Abstract
Background
Osteosarcoma (OS) is a malignant bone tumor that commonly occurs in children and adolescents under the age of 20. Dysregulation of microRNAs (miRNAs) is an important factor in the occurrence and progression of OS. MicroRNA miR-744-5p is aberrantly expressed in various tumors. However, its roles and molecular targets in OS remain unclear.
Methods
Differentially expressed miRNAs in OS were analyzed using the Gene Expression Omnibus dataset GSE65071, and the potential hub miRNA was identified through weighted gene co-expression network analysis. Quantitative real-time PCR (qRT-PCR) was used to detect the expression of miR-744-5p in OS cell lines. In vitro experiments, including CCK-8 assays, colony formation assays, flow cytometry apoptosis assays, and tube formation assays, were performed to explore the effects of miR-744-5p on OS cell biological behaviors. The downstream target genes of miR-744-5p were predicted through bioinformatics, and the binding sites were validated by a dual-luciferase reporter assay.
Results
The lowly expressed miRNA, miR-744-5p, was identified as a hub miRNA involved in OS progression through bioinformatic analysis. Nuclear factor I X (NFIX) was confirmed as a direct target for miR-744-5p in OS. In vitro studies revealed that overexpression of miR-744-5p could restrain the growth of OS cells, whereas miR-744-5p inhibition showed the opposite effect. It was also observed that treatment with the conditioned medium from miR-744-5p-overexpressed OS cells led to poorer proliferation and angiogenesis in human umbilical vein endothelial cells (HUVECs). Furthermore, NFIX overexpression restored the suppression effects of miR-744-5p overexpression on OS cell growth and HUVECs angiogenesis.
Conclusion
Our results indicated that miR-744-5p is a potential tumor-suppressive miRNA in OS progression by targeting NFIX to restrain the growth of OS cells and angiogenesis in HUVECs.
Similar content being viewed by others
Introduction
Osteosarcoma (OS) is a malignant tumor of bone that develops from primitive transformed cells of mesenchymal origin and usually occurs in the long diaphyseal region [1]. OS can occur in people of any age; however, children and adolescents have a high incidence of it [2]. Although advances achieved in OS treatment including neoadjuvant chemotherapy, radiotherapy, and surgical resection have improved survival rates in OS patients, about 30% of patients without metastatic diseases subsequently develop metastasis, which often results in poor prognosis [3, 4]. Therefore, it is urgent to explore the underlying molecular mechanism involved in OS progression, which might provide potential targets for the diagnosis and treatment of OS.
Currently, microRNAs (miRNAs) have drawn increasing attention in the research on OS progression [5, 6]. MiRNAs are short, single-stranded non-coding RNA molecules at a length of about 22 nucleotides [7]. They cannot encode proteins but can negatively regulate their target genes’ expression by binding to their 3’-untranslated regions. Due to this property, miRNAs are involved in a variety of biological processes [8,9,10]. Numerous studies have revealed that miRNAs play crucial roles in OS progression. MiR-188-5p has been revealed as a suppressive miRNA in OS by target-degrading cyclin T2 [11]. MiR-133b exerts suppressor activities in OS by directly targeting the fibroblast growth factor receptor 1 [12]. Therefore, miRNAs play essential roles in OS development and more investigations on their functional roles and molecular mechanisms are required.
Nowadays, many studies identify prognostic or therapeutic markers by analyzing high-throughput sequencing data. Besides, a variety of bioinformatic approaches are applied to understand the mechanisms underlying these data [13,14,15]. Weighted gene coexpression network analysis (WGCNA) is a commonly used bioinformatics tool for studying the structure and function of large-scale biomolecular networks. It can also be used to identify hub genes in networks and provide interpretation for molecular interactions [16]. For instance, Tian et al. identified OS metastasis-associated genes by WGCNA [17]. Therefore, the use of WGCNA can make contributions to exploring miRNAs that are critical for OS progression.
We screened miR-744-5p as a key miRNA in OS via WGCNA in the current study. The abnormal expression of miR-744-5p was recognized to be related to the formation and metastasis of ovarian cancer [18], lung squamous cell carcinoma [19], multiple myeloma [20] and many other cancers [21,22,23]. In OS, miR-744-5p showed tumor suppressive effects by negatively regulating transforming growth factor-beta 1 [24]. However, little is known about the role and molecular mechanism of miR-744-5p in OS progression.
Nuclear factor I X (NFIX), a nuclear factor I family member, has been reported to play important roles in carcinogenesis and tumor progression [25, 26]. NFIX could transcriptionally induce the Ezrin expression to the enhanced migration of glioblastoma cells [27]. Another study reported that NFIX could promote metastasis in lung cancer [28]. However, the role of NFIX in OS has not yet been reported. A previous study revealed that miR-744-5p promotes programmed cell death in ovarian cancer by targeting NFIX [18]. This suggested a molecular mechanism that miR-744-5p targets NFIX to suppress tumor growth in OS, although this has not been verified.
In this work, we identified miR-744-5p, which is downregulated in OS, through WGCNA. In vitro studies were designed to identify the function roles of miR-744-5p in OS cell growth and human umbilical vein endothelial cells (HUVECs) angiogenesis. Additionally, the regulation of miR-744-5p on NFIX in OS cells was also explored. Our study identified the miR-744-5p/NFIX axis as a novel regulator in OS progression. Therefore, targeting the miR-744-5p/NFIX axis may be a potential therapeutic approach for suppressing OS progression.
Methods
Microarray data
The miRNA microarray data came from the Gene Expression Omnibus dataset GSE65071 [29], which contains plasma samples from 20 OS patients and 15 healthy individuals. The expression profiling of miRNAs was obtained from the Exiqon miRNome platform GPL19631. The k-nearest neighbor algorithm was used to fill in the missing data. Then the data was normalized for subsequent WGCNA.
WGCNA
The OS-associated miRNAs were identified through the construction of miRNA co-expression networks using WGCNA [16]. First, a similarity matrix was constructed by calculating the absolute value of the Pearson correlation coefficient. The independence and the relevant degree of miRNA pairs in co-expression modules were calculated by the gradient method, with power values ranging from 1 to 20. A soft-thresholding power β was selected where the correlation index reached 0.9 (R2 value ≥ 0.90). Next, all adjacencies were transformed into a topological overlap matrix. Then, the community dissimilarity index of these miRNAs was analyzed and a hierarchical clustering tree was identified. Based on dynamic cut tree criteria, we set a cut height of 0.95 and a minimal module size of 3. After the classification of gene modules, the module eigenvectors of each module were calculated along with clinical traits and shown as a heatmap. The modules closely associated with the disease condition are considered to be the key modules. Gene significance (GS) represents the correlation of individual genes with disease traits. Module membership (MM) shows the relationship between module eigengenes and expression values of genes. The correlation factor of MM with GS was evaluated by Pearson correlation analysis to verify the intra-modular connectivity.
Enrichment analysis
The Gene Ontology (GO) functional and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed on miR-744-5p-targeted genes clusterProlifer [30]. The threshold was set at a P value and Q value < 0.05.
Cell culture
The 143B and U2OS human osteosarcoma cell lines, as well as the hFOB1.19 osteoblast cell line, were purchased from Shanghai FuHeng Biology (Shanghai, China). The HUVECs were purchased from the American Type Culture Collection (Manassas, USA). The 143B and U2OS cells were cultured in Dulbecco’s Modified Eagle Medium, the hFOB1.19 cells were cultured in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 with 1% penicillin/streptomycin and 10% fetal bovine serum, and the HUVECs were cultured in Endothelial Cell Growth Medium. All cells were maintained at 37℃ with 5% CO2.
Cell transfection
The miR-744-5p mimics, miR-744-5p inhibitor, NFIX overexpression plasmid, and their negative controls were obtained from GenePharma (Shanghai, China). Briefly, the miR-744-5p mimics were synthesized according to the mature miR-744-5p sequence (miRbase ID: MIMAT0004945), and the miR-744-5p inhibitor was the reverse complementary sequence of the mature miR-744-5p. As to the NFIX overexpression vector, the full-length NFIX coding sequence was amplified using polymerase chain reaction (PCR) with the following sequences: Forward: 5’ CGGGGTACCGCCACCATGTACTCCCCGTACT-3’ and Reverse: 5’-CCGCTCGAGTCAGAGGAACCAGGACTG-3’. Then, the amplification products were inserted into pcDNA3.1 vectors to construct the NFIX overexpression vector with the help of the KpnI and XhoI endonucleases.
For cell transfection, the cells were pre-incubated at 37℃ for 12 h and then transfected at 50–70% confluence. Cells were transfected with miR-744-5p mimics (50 nM), mimics-NC (50 nM), miR-744-5p inhibitor (100 nM), inhibitor-NC (100 nM), empty vector (1 µg), and NFIX overexpression vector (1 µg) using the Lipofectamine 3000 (Thermo Fisher Scientific, USA) under the manufacturer’s protocol. 48 h after the transfection, the cells were harvested for the following experiments.
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated and purified using QIAzol lysis solution (Qiagen, Germany) and miRNeasy kit (Qiagen). Then, cDNA was synthesized using the QuantiTect RT Kit (Qiagen) and miScript II RT Kit (Qiagen). RNA was detected by using the LightCycler 480 SYBR Green I Master kit (Roche, Switzerland) with primer sequences listed in Table 1. RNA was amplified through thermocycling with 10 min of initial denaturation at 95 °C, 40 cycles of amplification for 20 s per cycle, and 1 min of annealing at 60 °C. Data quantification was normalized by the 2−ΔΔCT method with GAPDH or U6 as internal references.
Cell viability
The Cell Counting Kit-8 (CCK-8) assay was applied to test cell viability. 143B and U2OS cells were split and transferred into 96-well plates with about 3 × 103 cells per well. For HUVECs, as previously reported [31], were mixed with OS cells conditioned medium (CM) and transferred into 96-well plates (2 × 103 cells/well) at 37℃, 5% CO2.
Cells were harvested at 0, 24, 48, 72, and 96 h after incubation. Subsequently, 10 µL of CCK-8 solution (Dojindo, Japan) was added to each well and cultured at 37℃ for 2 h. Cell viability was determined by measuring the optical density values at 450 nm using a microplate reader.
Colony formation assay
The treated 143B and U2OS cells were digested and transferred into 6-well plates with 1 × 10 3 cells per well. After incubation for 2 weeks, cells were fixed using methyl alcohol and stained to visualize colonies.
Cell apoptosis
The Annexin V Apoptosis Detection Kit (BD Biosciences, USA) was applied for cell apoptosis detection. Cell pellets were resuspended in 500 µL binding buffer and incubated with 5 µL FITC Annexin V and 5 µL Propidium Iodide for 1 h. Subsequently, the apoptotic cells were detected using flow cytometry.
Tube formation assay
HUVECs were cultured with the OS cells’ CM for 24 h before conducting the tube formation assay. A 96-well tray was coated by 50 µL Matrigel Basement Membrane Matrix (Corning, USA) per well at 37 ℃ for 30 min. The pre-treated HUVECs (2 × 104/well) were added into the pre-coated 96-well tray and incubated at 37℃ for 4 h. Finally, tube formation results were observed under a microscopy.
Dual-luciferase reporter assay
Partial sequences (300 bp) of NFIX 3’ untranslated regions (3’UTR) containing wild type (WT) or mutant (MUT) miR-744-5p binding sites were subcloned into psiCHECK-2 vector by GenePharma, to establish the NFIX WT vectors (C09005) and NFIX MUT vectors (C09006), respectively. Then, 143B cells in 24-well plates were co-transfected with 0.1 µg of corresponding reporter vector and 50 nM miR-744-5p mimics or mimic NC using Lipofectamine 3000 (Thermo Fisher Scientific). 48 h after transfection, cells were lysed and luciferase activities were measured using a Dual-Luciferase Reporter Assay System (Promega, USA).
Western blot
Briefly, total proteins from the cells were lysed in radioimmunoprecipitation assay buffer (Boster Bio, China) and quantified using a bicinchoninic acid assay kit (KeyGEN BioTECH, China). Next, 40 µg of protein was isolated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes (Millipore, USA). After being blocked with 5% non-fat milk, the membranes were incubated with primary antibodies at 4℃ overnight and corresponding secondary antibodies at 37℃ for 2 h. Finally, the protein bands were detected by chemiluminescence. The primary antibodies used were as follows: anti-NFIX (Nbp2-15038, 1:1000, Novus Biologicals); anti-GAPDH (AB9485, 1:2000, Abcam).
Statistical analysis
All data were expressed as mean ± standard deviation from at least three reduplicate experiments. Group comparisons were achieved by using Student’s t-test or one-way analysis of variance followed by the Tukey test. A significance level of P < 0.05 was considered statistically significant.
Results
Identification of WGCNA modules associated with OS
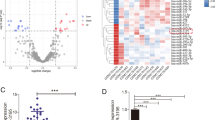
As WGCNA is recommended to try on a dataset of more than 15 samples (https://labs.genetics.ucla.edu/horvath/CoexpressionNetwork/Rpackages/WGCNA/faq.html), we used the dataset GSE65071 (35 samples) to establish the miRNA co-expression networks. Cluster analysis was carried out and the clustering tree was shown in Fig. 1A, where the hierarchical clustering dendrograms showed no outliers. The value of paired miRNA correlation was explored for network construction, followed by the construction of the adjacency matrix and topological overlap matrix. According to the result (Fig. 1B), the soft-thresholding power was set as 10, and 7 modules were obtained (Fig. 1C). From the analysis diagram of module eigengenes (Fig. 1D), we observed that the green module (r = 0.54, P = 9 × 10− 4), yellow module (r = -0.45, P = 0.007), turquoise module (r = -0.99, P = 3 × 10− 30), blue module (r = -0.6, P = 1 × 10− 4), and red module (r = -0.63, P = 6 × 10− 5) were closely associated with OS. The turquoise module showed the closest connection to OS and therefore presented clinical significance. MM and GS were also found to be positively correlated with disease status (Fig. 1E).
Dataset network construction and significant modules identification. (A) Hierarchical clustering plot of GSE65071. (B) Calculation of adjacency matrix weighting power. (C) Clustering dendrograms of co-expressed genes. (D) Module-trait associations of module’s color and disease trait. The correlation was qualified with the P-value presented at the bottom of each section. (E) A scatterplot of module membership and gene significance in the turquoise module
Identification of hub miRNA in turquoise module
A co-expression network of the turquoise module was constructed. It was found that miR-744-5p was the highly connected hub miRNA in the turquoise module (Fig. 2A). Further KEGG enrichment analysis results indicated that miR-744-5p was associated with some cancer-related pathways, such as the AMPK signaling pathway and the mTOR signaling pathway (Fig. 3A; Table 2). GO enrichment analysis also confirmed the involvement of miR-744-5p in cell junction, branching morphogenesis of epithelial cast, etc. (Fig. 3B; Tables 3, 4 and 5). These bioinformatics analysis results indicated that the hub miRNA miR-744-5p might play a regulatory role in OS progression.
Mir-744-5p inhibited the cell viability of osteosarcoma cells and the tube formation of HUVECs
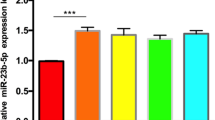
MiR-744-5p expression in OS cells was examined using qRT-PCR assay, which showed significantly reduced expression in OS cells compared with osteoblasts (Fig. 4A). Next, to investigate the involvement of miR-744-5p in OS carcinogenesis, we transfected 143B cells and U2OS cells with miR-744-5p mimics and inhibitors (Fig. 4B). After transfection, cell viability was determined by CCK-8 and colony formation assays. We observed that miR-744-5p overexpression observably reduced the cell viability of 143B and U2OS cells, while miR-744-5p inhibition observably promoted the cell viability (Fig. 4C-D). Additionally, cell apoptosis after transfection was detected. Overexpression of miR-744-5p promoted the apoptosis rates in OS cells, while the knockdown of miR-744-5p inhibited apoptosis in OS cells (Fig. 4E).
miR-744-5p inhibited the viability of osteosarcoma cells. (A) Expression levels of miR-744-5p in OS cells and normal endothelial cells. (B) The transfection efficiency was verified by qRT-PCR. (C) CCK-8 assay was applied to determine the viability of OS cells. (D) In vitro cell clonogenicity was examined by colony formation assay. (E) Cell apoptosis rate was evaluated by flow cytometry. *P < 0.05; ** P < 0.01
Considering that GO enrichment analysis showed the involvement of miR-744-5p in angiogenesis (Table 3), we investigated the effects of miR-744-5p on tube formation of HUVECs. As shown in Fig. 5A-B, treatment with CM from miR-744-5p-overexpressed OS cells inhibited the viability and tube formation of HUVECs, while treatment with CM from the miR-744-5p-suppressed OS cells showed promoting effects on the cell viability and tube formation of HUVECs. These results supported the bioinformatics analysis results and confirmed that miR-744-5p inhibited the cell viability of OS cells and the tube formation of HUVECs.
Mir-744-5p directly targeted NFIX
To explore the underlying mechanism of miR-744-5p, we used TargetScan, DIANA-Tarbase, and starBase for target prediction. By taking the intersection of the prediction results, NFIX and LDL receptor-related protein 3 (LRP3) were identified as potential targets of miR-744-5p (Fig. 6A). Their expressions in miR-744-5p overexpressed OS cells were detected. The results showed that miR-744-5p overexpression significantly decreased NFIX mRNA expression while having little effect on LRP3 mRNA expression (Fig. 6B). Thus, NFIX was selected as the target of miR-744-5p in OS cells. According to the predicted binding sequences, we conducted the dual-luciferase reporter experiments and confirmed that miR-744-5p targeted NFIX in 143B cells (Fig. 6C). Additionally, the protein expression of NFIX was markedly downregulated after the transfection with miR-744-5p mimics and markedly upregulated after the transfection with miR-744-5p inhibitor, indicating that miR-744-5p could negatively regulate NFIX at the protein level (Fig. 6D). It was also found that NFIX was highly expressed in OS cells (Fig. 6E). Taken together, these data suggested that miR-744-5p targeted NFIX in OS.
miR-744-5p directly targets NFIX. (A) TargetScan, DIANA-Tarbase, and starBase algorithms were applied to identify potential downstream targets of miR-744-5p. (B) NFIX mRNA expression in OS cells transfected with miR-744-5p mimics. (C) Wild type and mutant binding sequences of miR-744-5p on NFIX. The direct binding was verified by dual-luciferase reporter assay. (D) NFIX protein expression in OS cells transfected with miR-744-5p mimics and miR-744-5p inhibitor. (E) NFIX protein expression in OS cells. ** P < 0.01
NFIX attenuated the suppressive effects of mir-744-5p on the cell viability of OS cells and tube formation of HUVECs
Given the binding relationship between miR-744-5p and NFIX, whether miR-744-5p regulates the malignant phenotype of OS cells by targeting NFIX was further explored. To this end, we constructed the NFIX overexpression vector, which could promote the protein expression of NFIX in both 143B and U2OS cells (Fig. 7A). Then, the cells were co-transfected with miR-744-5p mimics and NFIX overexpression vector. As expected, the protein expression was suppressed in miR-744-5p-overexpressed cells, and co-transfection with NFIX overexpression vector restored the NFIX expression in OS cells (Fig. 7B). Subsequently, cell viability and apoptosis were detected. It was shown that the regulatory effects of miR-744-5p on cell viability and apoptosis were neutralized when restoring the expression of NFIX (Fig. 7C-E). Similarly, in HUVECs, transfection with miR-744-5p mimics suppressed the cell viability and tube formation of HUVECs, and this suppressive effect was weakened by the co-transfection of miR-744-5p mimics and NFIX overexpression vector (Fig. 7F-G). Above all, these results indicated that the regulatory role of miR-744-5p in OS cells and HUVECs was through mediating NFIX.
NFIX neutralized the effects of miR-744-5p on osteosarcoma cells. (A) Protein expression of NFIX in OS cells transfected with NFIX overexpression vector. (B) Protein expression of NFIX in different groups of OS cells. (C) OS cell viability was assessed by CCK-8 assay. (D) Cell proliferation was visualized by colony formation assay. (E) Flow cytometry assay was performed to determine cell apoptosis rate. (F) The viability of HUVECs was assessed by CCK-8 assay. (G) The effects of miR-744-5p and NFIX on the tube formation of HUVECs were detected by tube formation assay. *P < 0.05; ** P < 0.01
Discussion
Osteosarcoma is a malignant bone tumor that exhibits highly aggressive and rapid metastasis [32]. In the complicated pathogenesis of OS, miRNAs function as essential regulators, participating in various cellular processes such as cell proliferation, apoptosis, and angiogenesis [6, 33]. Therefore, we focused our attention on miRNAs in the current study.
To identify the key miRNA in OS progression, we used the miRNA dataset GSE65071 to conduct differential miRNA analysis and WGCNA. Ultimately, miR-744-5p was identified as the key miRNA in OS. As a multifunctional miRNA, miR-744-5p has been investigated in various cancer progressions. For example, it has been found that miR-744-5p is up-regulated in pancreatic neuroendocrine tumors (pNET) and enhances the proliferation and migration of pNET cells by inhibiting the expression of thrombospondin 2 [34]. Conversely, miR-744-5p is down-regulated in lung adenocarcinoma (LUAD). In vitro experiments have shown that miR-744-5p suppresses malignant behaviors of LUAD cells by targeting cell division cycle-associated protein 4 and MAF bZIP transcription factor G [35, 36]. Similarly, in glioma, miR-744-5p is expressed at low levels and acts as a tumor suppressor by inhibiting the polarization of macrophages M2 [37]. Additionally, recent studies have reported that miR-744-5p can reduce the resistance of gastric cancer cells to PD-L1 by inhibiting immune escape [38], and decrease the resistance of non-small cell lung cancer cells to cisplatin by promoting ferroptosis [39]. Here, we observed a down-regulated expression of miR-744-5p in OS cells. In addition, overexpression of miR-744-5p inhibited the cell proliferation of OS cells, which is consistent with the previous study [24]. Therefore, miR-744-5p is a prime candidate for tumor suppressive factors in OS.
GO analysis showed that miR-744-5p was enriched in angiogenesis, which is the process of developing new blood vessels and is essential for the progression of many types of tumors [40]. One of the characteristics of OS is the dense blood vessels [41]. Prior research has shown that miRNA miR-429 and miR-206 directly mitigate the angiogenesis of HUVECs to exert the anti-tumor effect in OS [42]. Another study reported that miR-877-3p attenuates the angiogenesis of HUVECs to exhibit a tumor-suppressive role in OS [43]. Although miR-744-5p has been reported to inhibit cell proliferation, migration, and invasion in various cancers, there have been no studies on its role in regulating the angiogenesis of HUVECs. In the current study, we confirmed the regulation of miR-744-5p on the cell viability and tube formation of HUVECs by treating the HUVECs with CM from the OS cells. We found that treatment with CM from miR-744-5p-overexpressed OS cells inhibited the viability and tube formation of HUVECs, whereas treatment with CM from miR-744-5p-suppressed OS cells showed opposite effects. To the best of our knowledge, this is the first study to provide evidence of the regulation of miR-744-5p on the angiogenesis of HUVECs.
During tumor-induced angiogenesis, the tumor-derived extracellular vesicles (EVs) play important roles [44, 45]. EVs are heterogeneous particles that are secreted by cells into the extracellular environment. Based on the size, biogenesis, and function, the EVs can be classified into exosomes, macrovesicles, and apoptotic bodies [46]. Recently, researchers isolated EVs from cells’ CM and found that EVs actively engage in intercellular communication through the transfer of intracellular cargos such as proteins and nucleic acids [47,48,49]. Specifically, miRNAs released by EVs can be uptake by other cells, thus influencing the various physiopathological conditions [44]. For example, EVs-derived miR-141-3p, miR-21-5p, and miR-210-3p are reported to facilitate endothelial cell angiogenesis [50,51,52]. Zhang et al. reported that miR-199a-5p is down-regulated in osteosarcoma plasma samples compared with normal plasma samples. Through exosomes, miR-199a-5p can be transported from OS cells to HUVECs, thereby inhibiting HUVEC proliferation, migration, and neovascularization [53]. These results suggest that the mechanism that miR-744-5p regulates the angiogenesis of HUVECs may be through the EVs-dependent manner. OS cells secreted miR-744-5p into the cell culture medium via EVs. Then, EVs containing miR-744-5p could enter HUVECs to regulate the proliferation and angiogenesis of HUVECs. Delivery of EVs-derived miR-744-5p, such as mesenchymal stem cell-derived EVs-miR-744-5p [37], may be a preventive strategy against angiogenesis in OS. However, caution should be exercised when targeting a tumor suppressor miRNA as a therapeutic target unless the miRNA can be delivered to a specific site [54]. Thus, further investigation on the EVs-miR-744-5p is needed to elucidate its complicated roles in OS initiation and progression.
Besides, in most cases, the regulatory mechanism of miRNAs is to bind to the 3’untranslated region of the target genes to inhibit their expression [55]. Previous studies have shown that miR-744-5p targets ADP ribosylation factor 1, SRY-box transcription factor 12, replication factor C subunit 2, and others to suppress the progression of several types of cancer [56,57,58]. In the current study, we used TargetScan, DIANA-Tarbase, and starBase algorithms to predict miR-744-5p targets and identified NFIX as the target gene of miR-744-5p in OS. Studies have identified the essential role of NFIX in hematopoiesis [59], muscle development [60], and brain development [61]. However, little is known about its role in tumor development. Previous research on lung cancer has found that NFIX inhibition reduces cell proliferation, migration, and invasion in vitro [28]. Another study reported that NFIX is mediated by miR-3196 to exacerbate the progression of pancreatic cancer [62]. In this study, we revealed that NFIX is overexpressed in OS cells. Moreover, NFIX overexpression neutralized the suppressive effects of miR-744-5p on the viability of OS cells and angiogenesis in HUVECs. These results are consistent with the previous observation that miR-744-5p negatively regulates NFIX to inhibit cell viability in ovarian cancer [18]. Our study revealed the association of miR-744-5p with NFIX in the OS progression and provided evidence for the importance of the miR-744-5p/NFIX axis in inhibiting OS cell growth and HUVECs angiogenesis. On considering the use of RNA interference technology in human diseases in recent years [63,64,65], targeting the miR-744-5p/NFIX axis may have potential application in the OS treatment.
This study had some limitations. We focused on the downstream regulatory mechanism of miR-744-5p in the current study. The mechanisms that suppress the miR-744-5p expression in OS have not been investigated. For example, long non-coding RNA MNX1-AS1 and LINC01116 bind to miR-744-5p and inhibit its expression in ovarian cancer and pituitary adenoma [57, 66]. Promoter methylation and transcriptional inhibition by transcription factors are also reported to be directly associated with suppressed miRNA expression [67, 68]. Additional investigations are warranted to unveil the mechanisms that regulate the expression of miR-744-5p in OS. Besides, we confirmed the roles of the miR-744-5p/NFIX axis on OS cells viability and HUVECs angiogenesis based on the in vitro assays. However, these results require further validation by clinical OS samples and in vivo studies such as subcutaneous injection or orthotopic models. Furthermore, the precise mechanism of miR-744-5p/NFIX in regulating the OS cells viability and HUVECs angiogenesis is still unclear, which needs further confirmation in the follow-up studies.
Conclusion
Taken together, we revealed the miR-744-5p/NFIX axis in osteosarcoma. MiR-744-5p overexpression could restrain osteosarcoma cell viability and HUVECs angiogenesis by targeting inhibition of NFIX. Our results highlight the significance of miR-744-5p as a potential target for osteosarcoma treatment.
Data availability
No datasets were generated or analysed during the current study.
References
Cortini M, Avnet S, Baldini N. Mesenchymal stroma: role in osteosarcoma progression. Cancer Lett. 2017;405:90–9.
Isakoff MS, Bielack SS, Meltzer P, et al. Osteosarcoma: current treatment and a collaborative pathway to Success. J Clin Oncol. 2015;33:3029–35.
van Maldegem AM, Bhosale A, Gelderblom HJ, et al. Comprehensive analysis of published phase I/II clinical trials between 1990–2010 in osteosarcoma and Ewing sarcoma confirms limited outcomes and need for translational investment. Clin Sarcoma Res. 2012;2:5.
Meltzer PS, Helman LJ. New Horizons in the Treatment of Osteosarcoma. N Engl J Med. 2021;385:2066–76.
Wang J, Liu S, Shi J, et al. The role of miRNA in the diagnosis, prognosis, and treatment of Osteosarcoma. Cancer Biother Radiopharm. 2019;34:605–13.
Zhang J, Yan YG, Wang C, et al. MicroRNAs in osteosarcoma. Clin Chim Acta. 2015;444:9–17.
Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14.
He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31.
Giordano L, Porta GD, Peretti GM, et al. Therapeutic potential of microRNA in tendon injuries. Br Med Bull. 2020;133:79–94.
Oliviero A, Della Porta G, Peretti GM, et al. MicroRNA in osteoarthritis: physiopathology, diagnosis and therapeutic challenge. Br Med Bull. 2019;130:137–47.
Wang F, Zhao QH, Liu JZ, et al. MiRNA-188-5p alleviates the progression of osteosarcoma via target degrading CCNT2. Eur Rev Med Pharmacol Sci. 2020;24:29–35.
Gao G, Tian Z, Zhu HY, et al. miRNA-133b targets FGFR1 and presents multiple tumor suppressor activities in osteosarcoma. Cancer Cell Int. 2018;18:210.
Lv Y, Wu L, Jian H, et al. Identification and characterization of aging/senescence-induced genes in osteosarcoma and predicting clinical prognosis. Front Immunol. 2022;13:997765.
Jiang Y, Wang J, Sun M, et al. Multi-omics analysis identifies osteosarcoma subtypes with distinct prognosis indicating stratified treatment. Nat Commun. 2022;13:7207.
Liu D, Wang H, Zhou Z, et al. Integrated bioinformatic analysis and experiment confirmation of the antagonistic effect and molecular mechanism of ginsenoside Rh2 in metastatic osteosarcoma. J Pharm Biomed Anal. 2021;201:114088.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559.
Tian H, Guan D, Li J. Identifying osteosarcoma metastasis associated genes by weighted gene co-expression network analysis (WGCNA). Med (Baltim). 2018;97:e10781.
Kleemann M, Schneider H, Unger K, et al. MiR-744-5p inducing cell death by directly targeting HNRNPC and NFIX in ovarian cancer cells. Sci Rep. 2018;8:9020.
Fan X, Sun Y, Guo X, et al. Long non-coding RNA LINC01116 regulated miR-744-5p/SCN1B axis to exacerbate lung squamous cell carcinoma. Cancer Biomark. 2020;28:473–82.
Guo B, Xiao C, Liu Y, et al. Mir-744-5p inhibits multiple Myeloma Proliferation, Epithelial Mesenchymal Transformation and Glycolysis by Targeting SOX12/Wnt/beta-Catenin signaling. Onco Targets Ther. 2021;14:1161–72.
Yuan Q, Fan Y, Liu Z, et al. Mir-744-5p mediates lncRNA HOTTIP to regulate the proliferation and apoptosis of papillary thyroid carcinoma cells. Exp Cell Res. 2020;392:112024.
Huang W, Chen Q, Dai J, et al. Mir-744-5p suppresses tumor proliferation and metastasis by targeting transforming growth factor-beta 1 (TGF-beta1) in hepatocellular carcinoma (HCC). J Gastrointest Oncol. 2021;12:1811–22.
Ma B, Ren G, Xu J, et al. LncRNA MNX1-AS1 contributes to laryngeal squamous cell Carcinoma Growth and Migration by regulating mir-744-5p/bcl9/beta-Catenin Axis. Cell Transpl. 2021;30:9636897211005682.
Liang H, Li L, Zhu S, et al. MicroRNA-744-5p suppresses tumorigenesis and metastasis of osteosarcoma through the p38 mitogen-activated protein kinases pathway by targeting transforming growth factor-beta 1. Bioengineered. 2022;13:12309–25.
Ribeiro V, Martins SG, Lopes AS et al. NFIXing Cancer: the role of NFIX in oxidative stress response and cell fate. Int J Mol Sci 2023, 24.
Chen KS, Lim JWC, Richards LJ, et al. The convergent roles of the nuclear factor I transcription factors in development and cancer. Cancer Lett. 2017;410:124–38.
Liu Z, Ge R, Zhou J, et al. Nuclear factor IX promotes glioblastoma development through transcriptional activation of Ezrin. Oncogenesis. 2020;9:39.
Rahman NIA, Abdul Murad NA, Mollah MM, et al. NFIX as a Master Regulator for Lung Cancer Progression. Front Pharmacol. 2017;8:540.
Allen-Rhoades W, Kurenbekova L, Satterfield L, et al. Cross-species identification of a plasma microRNA signature for detection, therapeutic monitoring, and prognosis in osteosarcoma. Cancer Med. 2015;4:977–88.
Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7.
Chen Z, Xu W, Zhang D, et al. circCAMSAP1 promotes osteosarcoma progression and metastasis by sponging mir-145-5p and regulating FLI1 expression. Mol Ther Nucleic Acids. 2021;23:1120–35.
Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13.
Sun L, Liu M, Luan S, et al. MicroRNA-744 promotes carcinogenesis in osteosarcoma through targeting LATS2. Oncol Lett. 2019;18:2523–9.
Jiao H, Zeng L, Zhang J, et al. THBS2, a microRNA-744-5p target, modulates MMP9 expression through CUX1 in pancreatic neuroendocrine tumors. Oncol Lett. 2020;19:1683–92.
Sui Y, Lin G, Zheng Y, et al. LncRNA MAFG-AS1 boosts the proliferation of lung adenocarcinoma cells via regulating miR-744-5p/MAFG axis. Eur J Pharmacol. 2019;859:172465.
Ren P, Chang L, Hong X, et al. Long non-coding RNA LINC01116 is activated by EGR1 and facilitates lung adenocarcinoma oncogenicity via targeting miR-744-5p/CDCA4 axis. Cancer Cell Int. 2021;21:292.
Liu L, Cheng M, Zhang T, et al. Mesenchymal stem cell-derived extracellular vesicles prevent glioma by blocking M2 polarization of macrophages through a miR-744-5p/TGFB1-dependent mechanism. Cell Biol Toxicol. 2022;38:649–65.
Shan H, Zhang X, Zhang X, et al. CircSCUBE3 reduces the anti-gastric Cancer activity of Anti-PD-L1. Mol Biotechnol. 2024;66:123–37.
Han B, Liu Y, Zhang Q, et al. Propofol decreases cisplatin resistance of non-small cell lung cancer by inducing GPX4-mediated ferroptosis through the miR-744-5p/miR-615-3p axis. J Proteom. 2023;274:104777.
Folkman J. The role of angiogenesis in tumor growth. Semin Cancer Biol. 1992;3:65–71.
Quan GM, Choong PF. Anti-angiogenic therapy for osteosarcoma. Cancer Metastasis Rev. 2006;25:707–13.
Zhu Y, Liu Z, Cao L, et al. FRS2 regulated by miR-429 and miR-206 promotes angiogenesis in osteosarcoma. Gene. 2024;898:148118.
Chen M, Li Z, Cao L, et al. Mir-877-3p inhibits tumor growth and angiogenesis of osteosarcoma through fibroblast growth factor 2 signaling. Bioengineered. 2022;13:8174–86.
Zhang L, Yu D. Exosomes in cancer development, metastasis, and immunity. Biochim Biophys. Acta Rev Cancer. 2019;1871:455–68.
Liu J, Ren L, Li S, et al. The biology, function, and applications of exosomes in cancer. Acta Pharm Sin B. 2021;11:2783–97.
Kang T, Atukorala I, Mathivanan SJN. New frontiers: extracellular vesicles. Biogenesis of extracellular vesicles; 2021. pp. 19–43.
Yang K, Zhang J, Bao C. Exosomal circEIF3K from cancer-associated fibroblast promotes colorectal cancer (CRC) progression via miR-214/PD-L1 axis. BMC Cancer. 2021;21:933.
Marton S, Miquel E, Acosta-Rodriguez J, et al. SOD1(G93A) astrocyte-derived Extracellular vesicles Induce Motor Neuron death by a miRNA-155-5p-Mediated mechanism. ASN Neuro. 2023;15:17590914231197527.
Choi JY, Seok HJ, Lee DH, et al. Tumor-derived mir-6794-5p enhances cancer growth by promoting M2 macrophage polarization. Cell Commun Signal. 2024;22:190.
Liao Z, Chen Y, Duan C, et al. Cardiac telocytes inhibit cardiac microvascular endothelial cell apoptosis through exosomal miRNA-21-5p-targeted cdip1 silencing to improve angiogenesis following myocardial infarction. Theranostics. 2021;11:268–91.
Wang H, Wang L, Zhou X et al. OSCC Exosomes Regulate miR-210-3p Targeting EFNA3 to Promote Oral Cancer Angiogenesis through the PI3K/AKT Pathway. Biomed Res Int 2020, 2020:2125656.
Masoumi-Dehghi S, Babashah S, Sadeghizadeh M. microRNA-141-3p-containing small extracellular vesicles derived from epithelial ovarian cancer cells promote endothelial cell angiogenesis through activating the JAK/STAT3 and NF-kappaB signaling pathways. J Cell Commun Signal. 2020;14:233–44.
Plouet J, Moro F, Bertagnolli S, et al. Extracellular cleavage of the vascular endothelial growth factor 189-amino acid form by urokinase is required for its mitogenic effect. J Biol Chem. 1997;272:13390–6.
O’Neill CP, Dwyer RM. Nanoparticle-based delivery of tumor suppressor microRNA for Cancer Therapy. Cells 2020, 9.
Valencia-Sanchez MA, Liu J, Hannon GJ, et al. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24.
Zhao LG, Wang J, Li J, et al. Mir-744-5p inhibits cellular proliferation and invasion via targeting ARF1 in epithelial ovarian cancer. Kaohsiung J Med Sci. 2020;36:799–807.
Shen Y, Lv M, Fang Y, et al. LncRNA MNX1-AS1 promotes ovarian cancer process via targeting the miR-744-5p/SOX12 axis. J Ovarian Res. 2021;14:161.
Fan F, Yao D, Yan P, et al. MicroRNA-744-5p inhibits glioblastoma malignancy by suppressing replication factor C subunit 2. Oncol Lett. 2021;22:608.
Walker M, Li Y, Morales-Hernandez A, et al. An NFIX-mediated regulatory network governs the balance of hematopoietic stem and progenitor cells during hematopoiesis. Blood Adv. 2023;7:4677–89.
Taglietti V, Angelini G, Mura G et al. RhoA and ERK signalling regulate the expression of the transcription factor nfix in myogenic cells. Development 2018, 145.
Fraser J, Essebier A, Gronostajski RM, et al. Cell-type-specific expression of NFIX in the developing and adult cerebellum. Brain Struct Funct. 2017;222:2251–70.
Ye L, Feng W, Weng H, et al. MAFG-AS1 aggravates the progression of pancreatic cancer by sponging miR-3196 to boost NFIX. Cancer Cell Int. 2020;20:591.
Gargano G, Oliviero A, Oliva F, et al. Small interfering RNAs in tendon homeostasis. Br Med Bull. 2021;138:58–67.
Gargano G, Oliva F, Oliviero A, et al. Small interfering RNAs in the management of human rheumatoid arthritis. Br Med Bull. 2022;142:34–43.
Gargano G, Asparago G, Spiezia F, et al. Small interfering RNAs in the management of human osteoporosis. Br Med Bull. 2023;148:58–69.
Huang T, Cai M, Chen C, et al. LINC01116 boosts the progression of pituitary adenoma via regulating miR-744-5p/HOXB8 pathway. Mol Cell Endocrinol. 2021;536:111350.
Yan Z, Ao X, Liang X, et al. Transcriptional inhibition of mir-486-3p by BCL6 upregulates snail and induces epithelial-mesenchymal transition during radiation-induced pulmonary fibrosis. Respir Res. 2022;23:104.
Ye S, Xiong F, He X, et al. DNA hypermethylation-induced miR-182 silence targets BCL2 and HOXA9 to facilitate the self-renewal of leukemia stem cell, accelerate acute myeloid leukemia progression, and determine the sensitivity of BCL2 inhibitor venetoclax. Theranostics. 2023;13:77–94.
Acknowledgements
Not applicable.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Lin Xie, Wei Li and Yu Li. The first draft of the manuscript was written by Lin Xie and Yu Li. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
None.
Consent to participate
None.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xie, L., Li, W. & Li, Y. mir-744-5p inhibits cell growth and angiogenesis in osteosarcoma by targeting NFIX. J Orthop Surg Res 19, 485 (2024). https://doi.org/10.1186/s13018-024-04947-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13018-024-04947-x