Abstract
Objective
Delayed fracture healing is a common complication of fractures that significantly impacts human health. This study aimed to explore the role of LINC00339 (lncRNA) in delayed fracture healing to provide new directions for its treatment.
Methods
This study included 82 patients with fractures healing in a normal manner and 90 patients experiencing delayed fracture healing. Levels of LINC00339, miR-16-5p, and osteogenic marker-related mRNAs were measured using RT-qPCR. The predictive potential of LINC00339 for delayed fracture healing was validated using ROC curve analysis. The interaction between LINC00339 and miR-16-5p was validated using dual-luciferase reporter assays and RIP experiments. CCK-8 was used to assess cell proliferation, and apoptosis rates were measured by flow cytometry.
Results
LINC00339 was significantly upregulated in delayed fracture healing patients and exhibited strong predictive ability for this condition. Overexpression of LINC00339 inhibited osteoblast proliferation, promoted apoptosis, and reduced mRNA levels of osteogenic markers (P < 0.05). miR-16-5p was recognized as a target mRNA of LINC00339, with LINC00339 exerting negative regulation on miR-16-5p, while overexpression of miR-16-5p mitigated the inhibitory effects of LINC00339 on fracture healing (P < 0.05).
Conclusion
This research indicated that LINC00339 may serve as a diagnostic marker for delayed fracture healing and revealed the function of the LINC00339/miR-16-5p axis on fracture healing by regulating osteoblasts.
Similar content being viewed by others
Introduction
Common types of fractures include femur fractures, hip fractures and patella fractures [1]. Research has shown that fractures have become a major threat to human health, with fragile fractures making significant contributions [2]. The healing of fractures is a highly complex physiological process influenced by various factors such as age, treatment methods, and chemical factors like hormones [3, 4], and there is a certain probability of delayed healing during the recovery process [5]. Furthermore, osteoporosis increases the risk of fractures and raises the probability of delayed healing [6, 7]. Osteoblast activity is crucial in the healing of fractures, aiding in the formation of callus at the fracture site, leading to new bone formation and the restoration of bone function [8]. Exploring the mechanisms of delayed fracture healing can establish a theoretical foundation for the prevention and treatment of delayed fracture healing.
Long non-coding RNAs (lncRNAs) are RNA molecules longer than 200 bp that are involved in numerous physiological processes, serving crucial functions [9]. It was also found that lncRNAs are involved in the regulation of the fracture healing process [10, 11]. Studies have found that knocking out lncRNA MIAT can promote osteoblast proliferation and differentiation [12]; while some lncRNAs have a positive effect on maintaining bone balance, for example, enhanced expression of lncRNA NORAD can inhibit cell apoptosis and accelerate fracture healing [13]. It is evident that different lncRNAs play varying roles in the fracture healing process. LINC00339 is a less understood lncRNA, with previous research mainly focusing on its role in cancer [14, 15]. Zhang et al. discovered an upregulation of LINC00339 expression in delayed fracture healing patients using lncRNA microarray analysis [16]. Chen et al. further found that LINC00339 negatively regulates the key regulatory factor of bone metabolism, CDC42 [17]. Nevertheless, the precise function of LINC00339 in delayed fracture healing has not been fully elucidated to this day.
Based on the aforementioned research, this study analyzed the levels of LINC00339 in delayed fracture healing patients and predicted the value of LINC00339 in delayed fracture healing. Through gain-of-function experiments, the functions of LINC00339 and miR-16-5p on osteoblast activity was validated. Additionally, examinations were conducted on the targeting relationship between LINC00339 and miR-16-5p, along with their roles in regulating delayed fracture healing.
Materials and methods
Inclusion of patients
The study included 172 patients subjects with fracture healing who were treated at author’s institution, including 82 patients with normal fracture healing and 90 patients with delayed fracture healing. All patients were aged 18 and above and were free from any history of previous fractures or osteoporosis. All patients involved in the study gave informed consent. The study was approved by the ethics committee of author’s institution.
Cultivation of cells
Human normal osteoblast hFOB1.19 cells (SIBCB, China) were cultured in DMEM medium containing 10% FBS, 100U/mL penicillin, and streptomycin. Ambient conditions were 37 °C with 5% CO2.
Transfection of cells
pcDNA3.1-LINC00339 overexpression transfection vector, small interference RNA (siRNA) against LINC00339 (si-LINC00339), as well as control groups pcDNA3.1 and si-NC, were constructed. miR-16-5p mimic, mimic NC, miR-16-5p inhibitor, and inhibitor NC were synthesized. hFOB1.19 cells were seeded and transfected with the above vectors using Lipofectamine 2000 when they reached the logarithmic growth phase.
RNA extraction and reverse transcriptase quantitative real-time PCR (RT-qPCR)
Total RNA was extracted from hFOB1.19 cells and serum using Trizol, and the RNA was reverse transcribed into cDNA. Real-time quantitative PCR analysis was performed on an MX3000p Real-time PCR instrument, using the miRNA cDNA synthesis Kit and HiFiScript cDNA Synthesis Kit following the manufacturer’s instructions. GAPDH and U6 were used as internal reference genes, and the relative RNA levels of LINC00339, miR-16-5p, and osteogenic markers (ALP, OCN, collagen I, and RUNX2) were calculated using the 2−ΔΔCt method.
Cell counting kit-8 (CCK-8) assay
hFOB1.19 cells were seeded in 96-well plates, and the Cell Counting Kit-8 reaction solution was added in proportion. After incubation for 1 h, the OD value was measured at a wavelength of 450 nm using a microplate reader.
Flow cytometry analysis of cell apoptosis
After reaching the logarithmic growth phase post-transfection, hFOB1.19 cells were washed with pre-cooled PBS and a binding buffer was added. Apoptosis was detected using the Annexin V-FITC/PI kit as per the instructions. Cell survival, necrosis, and early, and late apoptosis were analyzed using a flow cytometer, and the apoptotic rate was expressed as a percentage of early and late apoptotic cells combined.
Dual-luciferase reporter assay
The ENCORI, LncBook2, and DIANA databases predicted miR-16-5p as a target miRNA for LINC00339. To validate this prediction, LINC00339 sequence segments were cloned into the pcDNA3.1 vector to construct wild-type LINC00339-WT and mutant recombinant plasmid LINC00339-MT. The above vectors and miR-16-5p mimic or inhibitor were transfected into the hFOB1.19 cells. After 48 h of incubation, cells were lysed, and luciferase activity was measured.
RNA immunoprecipitation (RIP) assay
RIP was performed using a RIP kit to analyze the binding between LINC00339 and miR-16-5p. hFOB1.19 cells were lysed and incubated with magnetic beads containing anti-Ago2 or anti-IgG. Enrichment of LINC00339 and miR-16-5p was detected through RT-qPCR.
Nuclear and cytoplasmic RNA fraction isolations
Nuclear and cytoplasmic RNA in hFOB1.19 cells were isolated and collected using a SurePrep Nuclear or Cytoplasmic RNA Purification Kit. The subcellular localization expression of LINC00339 was detected using RT-qPCR, with U6 and GAPDH as the internal control.
Statistical analysis
All experimental data in this study were processed using GraphPad Prism 9.0 and SPSS 22.0. Data were presented as mean ± SD. Group comparisons were performed using ANOVA or Student’s t-test. The clinical predictive value of LINC00339 in fracture healing was analyzed by the Receiver operating characteristic curve (ROC). P < 0.05 was considered statistically significant.
Results
Serum LINC00339 is highly expressed in delayed fracture healing patients
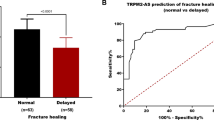
First, we evaluated the clinical data of 82 patients with normal fracture healing and 90 patients with delayed healing (Table 1) and found that there were no significant differences between the two groups in terms of age, BMI, gender, fracture side, smoking, and alcohol consumption (P > 0.05), suggesting comparability. As shown in Fig. 1A, the level of LINC00339 in the serum of delayed fracture healing patients was significantly higher than in normal patients (P < 0.001).
Analysis of the predictive value of serum LINC00339 levels for delayed fracture healing
As shown in Table 2, binary logistic regression analysis identified LINC00339 as a potential risk factor for delayed fracture healing (P < 0.001). Furthermore, ROC analysis revealed that at a cutoff value of 1.38, the area under the curve (AUC) for LINC00339 was 0.882 (95% CI: 0.832–0.932), with specificity and sensitivity of 74.39% and 87.78%, showing a high predictive ability for delayed fracture healing (P < 0.001, Fig. 1B). These findings indicated a possible correlation between LINC00339 and delayed fracture healing.
Regulation of osteoblast activity by abnormal expression of LINC00339
RT-qPCR analysis showed that with prolonged osteogenic differentiation time, the levels of osteogenic differentiation markers ALP, OCN, Collagen I, and RUNX2 mRNA significantly increased in Hfob1.19 cells (P < 0.001, Fig. 2A), while the level of LINC00339 decreased significantly (P < 0.001, Fig. 2B). Transfection of hFOB1.19 cells with pcDNA3.1-LINC00339 resulted in a critical increase in LINC00339 expression, whereas transfection with si-LINC00339 inhibited its expression (P < 0.001, Fig. 2C). Moreover, increased expression of LINC00339 inhibited proliferation and increases apoptosis in hFOB1.19 cells, while inhibiting LINC00339 expression had the opposite effect (P < 0.01, Fig. 2D and E). Additionally, overexpression of LINC00339 was found to suppress the expression of osteogenic differentiation markers mRNA, while silencing LINC00339 yielded contrasting outcomes. These findings indicated that LINC00339 may contribute to delayed fracture healing by inhibiting cell proliferation and promoting apoptosis.
The mRNA levels of markers of osteogenic differentiation increased with prolonged osteogenic differentiation time (A). and LINC00339 decreased (B). The levels of LINC00339 after the transfection (C). Effects of silencing or overexpression of LINC00339 on cell proliferation (D), apoptosis (E), and osteogenic marker-related mRNAs (F). *** P < 0.001 vs. 0 day osteogenic differentiation time; ** P < 0.01, *** P < 0.001 vs. Control group
LINC00339 targeted miR-16-5p directly
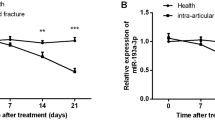
As shown in Fig. 3A, LINC00339 was primarily localized in the cytoplasm. miR-195-5p, miR-16-5p, miR-15b-5p, and miR-15a-5p were overlapping target miRNAs of LINC00339 as predicted by DIANA, LncBook2 and ENCORI databases (Fig. 3B), with a focus on miR-16-5p based its function. LINC00339 had a binding site to miR-16-5p (Fig. 3C). Luciferase reporter assays showed that miR-16-5p mimic suppressed the luciferase activity of the LINC00339-WT vector (P < 0.001), while it did not affect LINC00339-MT (Fig. 3D). RIP assays further confirmed the direct binding relationship between LINC00339 and miR-16-5p (Fig. 3E). Additionally, serum miR-16-5p levels were significantly lower in delayed fracture healing patients compared to those with normal healing (P < 0.001, Fig. 3F). The levels of LINC00339 and miR-16-5p in serum of patients with delayed fracture healing exhibited a notable negative correlation (r = -0.678, P < 0.001, Fig. 3G). Furthermore, the levels of miR-16-5p increased with prolonged osteogenic differentiation time (P < 0.001, Fig. 3H), and when hFOB1.19 cells were transfected with pcDNA3.1-LINC00339, there was a notable reduction in the miR-16-5p levels, while transfection with si-LINC00339 resulted in an increase of miR-16-5p expression. (P < 0.001, Fig. 3I).
Subcellular localization analysis of LINC00339 (A). LINC00339 target miRNAs predicted by the LncBook2, DIANA, and ENCORI databases are shown in Venn diagram (B). Binding site between LINC00339 and miR-16-5p (C). Dual luciferase gene reporter assay for interaction between miR-16-3p and LINC00339 (D). RIP experiment validation of LINC00339 targeting binding with miR-16-5p(E). miR-16-5p was significantly reduced in patients with delayed fracture healing (F). Serum LINC00339 was negatively correlated with miR-16-5p in delayed fracture healing patients (G). Impact of timing of osteogenic differentiation on miR-16-5p levels (H). Significant attenuation of miR-16-5p expression by LINC00339 (I). *** P < 0.001 vs. 0 day osteogenic differentiation time; *** P < 0.001 vs. Control group
Up-regulation of miR-16-5p attenuated the influence of LINC00339 on osteoblasts
In order to confirm the influence of miR-16-5p on LINC00339, we transfected hFOB1.19 cells with pcDNA3.1-LINC00339 and miR-16-5p mimic. The outcomes displayed a significant decrease in miR-16-5p levels in hFOB1.19 cells transfected with pcDNA3.1-LINC00339 (P < 0.001). Subsequent transfection with miR-16-5p mimic led to a restoration of miR-16-5p expression level (P < 0.001, Fig. 4A). Importantly, transfection with pcDNA3.1-LINC00339 resulted in decreased cell proliferation, increased apoptosis, and a notable reduction in the levels of osteogenic differentiation marker-related mRNAs (P < 0.01). However, co-transfection with pcDNA3.1-LINC00339 + miR-16-5p mimic caused a significant recovery in cell proliferation, expression levels of osteogenic differentiation marker-related mRNAs, and a reduction in apoptosis rate (P < 0.001, Fig. 4B-D).
Discussion
Delayed fracture healing, as a major complication of fractures, is becoming more prevalent with the increasing incidence of fractures [18], leading to a decline in patients’ quality of life [19]. Studies have shown that medical expenses for delayed fracture healing patients are more than twice as high as those for patients with normal healing [20, 21]. Therefore, further exploration of the process of delayed fracture healing is needed. Our study first validated the expression level of LINC00339 in delayed fracture healing patients and found that, in support of the results of Zhang et al., the expression level of LINC00339 in delayed fracture healing patients was remarkably improved. Subsequently, through binary logistic regression analysis, we identified LINC00339 as a possible contributor to delayed fracture healing, and ROC curve analysis also suggested that LINC00339 can predict delayed fracture healing. Nevertheless, the mechanism of action of LINC00339 in delayed fracture healing is unclear.
Previous studies have demonstrated that the activity of osteoblasts plays an essential role in fracture healing, and increasing the vitality of osteoblasts can promote fracture healing [22]. Therefore, we analyzed the impact of LINC00339 on osteoblasts. RT-qPCR results revealed that with prolonged osteogenic differentiation time, the expression level of LINC00339 in the serum gradually decreased, while the osteogenic differentiation markers levels related to osteoblast activity increased. Furthermore, overexpression of LINC00339 inhibited the proliferation of osteoblasts hFOB1.19 and promoted apoptosis, leading to a remarkable reduction in the levels of osteogenic differentiation markers in cells overexpressing LINC00339. These results suggested that LINC00339 can regulate the fracture healing process and may play a role in delayed fracture healing, aligning with the research findings by Chen et al. [17].
MicroRNAs (miRNAs), a class of short non-coding RNAs, are involved in the process of fracture healing, especially regulating physiological processes such as osteoblast differentiation [23, 24]. To further explore the specific mechanism of LINC00339 in delayed fracture healing, we predicted four target miRNAs of LINC00339 through the DIANA, ENCORI, and LncBook2 databases, including miR-16-5p. Although the effect of miR-16-5p in the fracture healing is controversial [25, 26], it has been demonstrated by researches that miR-16-5p has inhibitory effects on osteoclast genesis in Giant Cell Tumors of Bone [27] and its levels significantly decrease in lumbar vertebral tissues of osteoporotic patients [28]. Therefore, we focused on the targeted relationship between miR-16-5p and LINC00339 in the study. In our research, dual-luciferase reporter assays showed an interaction between miR-16-5p and LINC00339, which was further validated by RIP experiments. Levels of miR-16-5p were significantly decreased in delayed fracture healing patients, showing a clear negative correlation with LINC00339. Furthermore, miR-16-5p levels increased with prolonged osteogenic differentiation time and were negatively regulated by LINC00339.
From the above results, it is evident that silencing LINC00339 can elevate miR-16-5p levels, and miR-16-5p had a promoting effect on fracture healing, consistented with the study results of Duan et al. Additionally, the study also explored the impacts of transfecting miR-16-5p mimic on inhibiting delayed fracture healing caused by LINC00339. The results indicated that upregulating miR-16-5p can counteract the delayed fracture healing effect of LINC00339, restore osteoblast vitality, and enhance the levels of osteogenic differentiation markers mRNAs.
Our study has certain limitations. The biggest limitation of this study is the limited sample size. We intend to increase the sample size in the future. This will enhance the reliability of our findings and provide a strong basis for clinical application. Additionally, we will further elucidate the specific mechanisms by which LINC00339 functions in vivo using a fracture mouse model, promoting the clinical application of LINC00339 as a diagnostic biomarker for delayed fracture healing.
In conclusion, LINC00339 may serve as a novel predictive indicator for delayed fracture healing. LINC00339 demonstrated anti-osteogenic differentiation and pro-apoptotic effects and participated in delayed fracture healing by targeting and binding to miR-16-5p.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Melvin JS, Mehta S. Patellar fractures in adults. J Am Acad Orthop Surg. 2011;19(4):198–207.
Argote A, Mora-Hernandez O, Milena Aponte L, Barrera-Chaparro DI, Munoz-Ruiz LM, Giraldo-Mordecay L, et al. Cardiovascular Risk factors and carotid intima-media thickness in a Colombian Population with Psoriasis. Actas Dermosifiliogr. 2017;108(8):738–45.
Dhamangaonkar AC, Patankar HS. Salvage of delayed union of a phalangeal fracture with a hairpin wire. Hand Surg. 2013;18(3):431–3.
Daughaday WH, Hall K, Raben MS, Salmon WD Jr., van den Brande JL, van Wyk JJ. Somatomedin: proposed designation for sulphation factor. Nature. 1972;235(5333):107.
Jha S, Blau JE, Bhattacharyya T. Normal and delayed Fracture Healing: Symphony and Cacophony. Horm Metab Res. 2016;48(11):779–84.
Cheng Z, Li A, Tu CL, Maria CS, Szeto N, Herberger A, et al. Calcium-sensing receptors in chondrocytes and osteoblasts are required for Callus Maturation and Fracture Healing in mice. J Bone Min Res. 2020;35(1):143–54.
Committee of Accelerated Rehabilitation after Osteoporotic Fractures of China Association of Rehabilitation, Technology T, Promotion, Bone, Joint Group of Chinese Society of O, Bone Mineral R Osteoporosis Working Committee of Chinese Association of Orthopedic S. [Chinese expert consensus on treatment of osteoporotic fractures with teriparatide (2024 edition)]. Zhonghua Yi Xue Za Zhi. 2024;104(17):1456–65.
Szczesny G. Fracture Healing and its disturbances. A literature review. Ortop Traumatol Rehabil. 2015;17(5):437–54.
Yin Y, He Q, He J, Feng Y, Xu Y. Inhibition of LINC00958 hinders the progression of osteoarthritis through regulation of the miR-214-3p/FOXM1 axis. J Orthop Surg Res. 2024;19(1):66.
Zhou Z, Chen J, Huang Y, Liu D, Chen S, Qin S. Long noncoding RNA GAS5: a new factor involved in Bone diseases. Front Cell Dev Biol. 2021; 9807419.
Guo X, Zhang J, Han X, Wang G. LncRNA SNHG1 delayed Fracture Healing via modulating miR-181a-5p/PTEN Axis. J Invest Surg. 2022;35(6):1304–12.
Yu C, Chen B, Su H, Yang Y. Long non-coding RNA MIAT serves as a biomarker of fragility fracture and promotes fracture healing. J Orthop Surg Res. 2024;19(1):343.
Chen S, Ma H, Li M, Jia Z, Chen X, Bu N. Long noncoding RNA NORAD promotes fracture healing through interacting with osteoblast differentiation via Targeting miR-26a. Biomed Res Int. 2023; 20239950037.
Wu Z, Zhang S, Guo W, He Y. LINC00339: an emerging major player in cancer and metabolic diseases. Biomed Pharmacother. 2022; 149112788.
Guo J, Cai H, Liu X, Zheng J, Liu Y, Gong W et al. Long non-coding RNA LINC00339 stimulates glioma vasculogenic mimicry formation by regulating the miR-539-5p/TWIST1/MMPs Axis. Mol Ther Nucleic Acids. 2018; 10170–86.
Zhang Y, Zhang Y, Yang K, Guo W, Ma X, Ma X, et al. MALAT1 knockdown promoted cell viability and migration of LPS-treated MG-63 cells via sponging miR-212. Genes Genomics. 2021;43(5):523–31.
Chen XF, Zhu DL, Yang M, Hu WX, Duan YY, Lu BJ, et al. An osteoporosis risk SNP at 1p36.12 acts as an allele-specific enhancer to modulate LINC00339 expression via Long-Range Loop formation. Am J Hum Genet. 2018;102(5):776–93.
Zhang M, Xu F, Cao J, Dou Q, Wang J, Wang J et al. Research advances of nanomaterials for the acceleration of fracture healing. Bioact Mater. 2024; 31368–94.
Saleh K, Hak D. Nierengarten MJMOCu. Socioeconomic burden of traumatic tibial fractures: non union or delayed union. 2001; 1–22.
Antonova E, Le TK, Burge R, Mershon J. Tibia shaft fractures: costly burden of nonunions. BMC Musculoskelet Disord. 2013; 1442.
Dong Z, Hu B, Wang S, Wang M, Sun S, Liu X, et al. LncRNA MAGI2-AS3 promotes fracture healing through downregulation of miR-223-3p. J Orthop Surg Res. 2024;19(1):370.
Jiang M, Liu R, Liu L, Kot A, Liu X, Xiao W, et al. Identification of osteogenic progenitor cell-targeted peptides that augment bone formation. Nat Commun. 2020;11(1):4278.
Komatsu DE, Duque E, Hadjiargyrou M. MicroRNAs and fracture healing: pre-clinical studies. Bone. 2021; 143115758.
Nugent M. MicroRNAs and Fracture Healing. Calcif Tissue Int. 2017;101(4):355–61.
Yu T, You X, Zhou H, He W, Li Z, Li B, et al. MiR-16-5p regulates postmenopausal osteoporosis by directly targeting VEGFA. Aging. 2020;12(10):9500–14.
Sun Y, Xiong Y, Yan C, Chen L, Chen D, Mi B, et al. Downregulation of microRNA-16-5p accelerates fracture healing by promoting proliferation and inhibiting apoptosis of osteoblasts in patients with traumatic brain injury. Am J Transl Res. 2019;11(8):4746–60.
Sang S, Zhang Z, Qin S, Li C, Dong Y. MicroRNA-16-5p inhibits Osteoclastogenesis in Giant Cell Tumor of Bone. Biomed Res Int. 2017; 20173173547.
Duan J, Li H, Wang C, Yao J, Jin Y, Zhao J et al. BMSC-derived extracellular vesicles promoted osteogenesis via Axin2 inhibition by delivering MiR-16-5p. Int Immunopharmacol. 2023; 120110319.
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Conceptualization, Y.L., Y.S., K.M. and Z.W.; Data curation, Y.L., Y.S., K.M. and S.W.; Formal analysis, K.M. and S.W.; Funding acquisition, Z.W. and L.H.; Investigation, K.M. and S.W.; Methodology, K.M. and S.W.; Project administration, Z.W. and L.H.; Resources, K.M. and S.W.; Software, K.M. and S.W.; Supervision, Z.W. and L.H.; Validation, K.M. and S.W.; Visualization, Z.W. and L.H.; Roles/Writing - original draft, Y.L., Y.S. and K.M.; Writing - review & editing, Z.W. and L.H.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of Rizhao Central Hospital before the study began. The participants’ right to be informed about the study was ensured and agreed to participate in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, Y., Sun, Y., Ma, K. et al. Functional mechanism and clinical implications of LINC00339 in delayed fracture healing. J Orthop Surg Res 19, 511 (2024). https://doi.org/10.1186/s13018-024-04998-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13018-024-04998-0