Abstract
Background
Nonrandom clonal chromosomal aberrations can be detected in approximately 55% of adult patients with acute myeloid leukemia (AML). Recurrent cytogenetic abnormalities play an important role in diagnosis, classification and prognosis of AML. However, several chromosomal abnormalities have not been completely determined or characterized, primarily because of their low incidence and limited amount of data.
Results
We characterized an AML patient with a novel apparently balanced insertion ins(18;5)(q21;q31.2q35.1) that was cryptic by G-banding. The rearrangement was further examined by molecular cytogenetic methods and oligobased high-resolution array CGH (oaCGH) analysis. We show that an approximately 31.8 Mb large segment from chromosome 5 bands q31.2 to q35.1 has been inserted, by a direct mechanism, into chromosome 18 between bands q12.3 and q21.1. The insertion was unbalanced with concurrent submicroscopic deletions at 5q31.2 (approximately 0.37 Mb in size), 5q35.1q35.2 (approximately 1.98 Mb in size), and 18q12.3q21.1 (approximately 2.07 Mb in size). The microdeletions affect genes on 5q and 18q that have been associated with hematological malignancy and other cancers. A novel juxtaposition of the genes NPM1 and HAUS1 at 5q35.1 and 18q21.1, respectively, was detected by FISH analysis. Searching the literature and the Mitelman database revealed no previously reported ins(18;5) cases. Interestingly, however, two AML patients with translocation t(5;18)(q35;q21) encompassing the 5q35 and 18q21 breakpoint regions as detected in our present ins(18;5) patient have been reported.
Conclusions
It is well-known that cytogenetic abnormalities on the long arm of chromosome 5 affect hematopoiesis. However, the precise mechanism of their involvement in myeloid transformation is elusive. Our present data shed new light onto the frequent abnormalities on 5q as well as to the less frequent abnormalities observed on 18q in myeloid malignancies. In addition, we show that oaCGH analysis is a useful adjunct to revealing submicroscopic aberrations in regions of clinical importance. Reporting rare and nonrandom chromosomal abnormalities contribute to the identification of the whole spectrum of cytogenetic abnormalities in AML and their prognostic significance.
Similar content being viewed by others
Background
In acute myeloid leukemia (AML) recurrent nonrandom chromosomal aberrations occur in approximately 55% of the patients. Until now about one hundred different chromosomal rearrangements have been uncovered in AML. The rearrangements mostly include balanced translocations, inversions, deletions, amplifications, monosomies and trisomies [1]. It is well established that cytogenetic analysis is an important prognostic factor that influences therapeutic decision-making and disease outcome because the various chromosomal rearrangements play critical roles in the molecular pathogenesis [2]-[4].
Myeloid malignancies are subdivided into distinct disease entities on the basis of specific cytogenetic or molecular genetic abnormalities [5]. Cytogenetic characterization defines three different risk groups: favorable, intermediate, and adverse [6]. Molecular characterization has revealed that mutations in FLT3 and NPM1 define molecular subgroups with prognostic relevance [7]. AML patients that do not fulfill WHO criteria for other categories are grouped together in the "AML, not otherwise specified (NOS)" category, which do not provide prognostic information. AML is a heterogeneous disease with respect to clinical and biological features. Hence, it is very important to better define less frequent chromosomal rearrangements in AML patients to identify the full spectrum of molecular prognostic factors.
Here we report the characterization of a novel cryptic insertion ins(18;5)(q21.1;q31.2q35.1) in a patient with de novo AML, who, as detected by oligobased high-resolution array CGH (oaCGH) analysis, also harbored three concurrent submicroscopic microdeletions 5q31.2, 5q35.1q35.2, and 18q12.3q21.1 in his leukemic cells. Two previous AML patients with the translocation t(5;18)(q35;q21), and similar breakpoints as observed in our patient, have been reported. We review these patients and discuss the possibility that the ins(18;5) detected in our present patient is a variant of this rare non-random chromosomal abnormality t(5;18).
Case presentation
A 37-year-old male Caucasian, previously well, presented with 4-5 weeks of fatigue, increasing paleness and dyspnea. In this period and on admission there were no febrilia, infections, or signs of bleeding except for one occasion of melaena 3 weeks prior to admission. He had an unintended weight loss of five kg from 91 kg. Bone marrow (BM) examination showed marked hypercellularity with medium-sized mononuclear blasts and an 80% proportion of highly proliferative blasts, staining CD4+, CD7+, CD13+, CD43+, CD117+, CD123+, CD34-, HLA-DR+, CD56-, and TdT-. Hematological examination included a total white blood cell count of 4.49 × 109/L, hemoglobin of 5.1 mmol/L and, platelets of 24 × 109/L. Segmented neutrophil count was 0.70 × 109/L. The patient's father's cousin and great grandmother in his mother's line had leukemia. The patient had no comorbidity and had no previous history of being treated with chemotherapy or exposed to radiation. He had been smoking until 3 years prior to his AML diagnosis with an estimated pack years of 15. There was no information on possible occupational hazards.
Our patient entered the AML-17 treatment protocol (Trial reference ISRCTN55675535). This protocol is a randomized multi-arm Phase III study designed by the AML Working Group of the National Cancer Research Institute (NCRI) and the Hematology Oncology Study Group in Acute Myeloid Leukemia and high risk Myelodysplastic Syndrome (MDS) in adults (http://aml17.cardiff.ac.uk/). In this interventional treatment protocol, AML and high risk MDS patients are randomized to one of five subgroups for induction therapy, then risk assessed, and randomized to FLT3 inhibitor if mutated or high risk chemotherapy with or without mTOR inhibition. According to the protocol our patient was initially treated with DA because of intermediate-risk cytogenetics. Molecular genetic analysis of his bone marrow cells at diagnosis showed an internal tandem duplication mutation in FLT3 (FLT3-ITD) and NPM1 wt, and was then assigned to high-risk leukemia. He received FLAG-IDA treatment according to AML-17 and obtained complete remission 28 days after admission as evaluated by pathology, flowcytometry, cytogenetics and molecular genetics.
Results
Cytogenetic and multicolor FISH analyses
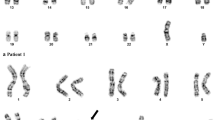
Karyotyping by G-banding of unstimulated cultured BM cells at initial diagnosis was interpreted as an apparently unbalanced male karyotype 46,XY,del(5)(q31q35),add(18)(q23)[25] (Figure 1A). To further characterize these chromosomal aberrations we next performed 24-color karyotyping using 24XCyte human multicolor FISH (mFISH) probe kit which revealed the insertion ins(18;5) and that there were no other structural abnormalities (Figure 1B). To further define the chromosome 5 segment that was cut out and where it was inserted into chromosome 18 we performed mBanding with XCyte probes for chromosomes 5 and 18 (Figure 2). These analyses showed that the chromosome segment 5q31q35 was inserted into chromosome 18 at band region q21 by a direct mechanism. By combining the obtained results a revised karyotype 46,XY,ins(18;5)(q21;q31q35)[25] could be made. Analysis of PHA-stimulated cultures of blood lymphocytes revealed a normal male karyotype, as did analysis of bone marrow cells after one induction series (data not shown). These data exclude the possibility that the observed ins(18;5) in the patient's bone marrow cells at diagnosis was constitutional.
mBanding analyses of chromosomes 5 and 18. The single color gallery tool in the ISIS software shows assigned false colors (FC) representing individual color schemes of labeled chromosomes arranged in its capture sequence (fluorescein isothiocyanate) FITC, (spectrum orange) SpO, (texas red) TR, (cyanine 5) Cy5, (7-diethylaminocoumarin-3-carboxylic acid, succinimidyl ester) DEAC, together with an inverted gray scale image of the DAPI image (Inv). Panel A. mBanding analysis using the XCyte 5 probe. The top panel shows a normal chromosome 5, the middle panel shows the deleted chromosome 5, and the lower panel shows the 5q-insertion on der(18)ins(18;5). To the right is a schematic representation of the localization of the different multicolor probes of XCyte 5 relative to the ideogram of chromosome 5 together with breakpoints marked by black horizontal lines. Panel B. mBanding analysis using the XCyte 18 probe. The top panel shows a normal chromosome 18 and the lower panel the der(18)ins(18;5). To the right is a schematic representation of the localization of the different multicolor probes of XCyte 18 relative to the ideogram of chromosome 18 together with the breakpoint marked by a black horizontal line.
oaCGH analysis
To search for possible copy number abnormalities involved in the ins(18;5) rearrangement we performed oaCGH analysis using the CytoChip Cancer 4×180K v2.0 (BlueGnome, Cambridge, UK). The oaCGH analysis detected four somatic copy number alterations in the form of three microdeletions at 5q31.2, 5q35.1-q35.2 and 18q12.3-q21.1 (Figure 3), and a single microamplification at 12q21.1. The microamplification had a maximal size of 158.3 kb (pos. 72,596,354-72,754,669) (Max: A_16_P19594168: 72,596,354 to A_16_P19594509: 72,754,669) and a minimum size of 107.0 kb (Min: A_16_P02650455: 72,625,008 to A_16_P19594474: 72,732,027) but this region contains no known genes (data not shown).
Genome analysis using high resolution 180 K oligo-based array CGH analysis. Panel A, upper panel. Chromosome 5 ideogram showing two submicroscopic deletions detected at the long arm of chromosome 5 at 5q31.2 and 5q35.1-q35.2. Lower panel, zoom view of genomic profile at chromosome 5 where the deleted regions on 5q are indicated by red shade. Panel B, upper panel. Chromosome 18 ideogram showing a submicroscopic deletion detected at the long arm of chromosome 18 at 18q12.3-q21.1. Lower panel, zoom view of genomic profile at chromosome 18 where the deleted region on 18q is indicated by red shade. Vertical blue lines in the zoom view indicate log2 ratios +0.24 and +0.60 and red lines indicate log2 ratios +0.24 and +1.0. The X-axis at the bottom indicates chromosomal position. The insert at the top of the genome profile indicates the chromosomal bands in the zoomed region and their relative position. The relative positions of the different FISH probes used for validation are indicated in different colors according to the direct fluorescent label used. The genes located in the minimum deleted regions are listed according to their relative genomic position (centromeric to telomeric orientation), and the asterisk (*) denotes that the genes separated by a slash represent different transcripts from the same transcriptional unit.
The maximum regions of microdeletions involved in the other break point regions are: 1) at chromosome band 5q31.2 the microdeletion encompasses the oligonucleotide probes A_16_P37384388 to A_16_P17320593 mapping from 138,390,821 to 138,769,054; 2) at chromosome bands 5q35.1-q35.2 the microdeletion encompasses the oligonucleotide probes A_16_P37464643 to A_16_P17402538 mapping from 170,768,753 to 172,758,763; and 3) at chromosome bands 18q12.3-q21.1 the microdeletion encompasses the oligonucleotide probes A_16_P41020231 to A_16_P03359511 mapping from 39,887,338 to 41,970,952. The minimum region of microdeletions in the involved break point regions are: 1) at chromosome band 5q31.2 the microdeletion encompasses the oligonucleotide probes A_16_P17319737 to A_16_P17320525 mapping from 138,403,448 to 138,746,932; 2) at chromosome bands 5q35.1-q35.2 the microdeletion encompasses the oligonucleotide probes A_18_P15493901 to A_16_P01397609 mapping from 170,771,533 to 172,741,295; and 3) at chromosome bands 18q12.2-q21.1 the microdeletion encompasses the oligonucleotide probes A_16_P20842291 to A_16_P20847704 mapping from 39,899,537 to 41,952,135. From these results the respective estimated minimum to maximum deletion sizes are: 1) at 5q31.2: 343.5-378.2 kb; 2) at 5q35.1-q35.2: 1,969.8-1,990.0 kb; and 3) 18q12.3-q21.1: 2,052.6-2,083.6 kb. The genes located in the minimal deleted regions are summarized in Figure 3.
FISH analyses
To validate the microdeletions, FISH analyses were performed using several BAC-based custom made probes. These were co-hybridized with subtelomeric probes from 5qter and 18qter, and compared to dual color whole chromosome painting with probes for chromosomes 5 and 18 and FISH analysis with the EGR1(5q31)/D5S23,D5S721(5p15.2) dual color probe (Figure 4A). The experiments showed that: 1) the EGR1 gene is not part of the proximal microdeletion as expected from the oaCGH analysis; 2) the BAC-based probes RP11-118P24 (5q31.2), RP11-145P20 (5q35.1) and RP11-9H20 (18q12.3) all showed mono-allelic deletions confirming the microdeletions as suggested by the oaCGH analysis; and 3) the microdeletions on 5q are located on the same short derivative homologue of chromosome 5. Counting 200 interphase nuclei using each of the BAC-probes showed that approximately 90% of the interphase nuclei contained the microdeletions. Using the BAC-probe RP11-184C9 (5q35.1) it was confirmed that it is not part of the deleted region, as expected from the oaCGH analysis, but was part of the 5q fragment that was inserted onto chromosome 18. Analyzing 200 interphase nuclei with this probe a normal signal pattern of 2R2G was observed in all of the examined cells, confirming that this probe is not part of the deleted region.
FISH analyses for validation of array findings and a model for the generation of ins(18;5)(q21.1;q31.2q35.1). Panel A. Partial karyograms of chromosome pairs 5 (upper row) and 18 (lower row) showing FISH results after hybridization using the respective probes as indicated at the top. The aberrant chromosomes are positioned to the left. The relative positions of the RP11-based BAC probes are indicated in Figure 3. For the probes RP11-117 L6 and RP11-474O19 their relative positions are indicated in Panel D by red and green boxes, respectively. Panel B. Model of the chromosomal rearrangement showing the localization of the breakpoints on the ideograms of chromosomes 5 and 18. Panel C. Schematic representation indicating the regions that are deleted, joined and inserted. Panel D. Schematic representation of genes (light brown boxes) mapping in correspondence to the breakpoint regions and each gene are indicated with respect to it genomic orientation by (+) or (-). Upper panel shows the joined region of 5q31.2 and 5q35.2 and the lower panel shows part of the directly inserted 5q31.2 to 5q35.1 fragment into 18q12.3 and 18q21.1, respectively. The deleted chromosomal fragments are omitted and the genes located in these regions are listed in Figure 3. The axis at the bottom of each panel indicates the chromosomal position of the involved regions. The resolution of the array is limited to the kilobase pair level and the density of the oligo probes differ according to chromosomal regions with the highest density at known cancer genes. Vertical red and green bars indicate the relative genomic position of deleted (red) and not deleted (green) oligonucleotide probes in oaCGH analysis. The asterisk (*) marks three minor genes in the following order MZB1(-), PROB1(-) and SPATA24(-).
Taken together we have shown that an approximately 31.8 Mb large chromosomal segment encompassing the bands 5q31.2q35.2 was cut out and inserted by a direct mechanism between chromosome bands q12.3 and q21 on chromosome 18 (Figure 4B and C). The insertion was unbalanced with concurrent submicroscopic deletions at 5q31.2 (approximately 0.37 Mb in size), 5q35.1q35.2 (approximately 1.98 Mb in size), and 18q12.3q21.1 (approximately 2.07 Mb in size). As a result of this complex rearrangement the following band regions became juxtaposed: 5q31.2-5q35.2, 18q12.3-5q31.2, and 5q35.1-18q21.1. By in silico analysis of the involved regions it was found that the 5'-part of SIL1 (spanning exons 1-9) at 5q31.2 and the 5'-part of HAUS1 (spanning exons 1-4) were deleted. A fusion of the chromosomal regions 5q35.1 and 18q21.1 spanning the NPM1 and HAUS1 genes, respectively, was confirmed by FISH analysis with the RP11-117L6 and RP11-474O19 probes (Figure 4D).
Discussion
The insertion ins(18;5)(q21.1;q31.2q35.1) detected in the leukemic cells of the presented de novo AML patient is to the best of our knowledge a novel chromosomal abnormality. A systematic review of the literature and a search in the Mitelman database [1] did not reveal any previous reports on ins(18;5) patients with hematological or other cancers.
Insertions are very rare chromosomal abnormalities, not only in hematological malignancies but also in constitutional genetics where the incidence was estimated to be as low as 1:80,000 [8]. Despite their rarity, several instances of insertion variants of the traditional common translocations have been reported in myeloid malignancy, including ins(8;21)/ins(21;8) and t(8;21)(q22;q22)/RUNX1-RUNXT1[9], ins(3;5) and t(3;5)(q25;q35)/NPM1-MLF1[10], and ins(22;9) and t(9;22)(p24;q11.2)/BCR-JAK2[11]. Although the molecular mechanisms for generating the insertions variants are different compared to those of generating the traditional common translocations the insertion variants have similar aberrant fusion genes. Further, the AML patients with the insertion variants exhibit similar morphology and prognosis compared to their traditional translocation counterparts.
These observations prompted us to speculate whether the rearrangement observed in our patient could be a variant of a putative t(5;18). Interestingly, a revised search revealed two previously reported AML patients with translocation t(5;18)(q35;q21) [12],[13] involving the same cytogenetic bands as in our patient. The clinical and genetic findings are summarized in Table 1. No additional cases could be found after a search in our cytogenetic registry containing more than 2,400 sequential entries of AML since 1990. Although all three patients share similar cytogenetic break points, the rearrangements may still be very different at the molecular level. Since there are no information regarding molecular breakpoint mapping or studies of possible concurrent submicroscopic aberrations in the reported t(5;18) patients this question cannot be addressed. From a clinical point of view it was remarkable that both patients were AML FAB subtype M2 and considered high-risk patients. For patient 2 this was because of an accompanying FLT3-ITD mutation while the basis for this assignment in patient 1 was cryptic. Our patient had AML with FAB subtype M1 and considered high-risk because he had an accompanying FLT3-ITD mutation. Patient 1 had bone marrow transplantation (BMT) in first remission. A BMT in CR1 is planned in our patient but this has been postponed because of complicating fungal infections.
Together these observations suggest that t(5;18), and perhaps our possible variant ins(18;5), may be associated with a high risk AML FLT3-ITD subgroup although the contribution of each genotypic component is unknown. It is well known, however, that AML patients with a normal karyotype and a high burden of FLT3-ITD often present with a more aggressive disease; and more often relapse after remission [14]-[16]. The impact of FLT3-ITD among other cytogenetic subgroups is not clear [15],[17].
With the aid of oaCGH analysis we detected concurrent submicroscopic deletions at each of the cytogenetic breakpoints involved in the complex rearrangement, including 5q31.2, 5q35.1q35.2 and 18q12.3q21.1. Submicroscopic deletions surrounding the most common recurrent translocations breakpoints have been reported in various leukemia with incidences ranging from approximately 2% to 20%, including t(8;21)(q22;q22), t(9;22)(q34;q11), and t(15;17)(q22;q22) [18]-[21]. The clinical significance of these accompanying submicroscopic deletions is largely unknown because they in some cases were associated with poor therapy response and unfavorable outcome while they in others had no effect. It was hypothesized that deletion of critical genes could account for the possible difference in disease course, or alternatively, that such deletions reflect an underlying genomic instability that may predispose the malignant cells to acquire other genetic abnormalities that confer a worse prognosis.
Simple reciprocal translocations between two non-homologous chromosomes fundamentally require only two chromosomal double stranded DNA breaks (DSBs) followed by an exchange of the resulting fragments before sealing of the breaks. A simple insertion requires three chromosomal DSBs, transfer of the segment and then sealing of the three breaks. When a simple insertion rearrangement is complicated by accompanying submicroscopic deletions in the kilobase-to-megabase size at each of the breakpoints, as described in our patient (Figure 4B and 4C), at least six DSBs are required for the complex rearrangement to occur. The additional submicroscopic deletions could either be a by-product of the chromosomal repair mechanism or part of an initiating event. The major DSB repair pathways in mammalian cells are the homologous recombination (HR) and non-homologous end-joining (NHEJ) pathways, and depending upon the chosen repair pathway aberrant chromosomal rearrangements can be generated [22]. In our patient it is likely that the error-prone NHEJ pathway repaired the respective breakpoints.
Translocations involving chromosome 5q35 are rare clonal abnormalities in hematological cancers [23]. The most common recurrent 5q35 translocations with formation of aberrant fusion genes are: t(2;5)(p23;q35)/NPM1-ALK in anaplastic large cell lymphoma [24], t(3;5)(q25;q35)/NPM1-MLF1 in AML [10], t(5;17)(q35;q21)/NPM1-RARA in APL [25], in t(5;11)(q35;p15)/NSD1-NUP98 in childhood AML [26], and t(5;11)(q35;q12)/NSD1-FEN1 in AML-M5 [27]. The NPM1 and NSD1 at 5q35 are common translocation partners. The NSD1 gene encodes a nuclear protein involved in transcriptional regulation. No prognostic mutations have been ascribed to this gene in relation to leukemia. The NPM1 gene encodes a nuclear matrix phosphoprotein involved in nucleolar ribosome assembly and protein localization. In addition to being a translocation partner NPM1 can be affected by mutations at the DNA sequence level. A 4 bp insertion in exon 12 in NPM1 is one of the most frequent genetic changes known in AML patients with a normal karyotype AML; and its presence in those patients confers a better prognosis [28]. In our patient we detected no genetic abnormalities in NPM1 except for its juxtaposition to HAUS1 at 18q21.1. In the previously reported AML patients with t(5;18)(q35;q21) there is no information about possible aberrant fusion genes or concurrent additional abnormalities involving the NPM1 or NSD1 at 5q35.
Chromosomal abnormalities of the long arm of chromosome 18 are most often associated with lymphoid malignancies. However, the number of reports of genes on chromosome 18 involved in myeloid malignancy is increasing. The SETBP1 and its intronic MIR4319 at 18q12.3 were recently described to be new players in myeloid malignancy [29],[30]. SETBP1 was shown to be overexpressed in secondary AML patients bearing the t(12;18)(p13;q12) while the intronic MIR4319 was downregulated [30]. Although the function of SETBP1 is unknown it has been implicated as a transcriptional regulator of many genes. Recurrent somatic mutations promote leukemic cell proliferation [29] and appear to be a poor prognostic marker especially in elderly AML patients [31]. In our AML patient we found that eleven RefSeq at 18q12.3q21.1 were completely or partially deleted (Figure 3), and that the SETBP1 and MIR4319 genes were two of the deleted genes. In addition, we found that the 5'-part of HAUS1 at 18q21.1 was partially deleted and that it has become juxtaposed to NPM1 as a result of the complex insertion-deletion rearrangement. However, with the methods we used we cannot establish whether the NPM1 and HAUS1 genes formed an aberrant fusion gene. HAUS1 encodes one of eight subunits of the 390 kDa human augmin complex that is a microtubule-binding complex vital for mitotic spindle assembly [32]. There are no previous reports on chromosomal rearrangements involving this gene.
Monosomy 5 and interstitial deletions of 5q are common chromosomal abnormalities in myeloid malignancy. These aberrations occur in 5-10% of karyotypic abnormal adult AML and are usually associated with complex karyotypes, rapid disease progression and poor outcome [24],[33]. In our patient we detected two interstitial microdeletions at 5q, one at 5q31.2 (between 343,5-378,2 kb in size) and another at 5q35.1-q35.2 (between 1.969,8-1.990,0 kb in size). The minor 5q31.2 deletion overlaps with the centromeric commonly deleted region (CDR) of two previously identified CDRs in myeloid malignancies [33]-[35]. Of the CDRs the centromeric CDR at 5q31.1-q32.2 is common in high risk MDS and in AML, while the telomeric CDR at 5q33.1 is associated with 5q-syndrom. The identification of pathogenic genes on 5q has proven to be challenging because most patients have extensive deletions encompassing both CDRs [36]. In a large SNP-based study it was found that among 1,115 examined patients with myeloid malignancies 12% had 5q deletions with a median size of 71,4 Mb ranging from 1.9 Mb to 131.28 Mb (whole arm) [37].
Since no single gene on 5q has been proven to be responsible for high risk myeloid malignancies a haploinsufficiency model has been proposed, reviewed in [33]. According to this model, loss of a single allele of more than one gene on 5q may act in concert to alter hematopoiesis, promote self-renewal of hematopoietic stem and progenitor cells (HSPCs), induce apoptosis of hematopoietic cells, and disrupt differentiation [38],[39]. A number of candidate tumor-suppressor genes located at the centromeric CDR at 5q31 (including CDC25C, EGR1, HSPA9, CTNNA1, and DIAPH1) have been implicated in the development of high risk MDS/AML. In our patient we found that six and seventeen RefSeq genes at 5q31.2 and 5q35.1q35.2, respectively, were completely or partially deleted as illustrated in Figures 3 and 4D. It is noticed that the alpha-1 E-catenin gene, CTNNA1, at 5q31.2 is not involved in copy number alterations while the 5'-part of the downstream neighbor gene SIL1 is deleted. The CTNNA1 gene is a tumor suppressor gene that has been associated with progression and poor prognosis in leukemia [40]. The SIL1 gene has not previously been reported to be directly involved in myeloid malignancy although it often is one of the many genes that are deleted in high risk AML/MDS patients with del(5q). It encodes a nucleotide exchange factor that is important for the function of glucose-regulated protein 78 (GRP78). GRP78 is known as a stress-inducible endoplasmic reticulum (ER) chaperone protein and serves as a master initiator of ER stress signaling [41]. Accumulation of unfolded proteins results in GRP78 activation via SIL1 and subsequently activation of the unfolded protein response (UPR). Proteasome inhibitors, such as bortezomib, suppress the degradation of unfolded proteins and trigger ER stress leading to activation of UPR and subsequently apoptotic signals. Although bortezomib is mainly used for treatment of multiple myeloma [42] and mantle cell lymphoma [43] it has in some instances been shown to be an effective agent for treatment of 5q- MDS [44]. The combination of genes that are completely or partially deleted in myeloid malignancies with interstitial deletions of 5q might contribute to the heterogeneity of high risk AML/MDS patients.
Application of array-based CGH analysis has not only significantly improved the detection rate of chromosome aberrations in patients with hematological malignancy compared to traditional cytogenetics [45]-[47] but also uncovered concurrent microdeletions in patients with apparently balanced translocations [21],[45]. The present study, add to the knowledge of chromosomal aberrations and indicate that oaCGH is a useful adjunct to revealing submicroscopic aberrations in genomic regions of clinical importance.
Conclusions
The present study characterizes a high-risk de novo AML patient and reports on a novel rather complex insertion ins(18:15)(q21;q31.2q35.1) with concurrent submicroscopic deletions at 5q31.2, 5q35.1q35.2 and 18q12.3q21.1. The rearrangement might be a variant of the chromosomal translocation t(5;18)(q35;q21), which previously was reported in two cases with high-risk de novo AML. This study also highlights the clinical usefulness of oaCGH analysis to identify additional submicroscopic copy number aberrations. We have narrowed the 5q31.2 CDR in AML and provided new insight to the putative role of the 5q31.2 deletion in myeloid malignancy. In addition, we have uncovered a novel fusion of the chromosomal regions at 5q35 and 18q21.1 containing the genes NPM1 and HAUS1, respectively, as a result of the complex insertion-deletion rearrangement. This study contributes to the identification of the whole spectrum of cytogenetic abnormalities in AML and their prognostic significance.
Methods
G-banding analysis
Chromosome analysis was done on G-banded chromosomes prepared after short-term unstimulated culturing of cells obtained from bone marrow at diagnosis, and G-banding performed on PHA-stimulated peripheral blood cells as described [48]. Karyotypes were described according to ISCN [49].
Fluorescent in situ Hybridization (FISH) analysis
Human multicolor FISH were done according to manufacturer's instructions using the following XCyting multicolor FISH probes: 1) 24-color karyotyping was done with the 24XCyte consisting of 24 different chromosome painting probes, 2) mBanding with XCyte 5 and XCyte 18 probes consisting of a series of partial chromosome paints for sequential partially overlapping chromosome regions of a single chromosome (MetaSystems, Altlussheim, Germany). Each of the XCyte probes was labeled with one of five fluorochromes or a unique combination thereof (combinatorial labeling). Metaphases were counterstained with 4',6-diamidino-2-phenylindole (DAPI). Image capture was done with an automated Zeiss Axio Imager.Z2 equipped with a CCD-camera (CoolCube1) and appropriate filters using Isis software (MetaSystems). Karyotyping was done using the 24-color mFISH upgrade package, ISIS, including mBanding.
Whole chromosome painting and locus specific FISH analysis was done with the following directly labeled probes according to manufacturers' instructions: 1) whole chromosome painting probes for chromosomes 5 and 18 (Kreatech Diagnostics, Amsterdam, The Netherlands); 2) the LSI EGR1(5q31)/D5S23,D5S721(5p15.2) dual color probe set (Abbott Molecular, Wiesbaden, Germany); and 3) subtelomeric probes for 5pter, 5qter, 18pter and 18qter (Kreatech Diagnostics). Table 2 summarizes the custom made BAC-based probes (Empire Genomics, New York, USA) for validating the oaCGH findings. Chromosomes were counterstained with DAPI. FISH results were reported according to ISCN [49].
Oligobased array comparative genomic hybridization analysis
oaCGH analysis was performed using CytoChip Cancer 4x180K v2.0 (BlueGnome, Cambridge, UK) encompassing a 20 kb backbone with highest concentration of probes at 670 cancer genes. The analysis was done according to manufacturer's instructions using 0.5 µg patient DNA from bone marrow cells at initial diagnosis as described in [48]. After hybridization, washing and drying the oligo array was scanned at 2.5 µm with GenePix 4400A microarray scanner. Initial analysis and normalization was done with BlueFuseMulti v2.6. For analysis and visualization normalized log2 probe signal values were imported into Nexus Copy Number software v. 6.1 (BioDiscovery, California, USA) and segmented using FASST2 segmentation algorithm with a minimum of 3 probes/segment. Regions of gain or loss contained within copy number variable regions (CNVs) were discarded. Reference genome was NCBI build 36.1 (hg18). Bioinformatics analysis was performed by querying the UCSC database (http://genome.ucsc.edu).
Consent
The study conforms to the provisions of the Declaration of Helsinki. Written informed consent was obtained from the patient. A copy of the written consent is available for review by the Editor-in-Chief of this journal.
Abbreviations
- BAC:
-
Bacterial artificial chromosomes
- PHA:
-
Phytohemagglutinin
- WHO:
-
World Helath Organization
References
Mitelman F, Johansson B, Mertens FE: Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer (2014). Last update: May 13, 2014.., [http://cgap.nci.nih.gov/Chromosomes/Mitelman]
Kayser S, Zucknick M, Dohner K, Krauter J, Kohne CH, Horst HA, Held G, von Lilienfeld-Toal M, Wilhelm S, Rummel M, Germing U, Gotze K, Nachbaur D, Schlegelberger B, Gohring G, Spath D, Morlok C, Teleanu V, Ganser A, Dohner H, Schlenk RF: Monosomal karyotype in adult acute myeloid leukemia: prognostic impact and outcome after different treatment strategies. Blood 2012, 119: 551–558. 10.1182/blood-2011-07-367508
Grimwade D, Mrozek K: Diagnostic and prognostic value of cytogenetics in acute myeloid leukemia. Hematol Oncol Clin North Am 2011, 25: 1135–1161. vii 10.1016/j.hoc.2011.09.018
Mrozek K, Heinonen K, Bloomfield CD: Clinical importance of cytogenetics in acute myeloid leukaemia. Best Pract Res Clin Haematol 2001, 14: 19–47. 10.1053/beha.2000.0114
Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM, Hellstrom-Lindberg E, Tefferi A, Bloomfield CD: The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 2009, 114: 937–951. 10.1182/blood-2009-03-209262
Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, Wheatley K, Harrison CJ, Burnett AK: Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010, 116: 354–365. 10.1182/blood-2009-11-254441
Estey EH: Acute myeloid leukemia: 2013 update on risk-stratification and management. Am J Hematol 2013, 88: 318–327. 10.1002/ajh.23404
Van Hemel JO, Eussen HJ: Interchromosomal insertions. Identification of five cases and a review. Hum Genet 2000, 107: 415–432. 10.1007/s004390000398
Rucker FG, Bullinger L, Gribov A, Sill M, Schlenk RF, Lichter P, Dohner H, Dohner K: Molecular characterization of AML with ins(21;8)(q22;q22q22) reveals similarity to t(8;21) AML. Genes Chromosomes Cancer 2011, 50: 51–58. 10.1002/gcc.20830
Arber DA, Chang KL, Lyda MH, Bedell V, Spielberger R, Slovak ML: Detection of NPM/MLF1 fusion in t(3;5)-positive acute myeloid leukemia and myelodysplasia. Hum Pathol 2003, 34: 809–813. 10.1016/S0046-8177(03)00251-X
Xu Y, Yin J, Pan J, Wu C, Wang Q, Yao H, Wu D, Chen S, Sun A: A BCR-JAK2 fusion gene from ins(22;9)(q11;p13p24) in a patient with atypical chronic myeloid leukemia. Leuk Lymphoma 2013, 54: 2322–2324. 10.3109/10428194.2012.762648
Wang ES, Maslak P, Cathcart K, Jurcic JG: Acute myeloid leukemia with t(5;18)(q35;q21). Cancer Genet Cytogenet 2001, 127: 71–73. 10.1016/S0165-4608(00)00408-8
Daraki A, Bourantas LK, Manola KN: Translocation t(5;18)(q35;q21) as a rare nonrandom abnormality in acute myeloid leukemia. Cytogenet Genome Res 2013, 139: 289–294. 10.1159/000348786
Fathi AT, Chen YB: Treatment of FLT3-ITD acute myeloid leukemia. Am J Blood Res 2011, 1: 175–189.
Santos FP, Jones D, Qiao W, Cortes JE, Ravandi F, Estey EE, Verma D, Kantarjian H, Borthakur G: Prognostic value of FLT3 mutations among different cytogenetic subgroups in acute myeloid leukemia. Cancer 2011, 117: 2145–2155. 10.1002/cncr.25670
Schiller J, Praulich I, Krings Rocha C, Kreuzer KA: Patient-specific analysis of FLT3 internal tandem duplications for the prognostication and monitoring of acute myeloid leukemia. Eur J Haematol 2012, 89: 53–62. 10.1111/j.1600-0609.2012.01785.x
Thanasopoulou A, Tzankov A, Schwaller J: Potent cooperation between the NUP98-NSD1 fusion and FLT3-ITD mutation in acute myeloid leukemia induction. Haematologica 2014, 99: 1465–1471. 10.3324/haematol.2013.100917
Kolomietz E, Al-Maghrabi J, Brennan S, Karaskova J, Minkin S, Lipton J, Squire JA: Primary chromosomal rearrangements of leukemia are frequently accompanied by extensive submicroscopic deletions and may lead to altered prognosis. Blood 2001, 97: 3581–3588. 10.1182/blood.V97.11.3581
Bacher U, Schnittger S, Kern W, Hiddemann W, Haferlach T, Schoch C: The incidence of submicroscopic deletions in reciprocal translocations is similar in acute myeloid leukemia, BCR-ABL positive acute lymphoblastic leukemia, and chronic myeloid leukemia. Haematologica 2005, 90: 558–559.
Moon HW, Chang YH, Kim TY, Oh BR, Min HC, Kim BK, Ahn HS, Cho HI, Lee DS: Incidence of submicroscopic deletions vary according to disease entities and chromosomal translocations in hematologic malignancies: investigation by fluorescence in situ hybridization. Cancer Genet Cytogenet 2007, 175: 166–168. 10.1016/j.cancergencyto.2007.01.012
Dolan M, Peterson B, Hirsch B: Array-based comparative genomic hybridization characterizes a deletion associated with a t(15;17) in acute promyelocytic leukemia. Am J Clin Pathol 2008, 130: 818–823. 10.1309/AJCPENMUI47OGKRW
Goodarzi AA, Jeggo PA: The repair and signaling responses to DNA double-strand breaks. Adv Genet 2013, 82: 1–45. 10.1016/B978-0-12-407676-1.00001-9
Johansson B, Harrison CJ: Acute myeloid leukemia. In Cancer Cytogenetics. Edited by: Heim S, Mitelman F. John Wiley & Sons, Inc., Hoboken, New Jersey; 2009:45–139.
Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT: Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science 1994, 263: 1281–1284. 10.1126/science.8122112
Redner RL, Rush EA, Faas S, Rudert WA, Corey SJ: The t(5;17) variant of acute promyelocytic leukemia expresses a nucleophosmin-retinoic acid receptor fusion. Blood 1996, 87: 882–886.
Jaju RJ, Fidler C, Haas OA, Strickson AJ, Watkins F, Clark K, Cross NC, Cheng JF, Aplan PD, Kearney L, Boultwood J, Wainscoat JS: A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood 2001, 98: 1264–1267. 10.1182/blood.V98.4.1264
Itoh M, Okazaki T, Tashima M, Sawada H, Uchiyama T: Acute myeloid leukemia with t(5;11): two case reports. Leuk Res 1999, 23: 677–680. 10.1016/S0145-2126(99)00033-8
Thiede C, Koch S, Creutzig E, Steudel C, Illmer T, Schaich M, Ehninger G: Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML). Blood 2006, 107: 4011–4020. 10.1182/blood-2005-08-3167
Trimarchi T, Ntziachristos P, Aifantis I: A new player SETs in myeloid malignancy. Nat Genet 2013, 45: 846–847. 10.1038/ng.2709
Albano F, Anelli L, Zagaria A, Coccaro N, Casieri P, Minervini A, Specchia G: SETBP1 and miR_4319 dysregulation in primary myelofibrosis progression to acute myeloid leukemia. J Hematol Oncol 2012, 5: 48. 10.1186/1756-8722-5-48
Cristobal I, Blanco FJ, Garcia-Orti L, Marcotegui N, Vicente C, Rifon J, Novo FJ, Bandres E, Calasanz MJ, Bernabeu C, Odero MD: SETBP1 overexpression is a novel leukemogenic mechanism that predicts adverse outcome in elderly patients with acute myeloid leukemia. Blood 2010, 115: 615–625. 10.1182/blood-2009-06-227363
Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD: Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol 2008, 181: 421–429. 10.1083/jcb.200711053
Fuchs O: Important genes in the pathogenesis of 5q- syndrome and their connection with ribosomal stress and the innate immune system pathway. Leuk Res Treat 2012, 2012: 179402.
Eisenmann KM, Dykema KJ, Matheson SF, Kent NF, DeWard AD, West RA, Tibes R, Furge KA, Alberts AS: 5q- myelodysplastic syndromes: chromosome 5q genes direct a tumor-suppression network sensing actin dynamics. Oncogene 2009, 28: 3429–3441. 10.1038/onc.2009.207
Douet-Guilbert N, De Braekeleer E, Basinko A, Herry A, Gueganic N, Bovo C, Trillet K, Dos Santos A, Le Bris MJ, Morel F, Eveillard JR, Berthou C, De Braekeleer M: Molecular characterization of deletions of the long arm of chromosome 5 (del(5q)) in 94 MDS/AML patients. Leukemia 2012, 26: 1695–1697. 10.1038/leu.2012.9
Zhao N, Stoffel A, Wang PW, Eisenbart JD, Espinosa R 3rd, Larson RA, Le Beau MM: Molecular delineation of the smallest commonly deleted region of chromosome 5 in malignant myeloid diseases to 1-1.5 Mb and preparation of a PAC-based physical map. Proc Natl Acad Sci U S A 1997, 94: 6948–6953. 10.1073/pnas.94.13.6948
Jerez A, Gondek LP, Jankowska AM, Makishima H, Przychodzen B, Tiu RV, O'Keefe CL, Mohamedali AM, Batista D, Sekeres MA, McDevitt MA, Mufti GJ, Maciejewski JP: Topography, clinical, and genomic correlates of 5q myeloid malignancies revisited. J Clin Oncol 2012, 30: 1343–1349. 10.1200/JCO.2011.36.1824
Stoddart A, Fernald AA, Wang J, Davis EM, Karrison T, Anastasi J, Le Beau MM: Haploinsufficiency of del(5q) genes, Egr1 and Apc, cooperate with Tp53 loss to induce acute myeloid leukemia in mice. Blood 2014, 123: 1069–1078. 10.1182/blood-2013-07-517953
Ebert BL: Molecular dissection of the 5q deletion in myelodysplastic syndrome. Semin Oncol 2011, 38: 621–626. 10.1053/j.seminoncol.2011.04.010
Fu CT, Zhu KY, Mi JQ, Liu YF, Murray ST, Fu YF, Ren CG, Dong ZW, Liu YJ, Dong M, Jin Y, Chen Y, Deng M, Zhang W, Chen B, Breslin P, Chen SJ, Chen Z, Becker MW, Zhu J, Zhang JW, Liu TX: An evolutionarily conserved PTEN-C/EBPalpha-CTNNA1 axis controls myeloid development and transformation. Blood 2010, 115: 4715–4724. 10.1182/blood-2009-11-255778
Matsumura K, Sakai C, Kawakami S, Yamashita F, Hashida M: Inhibition of cancer cell growth by GRP78 siRNA lipoplex via activation of unfolded protein response. Biol Pharm Bull 2014, 37: 648–653. 10.1248/bpb.b13-00930
Richardson PG, Schlossman R, Hideshima T, Anderson KC: New treatments for multiple myeloma. Oncology 2005, 19: 1781–1792. discussion 1792, 1795-1787
O'Connor OA, Wright J, Moskowitz C, Muzzy J, MacGregor-Cortelli B, Stubblefield M, Straus D, Portlock C, Hamlin P, Choi E, Dumetrescu O, Esseltine D, Trehu E, Adams J, Schenkein D, Zelenetz AD: Phase II clinical experience with the novel proteasome inhibitor bortezomib in patients with indolent non-Hodgkin's lymphoma and mantle cell lymphoma. J Clin Oncol 2005, 23: 676–684. 10.1200/JCO.2005.02.050
Terpos E, Verrou E, Banti A, Kaloutsi V, Lazaridou A, Zervas K: Bortezomib is an effective agent for MDS/MPD syndrome with 5q- anomaly and thrombocytosis. Leuk Res 2007, 31: 559–562. 10.1016/j.leukres.2006.05.018
Shao L, Kang SH, Li J, Hixson P, Taylor J, Yatsenko SA, Shaw CA, Milosavljevic A, Chang CC, Cheung SW, Patel A: Array comparative genomic hybridization detects chromosomal abnormalities in hematological cancers that are not detected by conventional cytogenetics. J Mol Diagn 2010, 12: 670–679. 10.2353/jmoldx.2010.090192
Walter MJ, Payton JE, Ries RE, Shannon WD, Deshmukh H, Zhao Y, Baty J, Heath S, Westervelt P, Watson MA, Tomasson MH, Nagarajan R, O'Gara BP, Bloomfield CD, Mrozek K, Selzer RR, Richmond TA, Kitzman J, Geoghegan J, Eis PS, Maupin R, Fulton RS, McLellan M, Wilson RK, Mardis ER, Link DC, Graubert TA, DiPersio JF, Ley TJ: Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci U S A 2009, 106: 12950–12955. 10.1073/pnas.0903091106
Bullinger L, Kronke J, Schon C, Radtke I, Urlbauer K, Botzenhardt U, Gaidzik V, Cario A, Senger C, Schlenk RF, Downing JR, Holzmann K, Dohner K, Dohner H: Identification of acquired copy number alterations and uniparental disomies in cytogenetically normal acute myeloid leukemia using high-resolution single-nucleotide polymorphism analysis. Leukemia 2010, 24: 438–449. 10.1038/leu.2009.263
Kjeldsen E, Roug AS: A novel unbalanced de novo translocation der(5)t(4;5)(q26;q21.1) in adult T-cell precursor lymphoblastic leukemia. Mol Cytogenet 2012, 5: 21. 10.1186/1755-8166-5-21
An International System for Human Cytogenetic Nomenclature (2013). S. Karger and Cytogenetic and Genome Research, Basel; 2013.
Acknowledgements
The biotechnologists Bente Madsen and Pia Kristensen are greatly thanked for excellent technical assistance. The Danish Cancer Society supported the study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author declares that he has no competing interest.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Kjeldsen, E. A novel insertion ins(18;5)(q21.1;q31.2q35.1) in acute myeloid leukemia associated with microdeletions at 5q31.2, 5q35.1q35.2 and 18q12.3q21.1 detected by oligobased array comparative genomic hybridization. Mol Cytogenet 7, 63 (2014). https://doi.org/10.1186/s13039-014-0063-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-014-0063-x