogenic genome variations. Especially CNVs identified in prenatal cases need careful considerations and correct interpretation if those are harmless or harmful variants from the norm.
Case presentation
Herein, we reported a paternally inherited duplication of 7.6 Mb in 7q31.3 with, surprisingly, a favorable outcome. GTG-banding and CMA on the DNA derived from uncultured amniocytes revealed a karyotype: 46,XX.arr[GRCh37] 7q31.31q31.33(118,601,001_126,177,044) × 3. Ultrasound examination showed no dysmorphisms or intrauterine growth restriction in the fetus and the father was clinically normal as well.
Conclusion
Prenatal detection of a 7.6 Mb in 7q31.31 to 7q31.33 duplication in a female fetus turned out to be a yet unreported unbalanced chromosome abnormality. This is another example that parental testing and GTG-banding are necessary additional tests to be done in prenatal cases, before a reliable conclusion on the meaning of an aberration can be drawn.
Similar content being viewed by others
Background
Copy number variants (CNVs) are an important source of normal and pathogenic genome variations [1]. While many CNVs are known since almost 2 decades for European population, recent widespread application of chromosomal microarray analysis (CMA) in diagnosis in China revealed many new CNVs. Overall, clinical significance of these new CNVs needs to be still some considerations [2, 3].
Talking about prenatally detected CNVs in the range of several mega base pairs (Mb) in size the great majority of those will lead to adverse effects for the carrier. However, rarely there are so-called unbalanced chromosome abnormalities (UBCAs), which, irrespective of their size, do not cause any, or less than expected harm to their carriers [4,5,6,7].
Herein, we reported a family with a 7.6 Mb duplication in7q31.3 with a favorable outcome, being a new example of a UBCA.
Case report
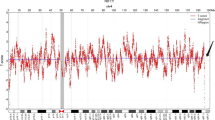
A 36-year-old gravida-1-para-0 pregnant women had amniocentesis at 18 weeks of gestation, due to advanced maternal age. She and her 39 years old husband reported no family history of birth defects or genetic diseases. CMA by Affymetrix CytoScan 750 K chip, which includes 550 k non-polymorphic markers and 200 k SNP markers performed on DNA derived from uncultured amniocytes identified a 7.6 Mb chromosomal duplication (Fig. 1), while GTG-banding of fetus and parents were normal (Fig. 2). The karyotype for the fetus was according to International System of Cytogenomic Nomenclature 2020 (ISCN 2020) [8] 46,XX.arr[GRCh37] 7q31.31q31.33(118,601,001_126,177,044) × 3. Parental CMA showed that the father had a duplication of the same region as the fetus.
Sonography revealed no hints on dysmorphisms or intrauterine growth restriction (IUGR) in the fetus, and a comprehensive physical examination of the parents, especially the father showed no abnormalities. After genetic counseling, the parents decided to continue the pregnancy. At 38 weeks of gestation, a female baby weighing 3,300 g was delivered vaginally. The baby received a complete physical examination, and the results were normal. At the 24-month checkup, the baby was developing normally.
Discussion
According to the literature [6] yet four UBCA regions with duplications are known along chromosome 7: 7pter to 7p22.3, 7p13 to 7p12.2, 7p11.2 to 7q11.22 and 7q32 to 7q36.1. This study adds one more UBCA region as 7q31.31 to 7q31.33.
While in many cases families with UBCA have a degree of phenotypic effects [6], the here reported family belongs to the smaller group of UBCA cases without any clinical signs or symptoms. Also the size of almost 8 Mb is unusually large even for UBCAs [6]. As being also typical for UBCAs there are also reports on similar duplications which show—in parts severe clinical consequences [9,10,11], like facial dysmorphism, moderate intellectual disability, autistic spectrum disorder, and epilepsy [11].
In the present case the duplicated region contains multiple genes: KCND2, TSPAN12, ING3, WNT16, FAM3C, PTPRZ1, AASS, FEZF1, CADPS2, TAS2R16, SLC13A1, NDUFA5, LMOD2, WASL, HYAL4, SPAM1, GPR37, POT1, and GRM8. Among these genes, CADPS2, WNT16, and GRM8 play essential roles in autism spectrum disorders. Besides, TSPAN12, ING3, FAM3C, PTPRZ1, FEZF1, WASL, HYAL4, GPR37, and POT1 are cancer-related genes [12, 13]. Still, yet no triplosensitivity genes in this region have been identified. The latter may be the reason for symptom freeness of the father and his daughter in the reported case.
Conclusion
Herein the first case of a (sub)chromosomal imbalance expressed as duplication in 7q31.31 to 7q31.33 is presented, which is per definition an UBCA without obvious clinical consequences for two carriers within the same family. The case highlights that prenatal detection of even large CNVs implicates parental testing to come to a well-funded estimation on the impact of the identified alteration.
Availability of data and materials
Please contact the corresponding author for data requests.
Abbreviations
- CNVs:
-
Copy number variants
- CMA:
-
Chromosomal microarray analysis
- UBCA:
-
Unbalanced chromosome abnormality
References
Sharp AJ. Emerging themes and new challenges in defining the role of structural variation in human disease. Hum Mutat. 2009;30:135–44.
Chen CP, Chern SR, Chen YN, Wu PS, Yang CW, Chen LF, Wang W. Mosaic trisomy 15 at amniocentesis: Prenatal diagnosis, molecular genetic analysis and literature review. Taiwan J Obstet Gynecol. 2015;54:426–31.
Yuan C, Lu Z, Guo T, Yue Y, Wang X, Wang T, Zhang Y, Hou F, Niu C, Sun X, Zhao H, Zhu S, Liu J, Yang B. A global analysis of CNVs in Chinese indigenous fine-wool sheep populations using whole-genome resequencing. BMC Genomics. 2021;22:78.
Barber JC. Directly transmitted unbalanced chromosome abnormalities and euchromatic variants. J Med Genet. 2005;42:609–29.
Barber J. Chromosome Anomaly Collection: http://www.ngrl.org.uk/Wessex/collection/ (accessed on 03.03.2022).
Liehr T. Cases with heteromorphisms. http://cs-tl.de/DB/CA/HCM/0-Start.html (accessed on 03.03.2022).
Liehr T. Benign & Pathological Chromosomal Imbalances, 1st Edition microscopic and submicroscopic copy number variations (CNVs) in genetics and counseling. Switzerland: Academic Press; 2014.
McGowan-Jordan J, Hastings RJ, Moore S. International system of cytogenomic nomenclature (ISCN 2020). Switzerland: Karger; 2020.
Liehr T, Schreyer I, Kuechler A, Manolakos E, Singer S, Dufke A, Wilhelm K, Jančušková T, Čmejla R, Othman MAK, Al-Rikabi AH, Mrasek K, Ziegler M, Kankel S, Kreskowski K, Weise A. Parental origin of deletions and duplications—about the necessity to check for cryptic inversions. Mol Cytogenet. 2018;9:11–20.
Leach NT, Chudoba I, Stewart TV, Holmes LB, Weremowicz S. Maternally inherited duplication of chromosome 7, dup(7)(p11.2p12), associated with mild cognitive deficit without features of Silver-Russell syndrome. Am J Med Genet A. 2007;143A:1489–93.
Pavone P, Corsello G, Marino SD, Martino R. Raffaele F. 7q31.32 partial duplication: First report of a child with dysmorphism, autistic spectrum disorder, moderate intellectual disability and epilepsy. Literature review. Epilepsy Res. 2019;158:223–8.
Tetsushi S, Yo S, Akira S, Hirotoshi I, Chiaki I, Makoto M, Ryosuke Y, Teiichi F. Mouse models of mutations and variations in autism spectrum disorder-associated genes: Mice expressing Caps2/Cadps2 copy number and alternative splicing variants. Int J Environ Res Public Health. 2013;10:6335–53.
Noriko S, Keiko S, Yuya T, Tsukasa O, Jun T, Toshiyuki Y. A 7q31.33q32.1 microdeletion including LRRC4 and GRM8 is associated with severe intellectual disability and characteristics of autism. Human Genome Variat. 2017;4:1–3.
Acknowledgements
The authors appreciate the patient and his parents for this study.
Funding
There was no funding available for this study.
Author information
Authors and Affiliations
Contributions
Yuexiang Feng conceived and designed the experiments. Huili Luo performed the experimental work. Linlin Liu analysed the data. Yuexiang Feng and Huili Luo contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Wuhan Hankou Hospital. The guardians of the patient gave informed consent for the study.
Consent for publication
All patient guardians gave informed consent to the publication of this manuscript.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Luo, H., Liu, L. & Feng, Y. Prenatal diagnosis of a novel 7q31.31q31.33 microduplication with a favorable outcome. Mol Cytogenet 15, 13 (2022). https://doi.org/10.1186/s13039-022-00589-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-022-00589-y