Abstract
Background
The side effects caused by Polygoni Multiflori Radix (PMR) and Polygoni Multiflori Radix Praeparata (PMRP) have often appeared globally. There is no research on the changes of endogenous metabolites among PMR- and PMRP-treated rats. The aim of this study was to evaluate the varying metabolomic effects between PMR- and PMRP-treated rats. We tried to discover relevant differences in biomarkers and endogenous metabolic pathways.
Methods
Hematoxylin and eosin staining and immunohistochemistry staining were performed to find pathological changes. Biochemical indicators were also measured, one-way analysis of variance with Dunnett’s multiple comparison test was used for biochemical indicators comparison among various groups. Metabolomics analysis based on ultra-high performance liquid chromatography-quadrupole time of flight mass spectrometry (UPLC-Q/TOF-MS) was performed to find the changes in metabolic biomarkers. Multivariate statistical approaches such as principal component analysis (PCA) and orthogonal partial least square-discriminant analysis (OPLS-DA) were applied to reveal group clustering trend, evaluate and maximize the discrimination between the two groups. MetaboAnalyst 4.0 was performed to find and confirm the pathways.
Results
PMR extracts exhibited slight hepatotoxic effects on the liver by increasing aspartate and alanine aminotransferase levels. Twenty-nine metabolites were identified as biomarkers, belonging to five pathways, including alpha-linolenic acid metabolism, taurine and hypotaurine metabolism, glycerophospholipid metabolism, arginine and proline metabolism, and primary bile acid biosynthesis.
Conclusion
This study provided a comprehensive description of metabolomic changes between PMR- and PMRP-treated rats. The underlying mechanisms require further research.
Similar content being viewed by others
Background
Polygoni Multiflori Radix (PMR) and Polygoni Multiflori Radix Praeparata (PMRP) are derived from the tuberous root of Polygonum multiflorum Thunb., and are the clinically used forms of P. multiflorum [1]. They are widely distributed worldwide and have been used as herbal drugs and healthcare products for centuries [2]. These extracts have a wide range of pharmacological activities including anti-aging [3, 4], anti-oxidant [5, 6], anti-tumor [7, 8], neuroprotective [9, 10], hair blacking [11], liver cirrhosis treatment [12], and lipid regulation effects [13,14,15]. Their functions are due to their flavonoid, phenolic acid, and 2,3,5,4′-tetrahydroxystilbene-2-O-β-d-glucoside (THSG) compositions [16]. As a commonly used Traditional Chinese Medicine (TCM), the side effects from PMR and its preparations have been observed clinically worldwide and include embryonic toxicity, nephrotoxicity, hepatotoxicity, lung toxicity, and hepatic adverse events such as acute toxic hepatitis [17,18,19,20,21]. PMR and PMRP may have paradoxical effects on the liver in terms of both hepatoprotection and hepatotoxicity [22], but the underlying mechanisms remain unknown.
Current research on the toxicity of PMR has mainly focused on the chemical constituents causing liver damage [23, 24]. Long-term use of TCM may lead to liver damage, mainly due to the accumulation of chemical components. However, the chemical composition is complex, and its content varies greatly; thus, it has been difficult to characterize the overall liver damage from single or multiple chemical components. There has been no research on the different metabolomic profiles between PMR- and PMRP-treated rats. Detection of the differences in endogenous metabolites following short-term administration can reveal the different types of metabolomic data.
Untargeted metabolomics methods have been used to simultaneously detect several classes of the metabolome, including changes in endogenous metabolites that are linked to toxicity. Non-biased detection platform is a powerful tool for metabolomic research [25]. To investigate the different endogenous metabolites between PMR and PMPR, ultra-high performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC/Q-TOF-MS) was performed. We also identified the changes in endogenous metabolites and elucidated the relative pathways.
Methods
Animals and ethical statement
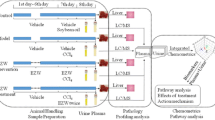
Male Sprague–Dawley rats (weighing 180–220 g) were purchased from the laboratory animal center of Academy of Military Medical Science (No. SCXK-(Jun) 2012–0004; Beijing, China). The total number of rats was 15. Rats were randomly divided into 3 groups of 5 each. All animals were handled under strict observation of rules outlined in the National Institutes of Health guide for the care and use of laboratory animals. All animals were kept housed in a barrier system with regulated temperature (25–28 °C), humidity (50–60%), and a light–dark cycle of 12 h per day. All rats were acclimated for 1 week before the experiment. All animal procedures were approved by the animal care and use committee of Academy of Military Medical Science (No. IACUC-AMMS-13-2017-012, May 2017).
Preparation of PMR and PMRP
PMR and PMRP were purchased from Beijing Tong Ren Tang Medicinal Materials Co., Ltd. (Beijing, China) (Fig. 1a, b). They were identified by Ma Baiping, a pharmacognosy professor from the Beijing Institute of Radiation Medicine (Beijing, China). PMRP was processed from PMR in strict accordance with the method of the Chinese Pharmacopoeia, 2015 edition [1]. After the PMR and PMRP were accurately weighed, a tenfold volume of 70% ethanol (including 30% distilled water) was added, and they were soaked for 1 h. Then they were boiled for 1 h and filtered, after which another tenfold volume of the same solution was added, following by boiling again for 1 h. Both set of filtrates were combined and vortexed to 1 g/mL. We adopted a dose of 1920 mg/kg/day (crude drug extract) for administration. Rats were administered the drugs orally for 28 days [26].
Sample collection and preparation
All rats were bled from the orbital venous plexus to collect blood samples after 28 days of drug administration. Blood samples were coagulated for 10 min at room temperature and centrifuged at 3000 rpm for 10 min. The supernatants were immediately stored at − 80 °C. Prior to analysis, a 200 μL aliquot of serum samples was thawed at 4 °C, followed by the addition of 800 μL methanol to precipitate the proteins. The solution mixture was vortexed for 30 s and centrifuged at 13,000 rpm for 15 min at 4 °C. The supernatant was transferred to an Eppendorf tube and evaporated to full dryness at 4 °C under stream of nitrogen. The residue was dissolved with 200 μL methanol, followed by vortexing for 60 s and centrifuging at 13,000 rpm for 15 min. Then 50 μL supernatant was transferred to a sampling vial for UPLC-Q/TOF-MS analysis.
Biochemical indices assay, HE (hematoxylin and eosin) staining and IHC (immunohistochemistry, IHC) staining
The activities of alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), lactate dehydrogenase (LDH), and total bilirubin (TBIL) were determined according to the manufacturer’s instructions using a biochemical analyzer (Rayto, Shenzhen, China). For HE staining, liver samples were stored in 4% paraformaldehyde solution. Three paraffin-embedded sections at 4–5 mm per specimen were prepared and stained with hematoxylin and eosin. For IHC Staining, the liver tissue blocks were cut into 4-mm slides and placed in an oven for 2 h at 65 °C. Xylene and graded concentrations of ethanol were used for sequential washing of the sections. Endogenous peroxidase activity and nonspecific staining were blocked by 3% H2O2 for 15 min and 3% bovine serum albumin (BSA; Roche) for 1 h, respectively. Incubation with the primary antibodies was performed at room temperature for 30 min and then at 4 °C overnight. The concentrations and sources of the antibodies used in this study were as follows: Anti-CD3 antibody (ab135372) (1:150) and Anti-CD4 antibody (ab183685) (1:200) were purchased from Abcam (Shanghai, China). Tissue samples were washed with PBS three times and stained with the secondary antibody (1:200) at 37 °C for 1 h, after which they were visualized by 3,3-diaminobenzidine staining, counterstained with 10% Mayer’s hematoxylin solution, dehydrated, mounted, dried and observed [27]. Each section was observed with an Olympus microscope and the Mshot Image Analysis System.
Quality control analysis
In order to monitor UPLC-Q/TOF-MS detecting system stability and reproducibility, we used quality control (QC) samples to test throughout the whole progress. QC samples were prepared by pooling equal volumes of each serum sample. The pretreat method of QC samples was the same as with the other samples. Three QC samples were injected at regular intervals among three samples throughout the analytical run. The features were selected based on their coefficients of variation (CVs) with QC samples; features with CVs over 15% were eliminated.
Metabolomics analysis
For the positive and negative ion mode, mobile phase A was acetonitrile/water (60/40), and mobile phase B was isopropanol/acetonitrile (90/10). Both A and B contained 0.1% formic acid and 10 mmol/L ammonium acetate. The column was HSS T3 column (2.1 × 100 mm, 1.8 µm of water) operated at 45 °C. For HILIC mode, mobile phase A was acetonitrile, and mobile phase B was water, both A and B contained 0.1% formic acid and 10 mmol/L ammonium acetate. The column was BEH amide column (2.1 × 100 mm, 1.7 µm water) operated at 40 °C. Raw data were obtained from MarkerView software (version 1.2.1.1; Applied Biosystems, Framingham, MA, USA) and were extracted by excluding missing values based on the 80% rule. Total peak area normalization was used for the retained peaks. Ions with relative standard deviations below 30% were input into SIMCA-P (version 11.0; Umetrics AB, Umea, Sweden) software. Principal component analysis (PCA) and orthogonal partial least square-discriminant analysis (OPLS-DA) modes were established. The OPLS-DA mode was assessed by the intercepts of R2 and Q2 in the permutation test to avoid overfitting. Metabolite identification was based on obtained results, database, and standards verification. Nonparametric tests were used to determine significantly changed ions (P < 0.05). For visualization of significant changes, clustering-heatmap analysis, correlational analysis, relative-intensities analysis, and pathway analysis were performed using MetaboAnalyst 4.0.
Statistical analysis
Serum biochemical data were analyzed using GraphPad Prism 6.0. To evaluate the statistically significant differences among multiple treatments for given parameters, one-way analysis of variance with Dunnett’s multiple comparison test was used for comparison among various groups. Differences with P values < 0.05 were considered statistically significant.
Results
Levels of biochemical indexes and histological results
Levels of ALT and AST are regarded as indicators of liver injury. Compared with the control group, ALT and AST levels were markedly increased in the PMR and PMRP groups (Fig. 2a, b). LDH was markedly increased in the PMR and PMRP group (Fig. 2c). In contrast, the levels of ALP and TBIL had no obvious changes (Fig. 2c, e). These results suggest that PMR and PMRP may have slight liver damaging effects. Histological analysis showed that liver tissues in the control group exhibited a normal cellular structure with neatly organized liver lobules, liver cords, liver sinusoids, and a clear three-pipeline structure of the portal area (Fig. 3a). Liver tissues in the PMR-treated group presented with morphological tissue degeneration including necrosis, and inflammatory cell infiltration (Fig. 3b). Liver tissues in the PMRP group of rats exhibited pyknosis, reduced intercellular space, blurred cell margins, and inflammatory cell infiltration (Fig. 3c). CD3 and CD4 cells are important T immune cells, they play an important role in chronic hepatitis B, hepatic infections, and immunosuppression [28,29,30]. If the liver tissue has an inflammatory response, the CD3 and CD4 cells will be highly expressed. Immunohistochemical results in this study showed that there was almost no expression of CD3 in liver tissue in the control group (Fig. 3d), and there was significant expression in the RPMP group (Fig. 3e), and higher expression in the PMR group (Fig. 3f). In terms of the expression of CD4, liver tissue in the control group showed a little expression (Fig. 3g), and there was significant expression in the RPMP group (Fig. 3h), and higher expression in the PMR group (Fig. 3i). The results showed that both PMR and PMR had mild liver damage, and the liver damage of PMR was stronger than PMRP.
Photomicrographs of representative sections of the livers of SD mice with H&E staining and IHC. a Control group (saline); b PMRP group; c PMR group. H&E staining (magnification: ×200). CD3 antibody: d control group (saline); e PMRP group; f PMR group, IHC staining (magnification: ×400); CD4 antibody: g control group (saline); h PMRP group, i PMR group. IHC staining (magnification: ×400)
Samples quality control assessment
Quality control (QC) and other experimental samples were analyzed using unsupervised PCA (principal component analysis). QC samples were the same ingredients and they should be brought together in a PCA score map. The PCA scores of ESI positive ion mode (Fig. 4a), negative ion mode (Fig. 4b), and HILIC mode (Fig. 4c) are showed. The relative clustering of the QC samples showed that the detecting system reproducibility was good, meaning the experimental data had good scientific quality and reliability.
Distinct metabolomic profiles between the PMR and PMRP groups
Total ion chromatography (TIC) was used to acquire the metabolomic profiles, and the TIC results revealed differences between the groups (Fig. 5). PCA was employed to reveal the metabolic changes. A PCA score plot showed a clear trend of group clustering. Three modes’ parameters were R2X of 0.761 and Q2 of 0.254 (Fig. 6a), R2X of 0.927 and Q2 of 0.798 (Fig. 7a), and R2X of 0.761 and Q2 of 0.254 (Fig. 8a). In the score plot obtained by PCA, the two groups were located further from each other, indicating a clear differentiation. We used OPLS-DA to evaluate and maximize the discrimination between the two groups. The variation values of OPLS-DA were R2Y of 0.976 and Q2 of 0.803 (Fig. 6b), R2Y of 0.998 and Q2 of 0.988 (Fig. 7b), and R2Y of 0.999 and Q2 of 0.970 (Fig. 8b). Plot distribution clearly displayed the observations with a high absolute value of p(corr) and absolute value of coefficients. OPLS-DA score plots indicated that clustering in the PMR group was well separated from the PMRP group. The permutation plot showed that all blue Q2 values to the left were lower than the original points to the right, indicating that the original models were valid (Figs. 6c, 7c, 8c). The parameter indicated that these metabolites had high sensitivity and specificity for mode identification, and thus could be used as biomarkers. Differences in variables between groups were revealed by the S-plot of the OPLS-DA mode (Figs. 6d, 7d, 8d). These results indicated that the test modes were valid and reliable.
Typical total ion chromatography. a Control group with positive mode, b PMR group with positive mode, c PMRP group with positive mode, d control group with negative mode, e PMR group with negative mode, f PMRP group with negative mode, g control group with Hilic mode, h PMR group with Hilic mode, i PMRP group with Hilic mode
Identification and analysis of metabolites
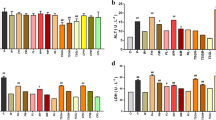
To identify potential metabolic biomarkers, the variable importance in projection (VIP) values of all metabolites from OPLS-DA were determined. Metabolites with a VIP value > 1 were selected as potential biomarkers. Comparisons of the peak intensity of potential biomarkers were performed (Fig. 9), and 29 serum homogenate metabolites were identified as biomarkers. In the positive ion mode, the following 17 biomarkers were identified: PC(14:0/18:2(9Z,12Z)), PC(18:3(6Z,9Z,12Z)/16:0), SM(d18:0/16:1(9Z)), SM(d18:0/18:1(11Z)), PC(20:4(8Z,11Z,14Z,17Z)/18:2(9Z,12Z)), PC(P-18:0/20:5(5Z,8Z,11Z,14Z,17Z)), PC(o-16:0/20:4(8Z,11Z,14Z,17Z)), PC(22:6(4Z,7Z,10Z,13Z,16Z,19Z)/18:0), PC(16:0/18:1(11Z)), PC(20:4(8Z,11Z,14Z,17Z)/18:0), PC(18:0/18:1(11Z)), SM(d18:1/22:1(13Z)), LysoPC(20:2(11Z,14Z)), LysoPC(20:1(11Z)), LysoPC(22:0), SM(d18:1/14:0), and LysoPC(24:0). In the negative ion mode, the following nine biomarkers were identified: chenodeoxycholic acid, myristic acid, alpha-linolenic acid, (Z)-9-heptadecenoic acid, 8,11,14-eicosatrienoic acid, oleic acid, heptadecanoic acid, adrenic acid, and eicosadienoic acid. In the Hilic mode, the three biomarkers identified were betaine, taurine, and ornithine. All of the identified biomarkers are summarized in Table 1.
Pathway analysis and biological interpretation
To determine the metabolic pathways, we performed pathway analysis using MetaboAnalyst 4.0. The P value and pathway impact were calculated from metabolic pathway enrichment analysis. The P value threshold was set at 0.01, and values above this threshold were filtered as significant pathways. To explore the possible different metabolic pathways, Human Metabolome Database (HMDB) numbers of the 29 biomarkers were imported into MetaboAnalyst 4.0 and the following five metabolic pathways were identified: alpha-linolenic-acid metabolism, taurine and hypotaurine metabolism, glycerophospholipid metabolism, arginine and proline metabolism, and primary bile acid (BA) biosynthesis (Tables 1, 2, Figs. 10, 11). To gain a better understanding of the interaction between metabolic pathways, a metabolite-to metabolite correlation analysis was performed, and the results are illustrated by correlation heatmap and hierarchal clustering (Figs. 9, 12). The results showed that the PMRP group had more metabolic changes. Relative intensity analysis is often used to investigate the magnitude of change in biomarkers. Compared with the PMR group, the levels in the PMPR group of PC(14:0/18:2(9Z,12Z)), PC(18:3(6Z,9Z,12Z)/16:0), SM(d18:0/16:1(9Z)), PC(20:4(8Z,11Z,14Z,17Z)/18:2(9Z,12Z)), PC(P-18:0/20:5(5Z,8Z,11Z,14Z,17Z)), PC(22:6(4Z,7Z,10Z,13Z,16Z,19Z)/18:0), PC(16:0/18:1(11Z)), PC(18:0/18:1(11Z)), SM(d18:1/22:1(13Z)), LysoPC(22:0), SM(d18:1/14:0), and LysoPC(24:0) were increased; whereas the levels of LysoPC(20:2(11Z,14Z)), LysoPC(20:1(11Z)), myristic acid, alpha-linolenic acid, (Z)-9-heptadecenoic acid, 8,11,14-eicosatrienoic acid, oleic acid, heptadecanoic acid, eicosadienoic acid, betaine, taurine, and ornithine were decreased (Fig. 13).
Five most impacted pathways. a Pathway of alpha-linolenic acid metabolism. b Pathway of taurine and hypotaurine metabolism. c Pathway of glycerophospholipid metabolism. d Pathway of “arginine and proline metabolism”. e Pathway of primary bile acid biosynthesis. Labels within small boxes correspond to KEGG identifiers for metabolites. In a the metabolites were PC(16:0/16:0) (C00157, HMDB0000564), alpha-linolenic acid (C06427, HMDB0001388). In b the metabolite was taurine (C00245, HMDB0000251). In c the metabolites were PC(16:0/16:0) (C00157, HMDB0000564), LysoPC(18:1(9Z)) (C04230, HMDB0002815). In d the metabolite was ornithine (C00077, HMDB0000214). In e the metabolites were taurine (C00245, HMDB0000251), chenodeoxycholic acid (C02528, HMDB0000518). Those markers were hit and colored in red
Discussion
As two common TCMs, PMR and PMRP have been used in clinical practice and in the food industry for many years in China and other countries. PMR has paradoxical effect in terms of both hepatoprotective and hepatotoxic effects [31]. Recently, many side effects of PMR have been reported worldwide [17,18,19, 32,33,34]. PMR may cause liver damage and even death, usually due to long-term use or overdose. Liver damage associated with PMR is reversible, and the majority of liver damage cases can be treated.
Different processing methods for PMR cause different effects on liver damage, as processing greatly changes the chemical composition of PMR and influences the distribution of compounds in vivo [31]. The order of toxicity is as follows: PMR ethanol extract > PMR water extract > PMRP ethanol extract > PMRP water extract. Liver toxicity may be associated with anthraquinone, emodin-O-(malonyl)-hex, emodin-O-glc, emodin, emodin-8-O-glc, emodin-O-(acetyl)-hex, and emodin-O-hex-sulphate [24]. Processing of PMR can reduce its effects on both cell proliferation and enzyme secretion from liver cells. Three major chemical constituents, including THSG, physcion, and emodin, have shown no cytotoxicity against L02 liver cell lines. Analysis of the relationship between the chemical constituents and cytotoxicity revealed that THSG and physcion likely have attenuating effects on emodin, but the mechanisms are still under investigation [35].
Although hepatotoxic cases linked to PMR have been frequently reported, the appropriate clinical diagnosis biomarkers are still unknown. The long-term use of high doses of PMR can potentially damage the liver [26]. One study identified BAs and urine T-β muricholic acid (MCA) as promising metabolic biomarkers to facilitate the clinical monitoring of PMR-induced hepatotoxicity, and urine T-β MCA served as a potential therapeutic target [36]. The perturbation of nine BAs is associated with PMR-induced liver injury, In addition, glycodeoxycholic acid in bile and hyodeoxycholic acid in serum may be potential biomarkers [37]. PMR can upregulate key enzymes for the biosynthesis of cholesterol and BA, which poses the risk of cholestatic liver injury [38]. Computational system toxicology approach can reveal the possible toxic components of TCM, which is helpful for discovering the hepatotoxic mechanisms. Using this method, seven compounds in PMR including emodin, quercetin, apigenin, resveratrol, gallic acid, kaempferol, and luteolin were found to be clearly associated with hepatotoxicity. Multiple interactions between apoptosis and metabolism may underlie PMR-induced liver injury including glutathione metabolism, cytochrome P450 metabolism, and the p53 pathway [39]. Computational toxicology methods is using software to analyze existing data, but the disadvantage is that the authenticity of the database and the lack of experimental verification of the calculation results. Analysis of urine metabolomics is the end point of drug action, absorption of drugs into the bloodstream through the stomach directly affects the changes in metabolites in the body, especially endogenous metabolites. Compared with other previous studies, our study focused on the comparative comparison of effects between PMR and PMRP on the metabolomics on rats. We adopted a non-targeted metabolomic method to analyze the endogenous metabolomics changes between the PMR and PMPR groups. A total of 29 biomarkers were confirmed, in five metabolic pathways, including alpha linolenic acid metabolism, taurine and hypotaurine metabolism, glycerophospholipid metabolism, arginine and proline metabolism, and primary BA biosynthesis pathways. Alpha-linolenic acid (18:3n-3) is essential in the human diet. It is the substrate for the synthesis of longer chain, more unsaturated n-3 fatty acids such as eicosapentaenoic acid (20:5n-3) and docosahexaenoic acid (22:6n-3), which are required for tissue function [40]. Our results showed that alpha linolenic acid metabolism and primary BA biosynthesis were two significantly different metabolic pathways between the PMR and PMPR groups. Coincidentally, clinical research has demonstrated that primary BA biosynthesis and alpha linolenic acid metabolic pathways are also related to the severity of drug-induced liver injury [41]. Taurine and hypotaurine metabolism is an important metabolic pathway. Taurine has a variety of physiological functions such as eye and brain development, immune function, reproduction, osmotic adjustment, and antioxidant and anti-inflammatory activities [42]. In our study, taurine and hypotaurine metabolism was a significantly different pathway between the PMR and PMPR groups. BAs can regulate lipid and glucose metabolism; modulate inflammation in the liver and other tissues; and serve important roles in cholesterol metabolism, lipid digestion, host-microbe interactions, and regulatory pathways in the human host [43, 44]. PMR can significantly injure bile-duct epithelial cells, intervene in liver cell functions, change bile compositions in rats, and induce cholestasis without severe liver injury. Cholestasis often occurs in PMR-induced hepatotoxicity in clinical, but the pathogenesis remains unknown. Betaine, taurine, and ornithine were identified as biomarkers. Primary BA biosynthesis was the most changed pathway. PMR can interfere with the process of synthesis and elimination of BAs, resulting in an overload of BA content in the liver, which leads to liver injury [38]. From the perspective of metabolomics, the results verified the effects of PMR on the metabolism of BAs.
Taurine and hypotaurine metabolism, glycerophospholipid metabolism, and arginine and proline metabolism were first reported to be differential pathways. Taurine can be bound by BAs, and is not present in a free state in the body. It has anti-oxidation properties and regulates osmotic pressure, BA binding, ion movement, and nerve transmission and plays an important role in BA metabolism [45,46,47]. Arginine participates in the ornithine cycle, promotes the formation of urea, and converts the ammonia produced into non-toxic urea through the ornithine cycle, which is discharged from urine. Proline is not only an ideal osmotic adjustment substance but also acts as a protective substance for membranes and enzymes and is a free radical scavenger to protect plants from growth under osmotic stress. Proline plays regulatory roles in cytoplasmic-osmotic balance and accumulation of important osmotic-adjustment substances in the vacuole [48]. Glycerol phospholipids are the most abundant phospholipids. In addition to forming biofilms, they are also the components of bile and membrane surfactants and participate in cell membrane recognition and signal transduction. Ten PCs, four Lyso PCs, and three SMs were identified as biomarkers, and confirmed to be important components of the five metabolic pathways.
Conclusions
UPLC-Q/TOF-MS was successfully applied to investigate the significant changes in serum between PMR- and PMRP-treated rats. Slight liver damage was induced by PMR, but was not observed in the PMRP group. Subsequently, mechanisms of differences in endogenous metabolites were investigated. A total 29 annotated metabolites were significantly changed, and identified as biomarkers. Furthermore, the five most related pathways were also determined by inputting the HMDB and KEEG numbers of these biomarkers into Metaboanalyst 4.0. This study provided a comprehensive description of metabolome changes between PMR- and PMRP-treated rats. However, the precise mechanism requires further study, which are currently. We plan to explore the effects of metabolite identification on pathway exploration to determine the mechanism underlying the hepatotoxicity caused by PMR and PMRP.
Data availability statement
All data used to support the findings of this study are available from the corresponding author upon request.
Abbreviations
- PMR:
-
Polygoni Multiflori Radix
- PMRP:
-
Polygoni Multiflori Radix Preparata
- DILI:
-
drug-induced liver injury
- HILI:
-
herb-induced liver injury
- UPLC-Q/TOF-MS:
-
ultra-performance liquid chromatography quadrupole time-of-flight mass spectrometry
- MS:
-
mass spectrometry
- ALT:
-
alanine aminotransferase
- AST:
-
aspartate aminotransferase
- ALP:
-
alkaline phosphatase
- LDH:
-
lactate dehydrogenase
- TBIL:
-
total bilirubin
- HE:
-
hematoxylin and eosin
- IHC:
-
immunocytochemistry
- QC:
-
quality control
- ANOVA:
-
one-way analysis of variance
- PCA:
-
principal component analysis
- PLS-DA:
-
partial least-squares-discriminate analysis
- OPLS-DA:
-
orthogonal to partial least-squares discriminate analysis
- TIC:
-
total ion chromatogram
- VIP:
-
variable importance in the projection
- MetPA:
-
metabolic pathway analysis
References
Committee CP. Chinese pharmacopoeia (2015 edition). Beijing: China Chemical Industry Press; 2015. p. 175–7.
Lin L, Ni B, Lin H, Zhang M, Li X, Yin X, Qu C, Ni J. Traditional usages, botany, phytochemistry, pharmacology and toxicology of Polygonum multiflorum Thunb.: a review. J Ethnopharmacol. 2015;159:158–83.
Niu S, Chen J, Duan F, Song Q, Qin M, Wang Z, Liu J. Possible mechanism underlying the effect of Heshouwuyin, a tonifying kidney herb, on sperm quality in aging rats. BMC Complement Altern Med. 2014;14:250.
Chen J, Wang Y, Hui C, Xi Y, Liu X, Qi F, Liu H, Wang Z, Niu S. Mechanisms of Heshouwuyin in regulating apoptosis of testicular cells in aging rats through mitochondrial pathway. BMC Complement Altern Med. 2016;16(1):337.
Liu LP, Liao ZP, Yin D, Li WD, Liu D, Li Q, Huang QR, Yang YF, He M. The protective effects of Polygonum multiflorum stilbeneglycoside preconditioning in an ischemia/reperfusion model of HUVECs. Acta Pharmacol Sin. 2010;31(4):405–12.
Chen HF, Chen YH, Liu CH, Wang L, Chen X, Yu BY, Qi J. Integrated chemometric fingerprints of antioxidant activities and HPLC-DAD-CL for assessing the quality of the processed roots of Polygonum multiflorum Thunb. (Heshouwu). Chin Med. 2016;11:18.
Zhu W, Xue X, Zhang Z. Ultrasonic-assisted extraction, structure and antitumor activity of polysaccharide from Polygonum multiflorum. Int J Biol Macromol. 2016;91:132–42.
Li H, Xiang L, Yang N, Cao F, Li C, Chen P, Ruan X, Feng Y, Zhou N, Wang X. Zhiheshouwu ethanol extract induces intrinsic apoptosis and reduces unsaturated fatty acids via SREBP1 pathway in hepatocellular carcinoma cells. Food Chem Toxicol. 2018;119:169–75.
Kim HN, Kim YR, Jang JY, Choi YW, Baek JU, Hong JW, Choi YH, Shin HK, Choi BT. Neuroprotective effects of Polygonum multiflorum extract against glutamate-induced oxidative toxicity in HT22 hippocampal cells. J Ethnopharmacol. 2013;150(1):108–15.
Li X, Matsumoto K, Murakami Y, Tezuka Y, Wu Y, Kadota S. Neuroprotective effects of Polygonum multiflorum on nigrostriatal dopaminergic degeneration induced by paraquat and maneb in mice. Pharmacol Biochem Behav. 2005;82(2):345–52.
Han MN, Lu JM, Zhang GY, Yu J, Zhao RH. Mechanistic studies on the use of Polygonum multiflorum for the treatment of hair graying. Biomed Res Int. 2015;2015:651048.
Huang C-H, Horng L-Y, Chen C-F, Wu R-T. Chinese herb Radix Polygoni Multiflori as a therapeutic drug for liver cirrhosis in mice. J Ethnopharmacol. 2007;114(2):199–206.
Wang M, Zhao R, Wang W, Mao X, Yu J. Lipid regulation effects of Polygoni Multiflori Radix, its processed products and its major substances on steatosis human liver cell line L02. J Ethnopharmacol. 2012;139(1):287–93.
Wang W, He Y, Lin P, Li Y, Sun R, Gu W, Yu J, Zhao R. In vitro effects of active components of Polygonum multiflorum Radix on enzymes involved in the lipid metabolism. J Ethnopharmacol. 2014;153(3):763–70.
Lin P, He YR, Lu JM, Li N, Wang WG, Gu W, Yu J, Zhao RH. In vivo lipid regulation mechanism of polygoni multiflori radix in high-fat diet fed rats. Evid Based Complement Altern Med. 2014;2014:642058.
Zhang M, Lin L, Lin H, Qu C, Yan L, Ni J. Interpretation the hepatotoxicity based on pharmacokinetics investigated through oral administrated different extraction parts of Polygonum multiflorum on rats. Front Pharmacol. 2018;9:505.
Cho HC, Min HJ, Ha CY, Kim HJ, Kim TH, Jung WT, Lee OJ, Bae IG. Reactivation of pulmonary tuberculosis in a patient with Polygonum multiflorum Thunb-induced hepatitis. Gut Liver. 2009;3(1):52–6.
Laird AR, Ramchandani N, Degoma EM, Avula B, Khan IA, Gesundheit N. Acute hepatitis associated with the use of an herbal supplement (Polygonum multiflorum) mimicking iron-overload syndrome. J Clin Gastroenterol. 2008;42(7):861–2.
Furukawa M, Kasajima S, Nakamura Y, Shouzushima M, Nagatani N, Takinishi A, Taguchi A, Fujita M, Niimi A, Misaka R. Toxic hepatitis induced by show-wu-pian, a Chinese herbal preparation. Intern Med. 2010;49(15):1537.
Li X, Qu C, He Q, Chen W, Zhang X, Liu X, Liu Y, Tang Y. Acute hepatitis induced by a Chinese herbal product Qibao Meiran Wan: a case study. Int J Clin Exp Med. 2015;8(7):11624–7.
Cardenas A, Restrepo JC, Sierra F, Correa G. Acute hepatitis due to shen-min: a herbal product derived from Polygonum multiflorum. J Clin Gastroenterol. 2006;40(7):629–32.
Lin L, Li H, Lin H, Zhang M, Qu C, Yan L, Yin X, Ni J. Application of iTRAQ-based quantitative proteomics approach to identify deregulated proteins associated with liver toxicity induced by Polygonum multiflorum in rats. Cell Physiol Biochem. 2017;43(5):2102–16.
Yang JB, Li WF, Liu Y, Wang Q, Cheng XL, Wei F, Wang AG, Jin HT, Ma SC. Acute toxicity screening of different extractions, components and constituents of Polygonum multiflorum Thunb. on zebrafish (Danio rerio) embryos in vivo. Biomed Pharmacother. 2018;99:205–13.
Lin L, Lin H, Zhang M, Ni B, Yin X, Qu C, Ni J. A novel method to analyze hepatotoxic components in Polygonum multiflorum using ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry. J Hazard Mater. 2015;299:249–59.
Cuykx M, Rodrigues RM, Laukens K, Vanhaecke T, Covaci A. In vitro assessment of hepatotoxicity by metabolomics: a review. Arch Toxicol. 2018;92(10):3007–29.
Xia XH, Yuan YY, Liu M. The assessment of the chronic hepatotoxicity induced by Polygoni Multiflori Radix in rats: a pilot study by using untargeted metabolomics method. J Ethnopharmacol. 2017;203:182–90.
Xue R, Chen L, Zhang C, Fujita M, Li R, Yan SM, Ong CK, Liao X, Gao Q, Sasagawa S, et al. Genomic and transcriptomic profiling of combined hepatocellular and intrahepatic cholangiocarcinoma reveals distinct molecular subtypes. Cancer Cell. 2019;35(6):932–47.
Ishii T, Yamaguchi J, Gu W, Hashimoto T, Yamamoto T, Kanematsu T. CD4 T-cell regulation of cytotoxic T cells during the induction phase of liver-induced spontaneous tolerance in rats. Surg Today. 2005;35(6):473.
Zhang JY, Zheng Z, Fang L, Zou ZS, Xu RN, Lei J, Fu JL, Feng S, Ming S, Wang HF. Interleukin-17-producing CD4+ T cells increase with severity of liver damage in patients with chronic hepatitis B. Hepatology. 2010;51(1):81–91.
Ulrike P, Maini MK, Knolle PA. Living in the liver: hepatic infections. Nat Rev Immunol. 2012;12(3):201–13.
Li H, Wang X, Liu Y, Pan D, Wang Y, Yang N, Xiang L, Cai X, Feng Y. Hepatoprotection and hepatotoxicity of Heshouwu, a Chinese medicinal herb: context of the paradoxical effect. Food Chem Toxicol. 2017;108(Pt B):407–18.
Park GJ, Mann SP, Ngu MC. Acute hepatitis induced by Shou-Wu-Pian, a herbal product derived from Polygonum multiflorum. J Gastroenterol Hepatol. 2010;16(1):115–7.
Mazzanti G, Battinelli L, Daniele C, Mastroianni CM, Lichtner M, Coletta S, Costantini S. New case of acute hepatitis following the consumption of Shou Wu Pian, a Chinese herbal product derived from Polygonum multiflorum. Ann Intern Med. 2004;140(7):W30.
Lei X, Chen J, Ren J, Li Y, Zhai J, Mu W, Zhang L, Zheng W, Tian G, Shang H. Liver damage associated with Polygonum multiflorum Thunb.: a systematic review of case reports and case series. Evid Based Complement Altern Med. 2015;2015:459749.
Yu J, Xie J, Mao XJ, Wang MJ, Li N, Wang J, Zhaori GT, Zhao RH. Hepatoxicity of major constituents and extractions of Radix Polygoni Multiflori and Radix Polygoni Multiflori Praeparata. J Ethnopharmacol. 2011;137(3):1291–9.
Zhao DS, Jiang LL, Fan YX, Dong LC, Ma J, Dong X, Xu XJ, Li P, Li HJ. Identification of urine tauro-beta-muricholic acid as a promising biomarker in Polygoni Multiflori Radix-induced hepatotoxicity by targeted metabolomics of bile acids. Food Chem Toxicol. 2017;108(Pt B):532–42.
Dong Q, Li N, Li Q, Zhang CE, Feng WW, Li GQ, Li RY, Tu C, Han X, Bai ZF, et al. Screening for biomarkers of liver injury induced by Polygonum multiflorum: a targeted metabolomic study. Front Pharmacol. 2015;6:217.
Jiang LL, Zhao DS, Fan YX, Yu Q, Lai YS, Li P, Li HJ. Transcriptome analysis to assess the cholestatic hepatotoxicity induced by Polygoni Multiflori Radix: up-regulation of key enzymes of cholesterol and bile acid biosynthesis. J Proteom. 2018;177:40–7.
Wang YY, Li J, Wu ZR, Zhang B, Yang HB, Wang Q, Cai YC, Liu GX, Li WH, Tang Y. Insights into the molecular mechanisms of Polygonum multiflorum Thunb-induced liver injury: a computational systems toxicology approach. Acta Pharmacol Sin. 2017;38(5):719–32.
Burdge GC. Metabolism of alpha-linolenic acid in humans. Prostaglandins Leukot and Essent Fatty Acids. 2006;75(3):161–8.
Xie Z, Chen E, Ouyang X, Xu X, Ma S, Ji F, Wu D, Zhang S, Zhao Y, Li L. Metabolomics and cytokine analysis for identification of severe drug-induced liver injury. J Proteome Res. 2019;18(6):2514–24.
Park E, Park SY, Cho IS, Kim BS, Schuller-Levis G. A novel cysteine sulfinic acid decarboxylase knock-out mouse: taurine distribution in various tissues with and without taurine supplementation. Adv Exp Med Biol. 2017;975(Pt 1):461–74.
Jiao N, Baker SS, Chapa-Rodriguez A, Liu W, Nugent CA, Tsompana M, Mastrandrea L, Buck MJ, Baker RD, Genco RJ, et al. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut. 2018;67(10):1881–91.
Staley C, Weingarden AR, Khoruts A, Sadowsky MJ. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl Microbiol Biotechnol. 2017;101(1):47–64.
Jacobsen JG, Smith LH. Biochemistry and physiology of taurine and taurine derivatives. Physiol Rev. 1968;48(2):424–511.
Danielsson H, Sjovall J. Bile acid metabolism. Annu Rev Biochem. 1975;44:233–53.
Hussy N, Deleuze C, Desarmenien MG, Moos FC. Osmotic regulation of neuronal activity: a new role for taurine and glial cells in a hypothalamic neuroendocrine structure. Progr Neurobiol. 2000;62(2):113–34.
Fichman Y, Gerdes SY, Kovacs H, Szabados L, Zilberstein A, Csonka LN. Evolution of proline biosynthesis: enzymology, bioinformatics, genetics, and transcriptional regulation. Biol Rev Camb Philos Soc. 2015;90(4):1065–99.
Acknowledgements
The authors would like to thank Dr. Longlong Xu and Mr. Xiaofei Luo for providing technical support for the UPLC-Q/TOF-MS analysis and data analysis.
Funding
This study was supported by the National Science and Technology Major Project (No. 2015ZX09501004-003-003).
Author information
Authors and Affiliations
Contributions
ZZ and YG designed the experiments. ZZ, LY and XH performed the experiments. ZZ analyzed the data, while the other authors assisted in experiments. ZZ wrote the manuscript. ZZ, LY, XH and YG discussed the results and revised the final manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhang, Z., Yang, L., Huang, X. et al. Metabolomics profiling of Polygoni Multiflori Radix and Polygoni Multiflori Radix Preparata extracts using UPLC-Q/TOF-MS. Chin Med 14, 46 (2019). https://doi.org/10.1186/s13020-019-0268-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13020-019-0268-3