Abstract
Background
Tumor microenvironment (e.g., cancer-associated fibroblast) plays a key role in cancer tumorigenesis and metastasis. However, the detailed mechanism of whether hypoxia promotes CRC progression via tumor microenvironment remains unclear.
Methods
In this study, circEIF3K exosome was examined by NanoSight Tracking Analysis and RT-qPCR. We used cell colony formation assay, transwell assay and tube formation assay to determine proliferation, invasion and metastasis of HCT116 or SW620 cells. Xenograft tumor assay was employed to show in vivo tumor growth of HCT116 cells.
Results
We found that hypoxia could induce secretion of circEIF3K exosome. Conditioned medium (CM) and exosome from circEIF3K knockdown CAF significantly attenuated proliferation, invasion and tube formation of HCT116 or SW620 cells, which could be reverted by miR-214 under hypoxia treatment. Besides, we observed that circEIF3K knockdown evidently impaired tumor growth in mice. TCGA dataset analysis showed that low expression of circEIF3K was observed in normal tissues and associated with prolonged survival time. Finally, PD-L1 was confirmed as important target for miR-214 in CRC.
Conclusion
In conclusion, our study reveals that a novel axis circEIF3K/miR-214/PD-L1 mediates hypoxia-induced CRC progression via CAF, providing the rationale for developing new targeted therapeutics to treat CRC.
Similar content being viewed by others
Background
Colorectal cancer is the third common tumor type and caused cancer-related mortality [1]. 5-year survival rate is 65% [2]. In general, most of CRC tumor will progress to liver metastasis state [3]. Tumor microenvironment plays a critical role in cancer progression. Of which, hypoxia is a common feature of solid tumors, which contributes to tumorigenesis and metastasis [4]. Recently, some studies reported that hypoxia regulated cancer progression via promoting secretion of circRNAs exosome [5, 6].
circRNA are originally regarded as non-functional junk RNA. However, some recent evidences showed that circRNAs are involved in a variety of pathological processes, such as tumors [7, 8]. In CRC, some circRNAs are found to regulate tumor phenotypes. CircCCDC66 regulated miR-3140/autophagy to promote CRC tumorigenesis [5]. Has_circ_0000826 was associated with CRC metastasis [6]. To date, circEIF3K was shown to modulate apoptosis and autophagy in bacterial cells [9]. Unfortunately, whether circEIF3K can affect cancer biology remains unknown.
miRNAs are usually 18-22 nt non-coding RNAs [10]. They regulate gene expression levels via binding 3′-UTR of mRNAs, which leads to aberrant expression of target genes [11]. A number of studies showed that miRNAs were dysregulated in various types of cancer cells [12, 13]. MiR-214 has been demonstrated to inhibit HCC [14], bladder cancer [15] and CRC [16] progression. Thus, we hypothesized that miR-214 might be potential target of circEIF3K in CRC. Programmed death ligand-1 (PD-L1, also known as CD274) is a negative immune regulator via blockage of T cell activation signaling [17]. Interestingly, PD-L1 is overexpressed in many tumors. Sun et al. revealed that miR-214 targeted PD-L1 in diffuse large B-cell lymphoma [18]. Based on these findings, we hypothesized that PD-L1 might act as downstream effector for circEIF3K in CRC.
In this study, the purpose is to illuminate the underlying mechanism of circEIF3K-regulated CRC tumorigenesis and metastasis. We attempted to investigate the role of miR-214/PD-L1 axis in CRC. The conclusions will advance our understanding of CRC pathogenesis and treatments.
Materials and methods
Cell culture and treatment
Human colorectal cancer cells HCT116 and SW620, and human embryonic kidney cell 293 T were all purchased from Shanghai cell bank of Chinese Academy of Sciences. Normal colon epithelial cell FHC were from ATCC. All cells were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with Fetal Bovine Serum (FBS, 10%). Human dermal lymphatic endothelial cells (HDLEC) (PromoCell) were cultured in endothelial cell growth medium at 5% CO2, 37 °C. For hypoxic treatment, the cells were cultured under hypoxia (94% N2, 5% CO2, 1% O2) or normoxia (20% O2, 5% CO2). 2 mg/ml Actinomycin D (Shanghai Haoran) was used to treat HCT116 cells for 24 h.
Generation of stable cell lines
sh-NC, sh-circEIF3K-1 and sh-circEIF3K-2 were introduced along with lentivirus packaging vectors (pVSVG and pPAX2) into 293 T cells by Lipofectamine 2000 (invitrogen). The viruses-containing media were collected after 48 h of transfection. The viruses were then used to infect CRC cells. To obtain stable cells, 2μg/ml puromycin was used to treat the cells for about 7 days. shNC denotes scramble vector.
-
sh-NC: UACCUUGAAGCCUUAAAACCU
-
sh-circEIF3K-1: GACUCAGCCGUACCAGUUCAA
-
sh-circEIF3K-2: GUUGACUCAGCCGUACCAGUU
Isolation of cancer-associated fibroblasts
CAFs were isolated according to previously described [19]. The cells were immortalized by transfecting with human telomerase reverse transcriptase. The medium of CAFs were termed as conditioned medium (CM). In this study, CM of CAFs treated with mock or hypoxia were named as mock-CM or hypoxia-CM. This study was approved by ethics committee of affiliated hospital of Jiangnan university, and informed consents were acquired from the patients before this study.
Cell transfection
We utilized Lipofectamine 2000 (invitrogen) to transfect miR-214 mimic or inhibitor (~ 50 uM) into HCT116 or SW620 cells. The cells were collected and used for subsequent analysis after 48 h of transfection.
Colony formation assay
CRC cells (2 × 104 cells) were seeded in 6-well plate and cultured for 2 weeks. Then, the cell colonies were fixed with 4% paraformaldehyde reagent. Next, 0.005% crystal violet was used to stain the colonies. For CM treatment, the old CM was replaced with fresh CM every 3 days.
Tube formation assay
In this experiment, 24-well plates were coated with Growth Factor Reduced (GFR) matrigel at 37 °C for 2–3 h. We seeded HDLEC cells (~ 20,000 cells/well) into coated plates. Formed tubes were observed and captured by microscopy (× 200 magnification). CM treatment was performed during this experiment.
Invasion assay
HCT116 or SW620 cells (1 × 105 cells) were suspended in 200 μL of media without FBS. Top chamber contains 8 μm pore membrane. Meanwhile, media with 10% FBS was added to the bottom chamber. After 24 h, invaded cells were stained with crystal violet. The invaded cell number was counted for statistical analysis.
RT-qPCR and RNase R treatment
Trizol (invitrogen) was used to lyse the cells, and RNAs were dissolved in RNase-free ddH2O. To differentiate circRNA and mRNA, RNAs were treated with RNase R (Epicenter) at 37 °C for 2–3 h. 1 μg of total RNAs were reverse transcribed using PrimeScript Kit (TAKARA). Real-time PCR was carried out using SYBR Green reagent. Thermocycling conditions were used: Initial denaturation at 95 °C for 5 min, followed by 40 cycles of 95 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s.
-
circEIF3K-F: TGACAGTGTGTCCAGCATCA
-
circEIF3K-R: AAGTCTGTGTGCGGCAAGTT
-
miR-214-F: TCTGCCTGTCTACACTTGCT
-
miR-214-R: TGTACAGGTGAGCGGATGTT
-
PD-L1-F: TGCCACCCACTGTCCTTTTA
-
PD-L1-R: GTTTTCCCCTCGCATCATCC
-
Actin-F: TGGCATCCACGAAACTACCT
-
Actin-R: TCTCCTTCTGCATCCTGTCG
Isolation of exosome
CAFs were seeded overnight, supernatants of media were collected and centrifuged at 10,000×g for 30 min to discard cell debris. Then, the supernatants were centrifuged at 120,000×g for 2 h to collect vesicles.
NanoSight tracking analysis
The CAFs were seeded in 12-well plate and cultured in DMEM for 24–36 h. Then, the supernatant were collected and centrifuged at 120,000×g for 30 min, 4 °C.Supernatants and exosomes were analyzed on NanoSight LM10.
Xenograft
We performed this experiment using 6 immunodecifient male NOD-SCID mice (3 mice per group). Mice were injected subcutaneously with HCT116 cells (2 × 106). Cells resuspended in 100 μL of PBS were injected into the tail vein. We measured tumor weight and volume. Tumor volume = 0.5 × length × width2. This study was approved by ethics committee of affiliated hospital of Jiangnan university.
TCGA
Datasets of CRC patients were downloaded from TCGA (275 tumors and 349 normal tissues). The circEIF3K expression levels were shown in normal and tumor tissues. In addition, overall survival and stage plot of CRC patients were also shown.
Statistical analysis
All data in this study were presented as mean ± SD. Chi-squared test, Student’s t test, One-way ANOVA were performed for comparisons. *P < 0.05 was statistically significant.
Results
Hypoxia stimulated exosomal circEIF3K secretion from CAF
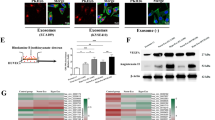
Hypoxia has been shown to enhance cancer progression; however, the detailed mechanism remains unclear. We hypothesized that hypoxia might contribute to cancer progression via tumor microenvironment (e.g., cancer-associated fibroblast). To validate, the CAFs were cultured and treated with normoxia or hypoxia. We found that CAFs could secrete exosomes under hypoxia treatment (Fig. 1A). More interestingly, we also observed that circEIF3K was existed in exosome. The level of exosomal circEIF3K in CAF media treated with hypoxia was higher (Fig. 1B). To confirm whether the exosomal RNA was circRNA, we used RNase R to treat circEIF3K. EIF3K mRNA (mEIF3K) was used as positive control (Fig. 1C). In addition, we used Actinomycin D to treat HCT116 cells and measured circEIF3K and EIF3K mRNA (Fig. 1D). The result showed that RNase R and Actinomycin D could decrease EIF3K mRNA, not circEIF3K.
Hypoxia stimulated exosomal circEIF3K secretion from CAF. A NanoSight quantification of solution or exosome from conditioned media of CAFs under hypoxia treatment. B RT-qPCR showed circEIF3K level in exosome fraction of mock-CM or hypoxia-CM of CAFs. ***p < 0.001. C RT-qPCR showed RNA level of circular RNA (circEIF3K) or mRNA (mEIF3K) treated with or without RNase R. ***p < 0.001. D RT-qPCR showed RNA level of circular RNA (circEIF3K) or mRNA (mEIF3K) in HCT116 cells treated with or without Actinomycin D. ***p < 0.001
circEIF3K depletion in CAF led to attenuated CRC progression
As circEIF3K could be released by CAF under hypoxia, we attempted to investigate the effect of circEIF3K knockdown in CAF on CRC cell tumorigenesis. RT-qPCR showed that circEIF3K was markedly decreased in sh-circEIF3K-1 and sh-circEIF3K-2 cells (Fig. 2A). Then, we performed cell colony formation assay to examine proliferation of HCT116 or SW620 cells incubated with media of CAF. circEIF3K knockdown significantly led to decrease in cell colony number (Fig. 2B and C). Transwell assay results demonstrated that CRC cells incubated with media of sh-circEIF3K CAF exhibited less invaded cells (Fig. 2D and E). Likewise, tube formation assay showed that tube length of HDLEC was attenuated by media of CRC cells preincubated with CM of sh-circEIF3K CAF (Fig. 2F and G). Eventually, we treated HCT116 or SW620 cells with exosomes derived from sh-NC or sh-circEIF3K CAFs. The results demonstrated that CRC cells treated with exosomes from sh-circEIF3K CAFs exhibited less cell colony number than CRC cells treated with exosomes from sh-NC CAFs (Fig. 2H and I). In conclusion, our data suggested that circEIF3K depletion in CAF inhibited CRC progression.
circEIF3K depletion in CAF led to attenuated CRC progression. A circEIF3K expression levels were shown in CAFs stably transfected with sh-NC, sh-circEIF3K-1, sh-circEIF3K-2 by RT-qPCR. ***p < 0.001. B and C Cell proliferation of HCT116 or SW620 cells incubated with CM of CAFs stably transfected with sh-NC, sh-circEIF3K-1, sh-circEIF3K-2 by cell colony formation assay under hypoxia. ***p < 0.001. D and E The invasion of HCT116 or SW620 cells incubated with CM of CAFs stably transfected with sh-NC, sh-circEIF3K-1, sh-circEIF3K-2 were determined by transwell assay under hypoxia. ***p < 0.001. F and G Tube lengths of HDLEC cells (sh-NC-CM, sh-circEIF3K-1-CM, sh-circEIF3K-2-CM) were measured by tube formation assay under hypoxia. ***p < 0.001. H and I Cell proliferation of HCT116 or SW620 cells incubated with exosomes derived from CAFs stably transfected with sh-NC, sh-circEIF3K-1, sh-circEIF3K-2 by cell colony formation assay under hypoxia. ***p < 0.001
circEIF3K modulated CRC progression in animal and patients
In order to investigate the role of circEIF3K in xenograft tumor growth, HCT116 cells were utilized to inoculate subcutaneously into NOD-SCID mice (Fig. 3A). Then, the mice were injected with exosomes from sh-NC or sh-circEIF3K CAFs. Tumor volume and weight data revealed that sh-NC exosome-treated tumors were larger than sh-circEIF3K exosome (Fig. 3B and C).
circEIF3K modulated CRC progression in animal and patients. A In vivo tumor growth of HCT116 cells treated with sh-NC exosome (sh-NC exo), sh-circEIF3K-1 exosome (sh-circEIF3K-1 exo) were examined in NOD-SCID mice (n = 3). B and C Tumor weights (B) and volumes (C) in Figure 3A were measured. **p < 0.01; ***p < 0.001. D RT-qPCR showed circEIF3K levels of normal colon epithelial cell FHC and CRC cell lines (HCT116 and SW620 cells). ***p < 0.001. E TCGA dataset analysis showed circEIF3K levels of CRC tumors (n = 275) and adjacent normal tissues (n = 349). F Overall survival of CRC patients with low circEIF3K level group (n = 135) and high circEIF3K level group (n = 135). p = 0.017. G Stage plot of CRC patients showed association of circEIF3K levels with stages. p = 0.0126 (stage IV vs stage I)
In addition, we evaluated circEIF3K levels in normal colon epithelial cell FHC and two CRC cell lines (HCT116 and SW620) by RT-qPCR. We found that circEIF3K was elevated in CRC cell lines compared to FHC (Fig. 3D). We analyzed circEIF3K levels in normal tissues and CRC tumors in TCGA datasets. Consistent with data in cell lines, we also observed increase in circEIF3K levels in CRC tumor patients (Fig. 3E). Patients with low level of circEIF3K exhibited prolonged survival rate in relative to high group (Fig. 3F). Moreover, expression levels of circEIF3K was higher in advanced stages (III + IV) in relative to low stages (I + II) (Fig. 3G). Together, circEIF3K exerted oncogenic role in animal and clinics.
miR-214 was key for hypoxia-induced tumorigenesis of CRC
To figure out the potential molecular mechanism of circEIF3K-promoted CRC, online predict program StarBase v2.0 was utilized to identify miR-214 as one of the downstream effectors (Fig. 4A). RT-qPCR data showed that circEIF3K knockdown increased miR-214 level in HCT116 cells (Fig. 4B). Because hypoxia could induce secretion of exosomal circEIF3K from CAF, we sought to explore whether miR-214 could rescue hypoxia-mediated phenotypes. We first examined miR-214 levels in HCT116 cells treated with normoxia, hypoxia or hypoxia plus miR-214 (Fig. 4C). Expectedly, we found that hypoxia evidently increased cell colony number, while miR-214 mimic reverted hypoxia-enhanced cell colony formation ability (Fig. 4D and E). Besides, transwell assay demonstrated that hypoxia induced invasion of CRC cells, which could be attenuated by miR-214 mimic (Fig. 4F and G). Finally, tube formation assay revealed that hypoxia substantially stimulated tube formation of HDLEC, whereas this enhancement was largely restored by miR-214 (Fig. 4H and I).
miR-214 was key for hypoxia-induced tumorigenesis of CRC. A StarBase 2.0 predicted the interaction between circEIF3K and miR-214. B miR-214 levels were examined by RT-qPCR in HCT116 cells stably transfected with sh-NC or sh-circEIF3K-1. ***p < 0.001. C miR-214 expression levels were detected by RT-qPCR in HCT116 cells treated with mock-CM, hypoxia-CM, hypoxia-CM + miR-214 mimic. ***p < 0.001. D and E Cell proliferation of HCT116 cells treated with mock-CM, hypoxia-CM, hypoxia-CM + miR-214 mimic were examined by cell colony formation assay. ***p < 0.001. F and G Invasion of HCT116 cells treated with mock-CM, hypoxia-CM, hypoxia-CM + miR-214 mimic were measured by transwell assay. ***p < 0.001. H and I Tube lengths of HDLEC cells (mock-CM, hypoxia-CM, hypoxia-CM + miR-214 mimic) were measured by tube formation assay. ***p < 0.001
miR-214 downregulated PD-L1 expression in CRC
Next, we asked how miR-214 inhibited CRC progression. Previous study showed that PD-L1 was regulated by miR-214 [18]. PD-L1 level was reduced in miR-214 mimic-transfected HCT116 cells, while its level was increased by miR-214 inhibitor (Fig. 5A). In addition, we employed RT-qPCR to determine PD-L1 mRNA levels in HCT116 cells transfected with sh-NC, sh-circEIF3K or sh-circEIF3K plus miR-214 inhibitor. The result displayed decreased PD-L1 expression and partial rescue of the attenuated PD-L1 expression (Fig. 5B). Then, RT-qPCR showed lower PD-L1 levels in HCT116 cells incubated with exosomes from sh-circEIF3K CAFs compared to sh-NC (Fig. 5C). In patients, TCGA dataset showed higher level of PD-L1 in CRC tumors compared to normal tissues (Fig. 5D). To sum up, these data indicated that PD-L1 might be an potential target for circEIF3K/miR-214 axis in CRC.
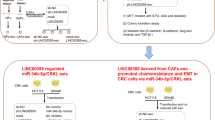
miR-214 downregulated PD-L1 expression in CRC. A PD-L1 expression levels were detected by RT-qPCR in HCT116 cells transiently transfected with NC mimic, miR-214 mimic, NC inhibitor, miR-214 inhibitor. ***p < 0.001. B PD-L1 mRNA levels were detected by RT-qPCR in HCT116 cells expressing sh-NC, sh-circEIF3K-1, sh-circEIF3K-1 plus miR-214 inhibitor. **p < 0.01; ***p < 0.001. C PD-L1 mRNA levels were detected by RT-qPCR in HCT116 cells incubated with exosomes derived from CAFs stably transfected with sh-NC, sh-circEIF3K-1, sh-circEIF3K-2. ***p < 0.001. D TCGA dataset analysis showed PD-L1 levels of CRC tumors (n = 275) and adjacent normal tissues (n = 349). E Working model of hypoxia-induced tumorigenesis of CRC by promoting circEIF3K exosomes secretion from CAFs
Discussion
Cancer-associated fibroblast was one of the stromal cells in tumor microenvironment that regulated proliferation, migration and metastasis of cancer cells [20]. During this process, some risk factors (e.g., hypoxia) might be important for CAFs-affected solid tumors [21]. For instances, CAFs were found to be implicated in breast cancer [22], pancreatic cancer [23], and gastric cancer [24]. However, few reports revealed the detailed and clear mechanism for explaining the role of hypoxia in promoting cancer progression via CAFs. In our study, we utilized a series of experiments to validate that hypoxia-treated CAFs could induce secretion of exosomal circEIF3K.
Exosomes are defined as secreted extracellular vesicles with 30–150 nm in diameter [25]. In which, some molecules (like DNAs, RNAs, proteins or lipids) were existed in exosomes with lipid bilayer [26]. The exosomes could transfer the molecules to surrounding cells and impact cell biology [27, 28]. We found that exosome secreted by hypoxia contained circEIF3K and transferred to CRC cells. Our findings were consistent with previous studies.
To date, circRNAs were involved in multiple biological processes. CircMTO1 regulated Wnt signaling to affect proliferation and invasion of CRC [29]. CircITGA7 suppressed CRC metastasis via regulating Ras pathway [30]. Zhang et al. showed circSMAD2 promoted CRC cells proliferation [31]. Besides, we also demonstrated that circEIF3K played a role in CRC proliferation, invasion and metastasis, which had never been reported before.
MiR-214 has been shown to inhibit cancer cell phenotypes through interacting with lncRNAs, circRNAs or others [32, 33]. In this study, miR-214 was confirmed to bind circEIF3K and could rescue circEIF3K-mediated CRC progression, which was in agreement with prior findings. On the other hand, Sun et al. showed that miR-214 targeted PD-L1 and downregulated its expression [18]. We finally confirmed this conclusion in CRC cells, which was first in CRC.
Conclusions
In conclusion, we originally propose that circEIF3K is implicated in CRC by regulating miR-214/PD-L1 (Fig. 5E). These findings, for the first time, show the relationship between circEIF3K and CRC. At molecular level, we also creatively link circEIF3K to miR-214 and PD-L1. Thus, our research will deepen the understanding of CRC pathogenesis and help develop novel therapeutic approaches.
Availability of data and materials
All data generated or analysed during this study were included in this article. TCGA dataset (https://tcga-data.nci.nih.gov) was used in the study.
Abbreviations
- CRC:
-
Colorectal cancer
- CAF:
-
Cancer-associated fibroblast
- RT-qPCR:
-
Reverse-transcription quantitative PCR
- GFR:
-
Growth Factor Reduced
- TCGA:
-
The Cancer Genome Atlas
- PD-L1:
-
Programmed death ligand-1
- HDLEC:
-
Human dermal lymphatic endothelial cells
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.
Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–64. https://doi.org/10.3322/caac.21601.
Rothbarth J, van de Velde CJ. Treatment of liver metastases of colorectal cancer. Ann Oncol. 2005;16(Suppl 2):ii144–9.
Deep G, Panigrahi GK. Hypoxia-induced signaling promotes prostate Cancer progression: exosomes role as messenger of hypoxic response in tumor microenvironment. Crit Rev Oncog. 2015;20(5-6):419–34. https://doi.org/10.1615/CritRevOncog.v20.i5-6.130.
Feng J, Li Z, Li L, Xie H, Lu Q, He X. Hypoxiainduced circCCDC66 promotes the tumorigenesis of colorectal cancer via the miR3140/autophagy pathway. Int J Mol Med. 2020;46(6):1973–82. https://doi.org/10.3892/ijmm.2020.4747.
Shi L, Tao C, Tang Y, Xia Y, Li X, Wang X. Hypoxia-induced hsa_circ_0000826 is linked to liver metastasis of colorectal cancer. J Clin Lab Anal. 2020;34:e23405.
Greene J, Baird AM, Brady L, Lim M, Gray SG, McDermott R, et al. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front Mol Biosci. 2017;4:38. https://doi.org/10.3389/fmolb.2017.00038.
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–4. https://doi.org/10.1038/cr.2015.82.
Yao L, Xu B, Li X. Neisseria gonorrhoeae-induced salpingitis is targeted by circular RNA EIF3K via miR-139-5p and regulating MAPK/NF-kappaB signaling pathway to promotes apoptosis and autophagy bacterial cells. Microb Pathog. 2020;142:104051. https://doi.org/10.1016/j.micpath.2020.104051.
Haider MT, Taipaleenmaki H. Targeting the metastatic bone microenvironment by MicroRNAs. Front Endocrinol. 2018;9:202. https://doi.org/10.3389/fendo.2018.00202.
Tian X, Chang J, Zhang N, Wu S, Liu H, Yu J. MicroRNA-429 acts as a tumor suppressor in colorectal cancer by targeting high mobility group box 3. Oncol Lett. 2021;21(4):250. https://doi.org/10.3892/ol.2021.12511.
Zhang H, Fu T, Zhang C. MicroRNA-1249 targets G protein subunit alpha 11 and facilitates gastric Cancer cell proliferation, motility and represses cell apoptosis. OncoTargets Ther. 2021;14:1249–59. https://doi.org/10.2147/OTT.S272599.
Zhu B, Mitheera V, Finch-Edmondson M, Lee Y, Wan Y, Sudol M, et al. miR-582-5p Is a Tumor Suppressor microRNA Targeting the Hippo-YAP/TAZ Signaling Pathway in Non-Small Cell Lung Cancer. Cancers. 2021;13:756.
Li Y, Li Y, Chen Y, Xie Q, Dong N, Gao Y, et al. MicroRNA-214-3p inhibits proliferation and cell cycle progression by targeting MELK in hepatocellular carcinoma and correlates cancer prognosis. Cancer Cell Int. 2017;17(1):102. https://doi.org/10.1186/s12935-017-0471-1.
Ecke TH, Stier K, Weickmann S, Zhao Z, Buckendahl L, Stephan C, et al. miR-199a-3p and miR-214-3p improve the overall survival prediction of muscle-invasive bladder cancer patients after radical cystectomy. Cancer Med. 2017;6(10):2252–62. https://doi.org/10.1002/cam4.1161.
Shan H, Zhou X, Chen C. MicroRNA214 suppresses the viability, migration and invasion of human colorectal carcinoma cells via targeting transglutaminase 2. Mol Med Rep. 2019;20(2):1459–67. https://doi.org/10.3892/mmr.2019.10325.
Chen J, Jiang CC, Jin L, Zhang XD. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol. 2016;27(3):409–16. https://doi.org/10.1093/annonc/mdv615.
Sun JR, Zhang X, Zhang Y. MiR-214 prevents the progression of diffuse large B-cell lymphoma by targeting PD-L1. Cell Mol Biol Lett. 2019;24(1):68. https://doi.org/10.1186/s11658-019-0190-9.
Gu C, Lu H, Qian Z. Matrine reduces the secretion of exosomal circSLC7A6 from cancer-associated fibroblast to inhibit tumorigenesis of colorectal cancer by regulating CXCR5. Biochem Biophys Res Commun. 2020;527(3):638–45. https://doi.org/10.1016/j.bbrc.2020.04.142.
Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–37. https://doi.org/10.1038/nm.3394.
Xu K, Zhan Y, Yuan Z, Qiu Y, Wang H, Fan G, et al. Hypoxia induces drug resistance in colorectal Cancer through the HIF-1alpha/miR-338-5p/IL-6 feedback loop. Mol Ther. 2019;27(10):1810–24. https://doi.org/10.1016/j.ymthe.2019.05.017.
Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121(3):335–48. https://doi.org/10.1016/j.cell.2005.02.034.
Richards KE, Zeleniak AE, Fishel ML, Wu J, Littlepage LE, Hill R. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene. 2017;36(13):1770–8. https://doi.org/10.1038/onc.2016.353.
Itoh G, Chida S, Yanagihara K, Yashiro M, Aiba N, Tanaka M. Cancer-associated fibroblasts induce cancer cell apoptosis that regulates invasion mode of tumours. Oncogene. 2017;36(31):4434–44. https://doi.org/10.1038/onc.2017.49.
Wang Y, Xie Y, Zhang A, Wang M, Fang Z, Zhang J. Exosomes: An emerging factor in atherosclerosis. Biomed Pharmacother. 2019;115:108951.
Boelens MC, Wu TJ, Nabet BY, Xu B, Qiu Y, Yoon T, et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159(3):499–513. https://doi.org/10.1016/j.cell.2014.09.051.
Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, et al. Suppression of Exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell. 2019;177(2):414–27 e13. https://doi.org/10.1016/j.cell.2019.02.016.
Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine. 2019;40:432–45. https://doi.org/10.1016/j.ebiom.2018.12.062.
Ge Z, Li LF, Wang CY, Wang Y, Ma WL. CircMTO1 inhibits cell proliferation and invasion by regulating Wnt/beta-catenin signaling pathway in colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22(23):8203–9. https://doi.org/10.26355/eurrev_201812_16513.
Li X, Wang J, Zhang C, Lin C, Zhang J, Zhang W, et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol. 2018;246(2):166–79. https://doi.org/10.1002/path.5125.
Zhang W, Wu G, Sun P, Zhu Y, Zhang H. circ_SMAD2 regulate colorectal cancer cells proliferation through targeting miR-1258/RPN2 signaling pathway. J Cancer. 2021;12(6):1678–86. https://doi.org/10.7150/jca.50888.
Tang Q, Zheng F, Liu Z, Wu J, Chai X, He C, et al. Novel reciprocal interaction of lncRNA HOTAIR and miR-214-3p contribute to the solamargine-inhibited PDPK1 gene expression in human lung cancer. J Cell Mol Med. 2019;23(11):7749–61. https://doi.org/10.1111/jcmm.14649.
Peng W, Zhu S, Chen J, Wang J, Rong Q, Chen S. Hsa_circRNA_33287 promotes the osteogenic differentiation of maxillary sinus membrane stem cells via miR-214-3p/Runx3. Biomed Pharmacother. 2019;109:1709–17.
Acknowledgements
Not applicable.
Funding
Our work was supported by Project of Wuxi Health Bureau (MS201924).
Author information
Authors and Affiliations
Contributions
K.Y. and J.Z. designed and performed experiments. J.Z. prepared figures. C.B. analyzed data and wrote the manuscript. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Animal experiment in this study was approved by the ethics committee of affiliated hospital of Jiangnan university. The study was carried out in compliance with the ARRIVE guidelines. All methods were carried out in accordance with relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
These authors declare no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yang, K., Zhang, J. & Bao, C. Exosomal circEIF3K from cancer-associated fibroblast promotes colorectal cancer (CRC) progression via miR-214/PD-L1 axis. BMC Cancer 21, 933 (2021). https://doi.org/10.1186/s12885-021-08669-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-021-08669-9