Abstract
Background
Reptiles are known to be asymptomatic carriers of Salmonella spp. in their gastrointestinal mucosa and a variety of Salmonella serovars including exotic serovars mainly associated with reptiles as well as human pathogenic serovars have been isolated. There are many case reports of reptile-associated Salmonella infections worldwide, including one case in Norway in 2000. In August 2017, there was a legislative change in Norway that allowed more permissive reptile ownership and legalized the keeping of 19 different reptile species by private persons. There has been a concern that this new legislation will lead to an increase in reptile-associated salmonellosis in Norway, however knowledge is lacking on the occurrence of Salmonella spp. in Norwegian reptiles. The aim of this study was therefore to investigate the prevalence of Salmonella spp. in captive reptile species in Norway, identify the serovars and evaluate their zoonotic potential. Thus, cloacal swabs were taken from 53 snakes, 15 lizards and 35 chelonians from three Norwegian zoos, and assessed for the presence of Salmonella spp. by culture, biochemical testing and serotyping.
Results
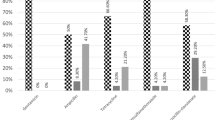
In total, 43% of the reptiles were shedding Salmonella spp., with a prevalence of 62%, 67% and 3% in snakes, lizards and chelonians, respectively. A total of 26 different serovars were found, including Salmonella enterica spp. enterica (40%) and S. enterica spp. arizonae (4%), both of which are considered to have a high zoonotic potential. S. enterica spp. diarizonae, salamae and houtenae were also identified, however these serovars are considered to have a lower zoonotic potential.
Conclusions
The current study demonstrates that captive Norwegian reptiles are carriers of potentially zoonotic Salmonella spp. Given the increasing popularity of reptiles as pets and the legislative change, reptile-associated salmonellosis could become an increasingly important public health concern in Norway. Adequate public information about the risk of Salmonella infection as well as preventive measures to avoid Salmonella transmission from reptiles to humans is needed. The risk of Salmonella infection is considered low when recommended precautions are taken and good hygiene exhibited.
Similar content being viewed by others
Background
Salmonella is a Gram-negative bacterium of the Enterobacteriaceae family that can survive for weeks in dry environments and for months in water [1]. The bacterium is generally considered a normal constituent of the reptilian intestinal microbiota. Some studies report prevalence of Salmonella spp. in reptiles up to 90%, with a wide variety of serovars being identified [2,3,4,5,6,7,8,9,10,11]. These includes several serovars primarily associated with reptiles as well as non-host adapted serovars, including well‐known zoonoses such as Salmonella serovar Enteritidis and S. ser. Typhimurium [12,13,14]. Reptiles excrete Salmonella spp. in their feces intermittently and the bacterial load they shed is reported to increase during periods of stress e.g. transportation, handling, illness, high animal density and otherwise suboptimal environments [10].
According to the World Health Organization [15], Salmonella is one of the major global causes of diarrheal diseases, and usually associated with consumption of contaminated food products of animal origin. In the USA, about 1.35 million cases of illness, 26 500 hospitalizations, and 420 deaths occur every year due to nontyphoidal Salmonella infection [16], and direct contact with animals is estimated to account for 11% of Salmonella enteritis cases [17]. Salmonellosis in mammals causes a range of symptoms from diarrhea, vomiting, and fever, to life threatening septicemia [18]. Infection in humans is most severe in young children, the elderly and those with a reduced immune system [15]. Most of the Salmonella isolates that cause disease in mammals belong to Salmonella enterica spp. enterica. A few serovars of this subspecies are strictly human pathogens without an animal reservoir. The remaining S. enterica spp. enterica serovars are considered zoonotic or potentially zoonotic [14, 19]. The most common serovars infecting humans worldwide are S. ser. Typhimurium and S. ser. Enteritidis [15]. Reptiles infected by Salmonella spp. do not usually develop disease, however clinical salmonellosis occurs in reptiles and is generally provoked by an underlying primary cause of disease, although primary disease can occur [20].
Exotic pet ownership has become increasingly popular worldwide [21, 22]. The European Union member states are among the largest importers of reptiles, and in the USA, 4.7 million households own a reptile [23, 24]. Simultaneously, reptiles and amphibians are estimated to account for 6% of all Salmonella infections in the USA and Europe and may be increasing [13, 25,26,27,28]. During 2006–2014, a total of 15 multistate outbreaks of turtle-associated salmonellosis in humans were reported in the USA [28]. Reptiles kept as pets are also potential sources of Salmonella infection for other companion animals, such as dogs and cats, which can contribute to the spread of this pathogen in the environment and increase the risk of infection for humans [29].
In Norway, the occurrence of human salmonellosis is low compared to other countries with about 1000 reported cases annually [30]. In August 2017, pet ownership of 19 different reptile species including snakes (nine species), lizards (seven species) and chelonians (three species) was legalized in Norway. Prior to this, permission to hold reptiles was given almost exclusively to zoos and aquaria, with privately owned reptiles being prohibited. Nevertheless, illegal hold of reptiles in private homes existed and a single case of reptile-associated salmonellosis had been reported in Norway [31]. There has been a concern that this new legislation will lead to increased occurrence of salmonellosis in Norway. The risk of reptile-associated salmonellosis in humans depends on several factors such as Salmonella spp. prevalence, serovar predominance and pathogenicity, as well as exposure and immunocompetence of the human [11, 13]. However, little is known about the prevalence and serovar predominance in Norwegian reptiles. Therefore, the aim of this study was to investigate the prevalence of Salmonella spp. in captive reptile species in Norway, identify the serovars isolated from this population, and evaluate their zoonotic potential.
Methods
Animal selection
During 2016, reptiles were sampled from three zoos (referred to as A, B and C) in Norway and examined for Salmonella. Exclusion criteria comprised animals (a) with a cloaca too small for swab insertion, (b) showing signs of disease (c) treated with antibiotics within the last 30 days, and (d) that shared cage with reptile(s) treated with antibiotics the last 30 days. Also, snakes that (e) showed signs of ecdysis, and (f) were fed the same day as sampling, were excluded from the study. All other reptiles in the three zoos were selected for sampling and included 35 chelonians, 15 lizards and 53 snakes representing 22 different species. The classification and numbers of reptiles sampled from each zoo are described in Table 1. All animals were considered healthy at the time of sampling based on daily observations by the zookeepers over the previous month, and physical examination by a veterinarian at time of sampling.
Sample collection and processing
Depending on animal size, regular or minitip bacteriology swabs of soft rayon were used for fecal sampling (Copan Diagnostics Inc., Murrieta, CA, USA). The live animals were physically restrained, and a swab was inserted into the cloaca and gently rotated longitudinally. Swabs were then placed into Amies agar gel medium with or without charcoal (Copan Diagnostics Inc.), stored at 4 °C and processed within 24 h.
All growth media and biochemical tests used were produced in-house at the Department of Food Safety and Infection Biology, Norwegian University of Life Sciences (Oslo, Norway). For recovery of Salmonella spp. cloacal swabs were direct-plated on selective Bromthymolblue-Lactose-Agar (BTBL) (Brolac, Cat.-No. 1639, Merck, Darmstadt, Germany) and incubated at 35 °C for 24 h. For enrichment of Salmonella spp., the swabs were first placed into 4 mL buffered peptone water (BPW, Merck) and cultivated at 35 °C for 24 h before 1 mL inoculum was transferred to 4 mL selenite broth (Difco™ Selenite Cystine Broth, BD Diagnostics, Sparks, MD, USA) and further incubated at 42 °C [32, 33]. Every day for three days, a sterile plastic bacterial loop was used to transfer 1 µL aliquot of enriched broth to a BTBL plate followed by incubation at 35 °C for 24 h. Oxidase-negative (Oxidase Strips, Oxoid, Cambridge, UK) and non-lactose-fermenting bacteria (blue), and thus suspected Salmonella colonies, were streaked onto urea agar (Oxoid) and triple sugar iron (TSI) agar (Difco, BD Diagnostics) and incubated at 35 °C for 24 h. Samples were considered positive for Salmonella based on a negative urea result and the production of hydrogen sulfide in the TSI test. Suspect colonies were also analyzed using the API20E kit after the manufacturer’s description (BioMérieux, Marcy-l’Étoile, France). One single colony were isolated from each BTBL plate for further identification when Salmonella was suspected. Presumptive colonies of Salmonella were restreaked onto blood agar plates (blood agar base no. 2 (Oxoid) supplemented with 5% bovine blood) and submitted to the Norwegian Veterinary Institute where the isolates were serotyped by agglutination tests with antisera (SIFIN, Berlin, Germany and Statens Serum Institut (SSI), Hillerød, Denmark) according to the White-Kauffmann scheme [34]. Salmonella subspecies I (S. enterica ssp. enterica) were identified as named serovars, except one sample.
Evaluation of zoonotic potential
A literature review was conducted by using publications indexed at PubMed as well as other Internet resources to evaluate the zoonotic potential of the Salmonella serovars isolated in the present study. Database searches were conducted from February 2016 to July 2019 with the search terms “reptile”, “zoonosis”, “Salmonella”, in addition to the name of specific serovars. Salmonella serovars that have been reported to cause illness in otherwise healthy adults with normal immune status were considered to have a high zoonotic potential. Serovars reported to cause disease in the immunonaieve or immunocompromised individuals were considered to be of moderate zoonotic potential, and those serovars that only have been reported to cause disease in a few individuals were considered to have a low zoonotic potential.
Statistical analysis
Confidence intervals (CI) for binominal distribution were calculated using online software available at Statpages.net [35]. A two-tailed P-value was calculated from a 2 × 2 contigency table by Fisher’s exact test using the GraphPad QuickCalcs software for statistical comparisons between groups for the prevalence of Salmonella [36]. A P-value ≤ 0.05 was considered statistically significant.
Results
Prevalence of Salmonella spp. from cloacal samples
A total of 44 out of 103 cloacal samples (43%, CI 33–53%) were Salmonella-positive, as determined by biochemical tests and serotyping. Salmonella spp. were isolated from 16 of the 22 different reptile species (73%) included in this study.
In snakes and lizards, 62% (CI 48–75%) and 67% (CI 38–88%) of samples were positive for Salmonella, respectively. In chelonians, Salmonella sp. was only identified in one sample (CI 0–0.15%) originating from a Hermann’s tortoise (Testudo hermanni). The prevalence of Salmonella was significantly lower in chelonians than in lizards and snakes (P < 0.001), and this difference remained even if the results of a large group of Trachemys scripta (n = 21) that were housed together were excluded.
Salmonella spp. were identified in 24% of the tested reptiles in Zoo A (CI 12–41%), 52% in Zoo B (CI 34–69%) and 55% in Zoo C (CI 36–72%). In Zoo A, all snakes and one lizard (50%) were Salmonella-positive. All 27 chelonians that were tested in the same zoo were found to be negative. In Zoo B, Salmonella spp. was identified in 37% of the snakes, 75% of the lizards and in 50% of the chelonians. In Zoo C, 67% of both the snakes and the lizards were Salmonella-positive, whilst all chelonians were negative.
Serotyping of Salmonella isolates
Of the samples that tested positive for Salmonella, a single isolate was identified from each sample except one, originating from a Royal python (Python regius), where one isolate was identified by direct-plating and another isolate after enrichment. Thus, 45 isolates of Salmonella spp. were identified from the 44 positive cloacal samples. In total, 26 different serovars were identified by serotyping. S. enterica spp. enterica was the most frequently identified subspecies, comprising 40% of positive samples, followed by S. enterica spp. diarizonae, 36%, S. enterica spp. salamae 11%, S. enterica spp. arizonae, 4%, and S. enterica spp. houtenae, 2%. Three Salmonella serovars (7%) were of unknown subspecies (Table 2). The number of serovars identified from each subspecies is listed in Table 2.
Whilst several of the Salmonella-positive reptiles shared their cage with other animals of the same species, cohabiting reptiles carrying the same serovars of Salmonella were only identified in 2 of 14 cages in this study, both cages holding snakes. In seven (50%) of the cages, both Salmonella-positive and Salmonella-negative animals were identified.
Zoonotic potential of identified Salmonella serovars
Salmonella serovars identified in this study and their zoonotic potential are listed in Table 3. Sixteen reptiles (15.5%) carrying Salmonella serovars with a high zoonotic potential were identified. These serovars were; S. ser. Paratyphi B var Java, S. ser. Muenchen, S. ser. Cotham, S. ser. Kottbus, S. ser. Hadar, S. enterica spp. arizonae 44:z4, z23:- and S. enterica spp. arizonae 51:z4z23:. Serovars of S. enterica spp. diarizonae and S. enterica spp. houtenae that were considered to have a moderate zoonotic potential were isolated from 17 reptiles (16%).
Discussion
Prevalence of Salmonella spp. in captive Norwegian reptiles compared to other countries
The overall prevalence of Salmonella in captive Norwegian reptiles (43%, CI 33–53%) is consistent with the spectrum of prevalence’s reported globally: Japan (74%) [6], Germany/Austria (54%) [3], Italy (51 and 57%) [4, 22], Australia (47%) [10], Denmark (35%) [11], Taiwan (31%) [8], Trinidad (31%) [2], Republic of Korea (30%) [7], Croatia (13%) [37] and New Zealand (11%) [9]. The variation in reported Salmonella prevalence amongst different reptile populations may represent a true different in infection status, for instance Scheelings et al. [10] found a higher prevalence of Salmonella in reptiles held in captivity (47%) compared to wild reptiles (14%), although this is yet to be confirmed by other studies. Unfortunately, whilst one can speculate about factors that may influence the true infection status, such as wild vs captive, season, climate, environment, other diseases and diet, little evidence is available on how these factors truly affects Salmonella infection. Further limiting the usefulness of comparing results between studies is the considerable variation in experimental design and the use of different diagnostic techniques. For instance, whilst we used cloacal swabs, other studies have used fecal samples, oral swabs and skin swabs [3, 4, 6, 37]. Additionally, the phenomenon of intermittent shedding probably accounts significantly for the variability in detection rates between authors [12]. In the studies from Croatia, New Zealand and Italy, sampling was performed by the animal’s owner, which could have given some more unreliable results and thus lower prevalence [4, 9, 22, 37].
In general, Salmonella prevalence is reported to be higher in snakes than in lizards or chelonias [3, 6, 8, 10, 11]. In this study, no significant difference in Salmonella prevalence was found between snakes and lizards, however this may have been due to inadequate sample size. In contrast, Salmonella prevalence in chelonians was significant lower compared to the two other groups, which is consistent with results from Germany/Austria, Australia, New Zealand and Taiwan [3, 8,9,10]. However, reported Salmonella prevalence in chelonians varies highly between studies (3–72%) [3, 6, 8,9,10,11, 22, 37]. The prevalence within Chelonia in this study may have been skewed by a very large group of Salmonella-negative Trachemys scripta that were housed together and constituted over half the total number Chelonia included in the study.
The occurrence of Salmonella spp. in Norwegian production and companion animals, as well as animal feeds and products is very low compared to most other countries [30]. This favorable situation does not however include the captive Norwegian reptiles. This study shows that the Salmonella prevalence in Norwegian reptiles is similar to the prevalence reported in other countries. Most captive reptiles in Norway are imported from other countries and might have been exposed to Salmonella spp. early in life when the intestinal microbiota is established, thus becoming permanent carriers of the bacteria. Only a few wild-living reptile species exist in Norway, however screening of these animals would be of great interest to further elucidate the relationship between Salmonella spp. and reptiles.
This study represents the first investigation into the prevalence of Salmonella in Norwegian reptiles. Ideally, investigation of the risk for reptile associated salmonellosis would be based upon a population of pet reptiles, not zoological collections. However, at the time of sampling, hold of reptiles in private households was illegal in Norway, thus making it complicated to access this population. Instead, reptiles kept in different zoos in Norway were studied to evaluate the risk of zoonotic transmission of Salmonella spp. to visitors and employees. Reptiles kept in Norwegian zoos often originate from private homes and end up being relocated to a zoo after confiscation by the Norwegian Food Safety Authority. As such, these results can serve as a proxy for Salmonella in reptiles in private homes. However, little is known about how the intestinal microbiota is influenced by housing conditions and other environmental factors. Also, although all zoos in this study invited their visitors, including children, to hold and/or touch the reptiles the interaction with animals is probably more intense in private holdings and precautions less than in zoos. Thus, the zoonotic risk of salmonellosis may be higher in private homes compared to zoos.
Salmonella serovars in Norwegian reptiles
In total, 45 Salmonella isolates were identified in 44 different individuals. Out of these, 18 (40%) and 16 (36%) isolates were of subspecies enterica and diarizonae, respectively, which is consistent with other studies [3, 6, 9,10,11]. S. bongori and S. enterica spp. indica. were not identified in this study, similar to previous reports [3, 6, 9, 10, 37]. The results documented in the present work as well as previous studies indicates that a great diversity of different S. enterica subspecies and serovars infect reptiles. Routinely, only one single colony was isolated from each sample, thus this investigation was not designed to detect a diversity of Salmonella subspecies and serovars in each single reptile’s intestinal microbiota. Nevertheless, two different subspecies were identified from the same animal on one occasion. A diversity of Salmonella subspecies and serovars in the reptilian intestine is previously described, and although a single serovar has been the most frequent finding, up to four different serovars have been reported from the same animal [3, 6].
This work does not clarify if Salmonella serovars transmit between the individual reptiles. Identical serovars of Salmonella in cohabiting reptiles were only identified in 2 of 14 cages in this study, however the study design does not exclude the possibility for unidentified Salmonella spp. in both the Salmonella-positive as well as the Salmonella-negative reptiles. In half of the cages, both Salmonella-positive and Salmonella-negative animals were identified. The fact that Salmonella excretion is intermittent represent a potential source of false negatives in prevalence studies, particularly if only one sample is taken [29]. Thus, a Salmonella-free status may have been a misinterpretation. By testing each individual multiple times, higher prevalence and diversity of Salmonella spp. could have been detected.
Zoonotic potential of identified Salmonella serovars
Salmonella is one of the most common and important zoonoses in the world. However, the pathogenicity and zoonotic potential of Salmonella varies between different subspecies, serovars and strains [38]. In the current work, no pathogenicity studies on the different Salmonella isolates were performed and the evaluations of zoonotic potential should therefore be regarded with caution. S. enterica spp. enterica and S. enterica spp. arizonae were the subspecies with the highest zoonotic potential found in this study. S. enterica spp. enterica causes 99% of all human Salmonella infections [1], however none of the most common serovars identified to cause human salmonellosis in Norway (S. Enteritidis, S. Typhimurium, S. ser. Stanley, S. ser. Newport and S. ser. Java) were isolated in this study [30]. Nevertheless, Salmonella serovars with a high zoonotic potential were identified in 15.5% of the reptiles (Table 3). S. Paratyphi B var Java [39,40,41], S. ser. Muenchen [42], S. ser. Cotham [43, 44] and S. enterica spp. arizonae [45] are reported to cause several incidences of reptile-associated salmonellosis in otherwise healthy humans with normal immune status. Salmonella Kottbus and S. Hadar are serovars often related to human cases of food poisoning and have not been identified with reptile-associated salmonellosis [46,47,48,49,50]. However, close contact with reptiles carrying these Salmonella serovars could probably increase the risk of salmonellosis.
Salmonella enterica spp. diarizonae is found in high prevalence in both wild and captive reptiles and is frequently identified to be the cause of reptile-associated salmonellosis [21]. This subspecies, as well as S. enterica spp. houtenae were identified in 16% of the reptiles and considered to have a moderate zoonotic potential as most human cases occur in immunosuppressed individuals or children (Table 3) [21, 51,52,53,54]. In total almost 1/3 of the reptiles were identified as carriers of highly or moderately zoonotic Salmonella serovars. These results underline that all reptiles should be considered to be potential sources of zoonotic Salmonella spp.
Conclusions
The current study demonstrates that captive Norwegian reptiles are carriers of potentially zoonotic Salmonella spp. Given the increasing popularity of reptiles as pets, reptile-associated salmonellosis could become an increasingly important public health concern in Norway. Adequate public information about the risk of Salmonella infection as well as preventive measures to avoid Salmonella transmission from reptiles to humans is needed.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- BTBL:
-
bromthymolblue-lactose-agar
- BPW:
-
buffered peptone water
- CDC:
-
centers for Disease Control and Prevention
- TSI:
-
triple sugar iron
- WHO:
-
world Health Organization
References
Brenner FW, Villar RG, Angulo FJ, Tauxe R, Swaminathan B. Salmonella nomenclature. J Clin Microbiol. 2000;38:2465–7.
Adesiyun AA, Caesar K, Inder L. Prevalence of Salmonella and Campylobacter species in animals at Emperor Valley Zoo Trinidad. J Zoo Wildl Med. 1998;29:237–9.
Geue L, Loschner U. Salmonella enterica in reptiles of German and Austrian origin. Vet Microbiol. 2002;84:79–91.
Corrente M, Madio A, Friedrich KG, Greco G, Desario C, Tagliabue S, et al. Isolation of Salmonella strains from reptile faeces and comparison of different culture media. J Appl Microbiol. 2004;96:709–15.
Ebani VV, Cerri D, Fratini F, Meille N, Valentini P, Andreani E. Salmonella enterica isolates from faeces of domestic reptiles and a study of their antimicrobial in vitro sensitivity. Res Vet Sci. 2005;78:117–21.
Nakadai A, Kuroki T, Kato Y, Suzuki R, Yamai S, Yaginuma C, et al. Prevalence of Salmonella spp. in pet reptiles in Japan. J Vet Med Sci. 2005;67:97–101.
Jang YH, Lee SJ, Lim JG, Lee HS, Kim TJ, Park JH, et al. The rate of Salmonella spp. infection in zoo animals at Seoul Grand Park Korea. J Vet Sci. 2008;9:177–81.
Chen CY, Chen WC, Chin SC, Lai YH, Tung KC, Chiou CS, et al. Prevalence and antimicrobial susceptibility of salmonellae isolates from reptiles in Taiwan. J Vet Diagn Invest. 2010;22:44–50.
Kikillus KH, Gartrell BD, Motion E. Prevalence of Salmonella spp. and serovars isolated from captive exotic reptiles in New Zealand. N Z Vet J. 2011;59:174–8.
Scheelings TF, Lightfoot D, Holz P. Prevalence of Salmonella in Australian reptiles. J Wildl Dis. 2011;47:1–11.
Hydeskov HB, Guardabassi L, Aalbaek B, Olsen KE, Nielsen SS, Bertelsen MF. Salmonella prevalence among reptiles in a zoo education setting. Zoonoses Public Health. 2013;60:291–5.
Pedersen K, Lassen-Nielsen AM, Nordentoft S, Hammer AS. Serovars of Salmonella from captive reptiles. Zoonoses Public Health. 2009;56:238–42.
Mermin J, Hutwagner L, Vugia D, Shallow S, Daily P, Bender J, et al. Reptiles, amphibians, and human Salmonella infection: a population-based, case-control study. Clin Infect Dis. 2004;38(Suppl 3):S253–S261261.
Baumler AJ, Tsolis RM, Ficht TA, Adams LG. Evolution of host adaptation in Salmonella enterica. Infect Immun. 1998;66:4579–87.
World Health Organization (WHO). Salmonella (non-typhoidal): World Health Organization. 2018. https://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal). Accessed 02 Jan 2020.
Centers for Disease Control and Prevention (CDC). Antibiotic resistance threats in the United States, 2019. CDC website. 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. Accessed 02 Jan 2020.
Hale CR, Scallan E, Cronquist AB, Dunn J, Smith K, Robinson T, et al. Estimates of enteric illness attributable to contact with animals and their environments in the United States. Clin Infect Dis. 2012;54(Suppl 5):S472–S479479.
Saphra I, Winter JW. Clinical manifestations of salmonellosis in man; an evaluation of 7779 human infections identified at the New York Salmonella Center. N Engl J Med. 1957;256:1128–34.
Jajere SM. A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet World. 2019;12:504–21.
Pasmans F, Blahak S, Martel A, Pantchev N. Introducing reptiles into a captive collection: the role of the veterinarian. Vet J. 2008;175:53–68.
Schroter M, Roggentin P, Hofmann J, Speicher A, Laufs R, Mack D. Pet snakes as a reservoir for Salmonellaenterica subsp. diarizonae (Serogroup IIIb): a prospective study. Appl Environ Microbiol. 2004;70:613–5.
Corrente M, Sangiorgio G, Grandolfo E, Bodnar L, Catella C, Trotta A, et al. Risk for zoonotic Salmonella transmission from pet reptiles: a survey on knowledge, attitudes and practices of reptile-owners related to reptile husbandry. Prev Vet Med. 2017;146:73–8.
Engler M, Parry-Jones R. Opportunity or threat: The role of the European Union in global wildlife trade. Brussels, Belgium: TRAFFIC Europe. 2007. https://www.traffic.org/site/assets/files/3604/opportunity_or_threat_eu_imports.pdf. Accessed 02 Jan 2020.
Amercan Pet Products Association (APPA). 2017–2018 APPA National pet owners survey statistics: pet ownership & annual expenses: Amercan Pet Products Association (APPA). 2018. https://www.americanpetproducts.org/press_industrytrends.asp. Accessed 02 Jan 2020.
Woodward DL, Khakhria R, Johnson WM. Human salmonellosis associated with exotic pets. J Clin Microbiol. 1997;35:2786–90.
de Jong B, Andersson Y, Ekdahl K. Effect of regulation and education on reptile-associated salmonellosis. Emerg Infect Dis. 2005;11:398–403.
Whitten T, Bender JB, Smith K, Leano F, Scheftel J. Reptile-associated salmonellosis in Minnesota, 1996–2011. Zoonoses Public Health. 2015;62:199–208.
Bosch S, Tauxe RV, Behravesh CB. Turtle-associated salmonellosis, United States, 2006–2014. Emerg Infect Dis. 2016;22:1149–55.
Ebani VV. Domestic reptiles as source of zoonotic bacteria: a mini review. Asian Pac J Trop Med. 2017;10:723–8.
Jørgensen H, Hauge K, Lange H, MacDonald E, Lyngstad TM, Heier B. The Norwegian Zoonoses Report 2017. Norwegian Veterinary Institute. 2018. https://www.vetinst.no/rapporter-og-publikasjoner/rapporter/2018/the-norwegian-zoonoses-report-2017. Accessed 02 Jan 2020.
Torfoss D, Abrahamsen TG. Salmonella infection from turtles [in Norwegian]. Tidskr Nor Legeforen. 2000;120:3670–1.
Juven BJ, Cox NA, Bailey JS, Thomson JE, Charles OW, Shutze JV. Recovery of Salmonella from artificially contaminated poultry feeds in non-selective and selective broth media. J Food Prot. 1984;47:299–302.
Selenite cystine broth. In: Corry JEL, Curtis GDW, Baird RM, editors. Progress in industrial microbiology. Amsterdam: Elsevier; 2003. p. 592–594.
Grimont PAD, Weill FX. Antigentic formulae of the Salmonella serovars. 9th ed. Paris: WHO Collaborating Centre for Reference and Research on Salmonella, World Health Organization/Institut Pasteur; 2007.
Pezzullo JC. Exact Binomial and Poisson Confidence Intervals. 2009. https://statpages.info/confint.html. Accessed 02 Jan 2020.
GraphPad QuickCalcs. Analyze a 2x2 contigency table. https://www.graphpad.com/quickcalcs/contingency1/. Accessed 02 Jan 2020.
Lukac M, Pedersen K, Prukner-Radovcic E. Prevalence of Salmonella in captive reptiles from Croatia. J Zoo Wildl Med. 2015;46:234–40.
Wei S, Huang J, Liu Z, Wang M, Zhang B, Lian Z, et al. Differential immune responses of C57BL/6 mice to infection by Salmonella enterica serovar Typhimurium strain SL1344, CVCC541 and CMCC50115. Virulence. 2019;10:248–59.
Hernandez E, Rodriguez JL, Herrera-Leon S, Garcia I, de Castro V, Muniozguren N. Salmonella Paratyphi B var Java infections associated with exposure to turtles in Bizkaia, Spain, September 2010 to October 2011. Euro Surveill. 2012;17:20201.
Harris JR, Bergmire-Sweat D, Schlegel JH, Winpisinger KA, Klos RF, Perry C, et al. Multistate outbreak of Salmonella infections associated with small turtle exposure, 2007–2008. Pediatrics. 2009;124:1388–94.
Krishnasamy V, Stevenson L, Koski L, Kellis M, Schroeder B, Sundararajan M, et al. Notes from the field: investigation of an outbreak of Salmonella Paratyphi B variant L (+) tartrate+(Java) associated with ball python exposure—United States, 2017. MMWR Morb Mortal Wkly Rep. 2018;67:562–3.
Outbreak of Salmonella Muenchen linked to pet crested geckos 2014–2015. Foodborn Illness Outbreak Database. The Marler Clark Network. 2015. https://outbreakdatabase.com/details/2014-2015-outbreak-of-salmonella-muenchen-linked-to-pet-crested-geckos/?outbreak=reptile&organism=Salmonella. Accessed 02 Jan 2020.
Salmonella Cotham. Confluence wiki, Cornell University. 2014. https://confluence.cornell.edu/display/FOODSAFETY/Salmonella+Cotham. Accessed 02 Jan 2020.
Centers for Disease Control and Prevention (CDC). Multistate outbreak of human Salmonella Cotham. CDC website. 2014. https://www.cdc.gov/media/releases/2014/a0424-dragon-salmonella.html. Accessed 02 Jan 2020.
Lee YC, Hung MC, Hung SC, Wang HP, Cho HL, Lai MC, et al. Salmonella enterica subspecies arizonae infection of adult patients in Southern Taiwan: a case series in a non-endemic area and literature review. BMC Infect Dis. 2016;16:746.
Mohle-Boetani J, Anderson S, Komatsu K, Tapp K, Peterson N, Painter J. Outbreak of Salmonella serotype Kottbus infections associated with eating alfalfa sprouts—Arizona, California, Colorado and New Mexico, February–April 2001. MMWR Morb Mortal Wkly Rep. 2002;51:7–9.
Ryder RW, Crosby-Ritchie A, McDonough B, Hall WJ. Human milk contaminated with Salmonella Kottbus. A cause of nosocomial illness in infants. JAMA. 1977;238:1533–4.
Rowe B, Hall ML, Ward LR, de Sa JD. Epidemic spread of Salmonella Hadar in England and Wales. Br Med J. 1980;280:1065–6.
Papadopoulos T, Petridou E, Zdragas A, Nair S, Peters T, de Pinna E, et al. Phenotypic and molecular characterization of multidrug-resistant Salmonella enterica serovar Hadar in Greece, from 2007 to 2010. Clin Microbiol Infect. 2015;21(149):e1–4.
Salmonella Hadar. Confluence wiki, Cornell University. 2013. https://confluence.cornell.edu/display/FOODSAFETY/Salmonella+Hadar. Accessed 02 Jan 2020
Lourenco MC, dos Reis EF, Valls R, Asensi MD, Hofer E. Salmonella enterica subsp. houtenae serogroup O:16 in a HIV positive patient: case report. Rev Inst Med Trop Sao Paulo. 2004;46:169–70.
Wybo I, Potters D, Plaskie K, Covens L, Collard JM, Lauwers S. Salmonella enterica subspecies houtenae serotype 44:z4, z23: as a rare cause of meningitis. Acta Clin Belg. 2004;59:232–4.
Chong Y, Kwon OH, Lee SY, Chung KS, Shimada T. Salmonella enterica subspecies diarizonae bacteremia in an infant with enteritis—a case report. Yonsei Med J. 1991;32:275–8.
Horvath L, Kraft M, Fostiropoulos K, Falkowski A, Tarr PE. Salmonella enterica subspecies diarizonae maxillary sinusitis in a snake handler: first report. Open Forum Infect Dis. 2016;3:66ofw.
Basler C, Nguyen TA, Anderson TC, Hancock T, Behravesh CB. Outbreaks of human Salmonella infections associated with live poultry, United States, 1990–2014. Emerg Infect Dis. 2016;22:1705–11.
Toboldt A, Tietze E, Helmuth R, Junker E, Fruth A, Malorny B. Molecular epidemiology of Salmonella enterica serovar Kottbus isolated in Germany from humans, food and animals. Vet Microbiol. 2014;170:97–108.
Bockemuhl J. Salmonellosis and shigellosis in Togo (West Africa), 1971–1973. II. Infections in the urban population of Lome. Tropenmed Parasitol. 1977;28:377–83.
Morbidity and mortality weekly report (MMWR). Lizard-associated salmonellosis—Utah. Report No. 0149–2195. MMWR Morb Mortal Wkly Rep. 1992;41:610–11.
Abbott SL, Ni FC, Janda JM. Increase in extraintestinal infections caused by Salmonella enterica subspecies II–IV. Emerg Infect Dis. 2012;18:637–9.
Nair S, Wain J, Connell S, de Pinna E, Peters T. Salmonella enterica subspecies II infections in England and Wales—the use of multilocus sequence typing to assist serovar identification. J Med Microbiol. 2014;63:831–4.
Acknowledgements
The authors would like to acknowledge the animal carers from the three zoos and Gaute Skogtun (Department of Food Safety and Infection Biology, Faculty of Veterinary Medicine, Norwegian University of Life Sciences) for technical assistance and valuable advices.
Prior publication
Data have not been published previously.
Funding
This study was funded by the Faculty of Veterinary Medicine, Norwegian University of Life Sciences.
Author information
Authors and Affiliations
Contributions
All authors contributed in designing the study. LMS, MMS, LS and JJD sampled from animals, and LMS, MMS, LS and AMB performed the bacteriological diagnostic work and analysis. All authors contributed in interpretation of data. AMB was the major contributor in writing the manuscript with contribution from all authors. Further revisions of manuscript were done by AMB and JJD. AMB submitted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study did not require official or institutional ethical approval. The animals were handled according to high ethical standards and national legislation.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Bjelland, A.M., Sandvik, L.M., Skarstein, M.M. et al. Prevalence of Salmonella serovars isolated from reptiles in Norwegian zoos. Acta Vet Scand 62, 3 (2020). https://doi.org/10.1186/s13028-020-0502-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13028-020-0502-0