Abstract
Background
Osteosarcoma is the most prevalent malignant osseous sarcoma in children and adolescents, whose prognosis is still relatively poor nowadays. Recent studies have shown the critical function and potential clinical applications of circular RNAs (circRNAs) in osteosarcoma. Our review aimed to perform an updated meta-analysis to explore their clinicopathologic significance and prognostic value.
Methods
The structured literature was conducted via eight electronic databases and four gray literature sources until 20 Feb 2021 to identify eligible studies. The data was extracted directly from the articles or reconstructed based on Kaplan-Meier curves. The Newcastle-Ottawa Scale (NOS) tool was used to assess study quality. The clinicopathologic significance of circRNAs was measured through odds ratios (ORs) and their 95% confidence intervals (CIs), while the prognostic value was evaluated through hazard ratios (HRs) and their 95% CIs of overall survival (OS) and disease-free survival (DFS). Heterogeneity and publication bias were assessed. Sensitivity analyses were conducted. Subgroup analyses were performed according to study characteristics. An additional analysis was performed to investigate the relation between circ_0002052 and osteosarcoma.
Results
Fifty-two studies were identified, in which 38 on clinicopathologic features and 36 on survival prognosis were included in quantitative analysis. The overall study quality was moderate with a median NOS score of 5.5 stars (range 3 to 8). For clinicopathologic features, dysregulated circRNAs were related to larger tumor size (OR 2.122, 95%CI 1.418–3.175), advanced clinical stage (OR 2.847, 95%CI 2.059–3.935), and present of metastasis (OR 2.630, 95%CI 1.583–4.371). For chemotherapy, dysregulated circRNAs suggest a better response (OR 0.443, 95%CI 0.231–0.849), but a higher probability of resistance (OR 9.343, 95%CI 5.352–16.309). For survival prognosis, dysregulated circRNAs were significantly correlated with poor OS (HR 2.437, 95%CI 2.224–2.670) and DFS (HR 2.125, 95%CI 1.621–2.786). The results did not show differences among subgroups. Higher circ_0002052 expression showed a relation with poor OS (HR 3.197, 95%CI 2.054–4.976).
Conclusions
Our review demonstrated that abnormally expressed circRNAs have a relation with advanced clinicopathologic features and better response, but a higher probability of resistance and poor survival prognosis in osteosarcoma patients. However, more studies are encouraged to provide more robust evidence to translate circRNAs into clinical practice.
Trial registration
PROSPERO ID: CRD42021235031
Similar content being viewed by others
Background
Osteosarcoma is a malignant bone tumor characterized by neoplastic bone formation directly from tumor cells [1], which presents the most common primary osseous sarcoma in children and adolescents [2]. The diagnostic work-up of osteosarcoma usually started with radiological examinations for detecting the local diseases, followed by checkup for distant metastases, and finalized with a biopsy to reach a histology diagnosis [2,3,4]. Although this approach can guide the clinician to an appropriate treatment plan, the 5-year survival rate is still unsatisfying and the etiology of osteosarcoma remains unclear [1, 5]. Current clinicopathologic features and regular tests show potentials in patient prognosis prediction [6], but are unable to reveal the pathogenesis of osteosarcoma. Therefore, it is urgent to identify new biomarkers related to prognosis and clinicopathologic features.
With the development of sequencing technologies, several non-coding RNAs were discovered. Non-coding RNAs participate and regulate the transcription and translation of genes and sometimes play significant roles during dysregulated gene expression in cancer [7, 8]. Circular RNA (circRNA) is one of the non-coding RNAs with a closed loop that is generated by the back-splicing of pre-RNA with covalent bonding in between, functions as a sponge for microRNA, or directly regulates transcription and interfering with splicing mechanisms [9]. Studies have shown that circRNA can serve as diagnostic, prognostic, and predictive biomarkers [10,11,12]. Further, circRNA may be a more detectable biomarker for cancer, since it has the characteristics of a stable structure that is resistant to degradation by most RNA decay machinery [13,14,15].
The relation between circRNAs and osteosarcoma has been present in several reviews [16,17,18,19,20]. CircRNAs play oncogenic roles or show tumor-suppressive effects in the pathogenesis and progression of osteosarcoma including cell apoptosis, invasion, growth, differentiation, and migration. They are also involved in malignant phenotypes of osteosarcoma, such as treatment resistance and metastasis. Further quantitative analysis showed the potential of circRNAs in clinical implication as diagnostic or prognostic biomarkers [21, 22]. However, the previous meta-analyses included a number of studies that did not analyze the relation between circRNAs and treatment response and failed to pool repeatably studied circRNAs. Therefore, our systematic review and meta-analysis aimed to provide a more up-to-date and comprehensive summary of the clinicopathologic significance and prognostic value of circRNAs in osteosarcoma.
Methods
Protocol and registry
The reporting of our review followed Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) statement and several extensions [23]. A checklist was presented as Additional file 1. A protocol has been drafted before our review started and has been registered and updated on the International Prospective Register of Systematic Reviews (PROSPERO) [24] as CRD42021235031.
Literature search
Our systematic literature search was performed by two independent reviewers until 20 Feb 2021 following the Peer Review of Electronic Search Strategies (PRESS) guideline [25]. We searched eight electronic databases including PubMed, Embase, The Cochrane Library, Web of Science, Scopus, SinoMed, China National Knowledge Infrastructure (CNKI), and WanFang databases, as well as four gray literature sources namely OpenGrey, British Library Inside, ProQuest Dissertations & Theses Global, and BIOSIS preview. A search string was firstly developed in PubMed using two key terms, namely circular RNA and osteosarcoma in free words, Medical Subject Headings (MeSH) and/or Emtree words. The search string used in PubMed was (“RNA, Circular”[Mesh] OR circRNA OR ciRNA OR (circular AND RNA) OR “circular ribonucleic acid”) AND (“osteosarcoma”[Mesh] OR osteosarcoma OR (osseous AND sarcoma) OR (osteogenic AND sarcoma)). Then, the search strings were modified into other data sources (Additional file 1). There was no limitation for the time period, study design, or languages during the literature search. Duplicates were excluded through a rigorous and reproducible method via Endnote software version X9.2 (Clarivate Analytics, Philadelphia, PA, USA) [26].
Study selection
Two reviewers separately screened the titles and abstracts of records from electronic databases after deduplication. The records from gray literature sources were directly screened online to identify additional relevant records. The full texts and supplementary materials of potentially eligible records were obtained by two same reviewers and further assessed for eligibility. The reference lists of included studies and relevant reviews were screened to identify additional eligible studies. In the case of uncertainties, a final consensus was reached through discussion or help from a third reviewer.
Our study inclusion criteria included (1) study with histologically diagnosed osteosarcoma patients; (2) circRNA expression detected using tissues, serum, or plasma; (3) analysis about circRNA on clinicopathologic features or survival prognosis performed. Our study exclusion criteria were (1) ex vivo study or animal study; (2) duplicate studies; (3) reviews, conference abstracts, book chapters, editorials, letters, case reports, and other unsuitable article types; (4) reported in a language other than English, Japanese, Chinese, German, or French.
Data extraction
Data extraction was independently completed by two reviewers with our standardized sheet. The data extraction sheet contains the following items: (1) bibliographic data: author, publication year, study country; (2) circRNA characteristics: circRNA type, regulation pattern, sample size, specimen type, detection method, cutoff value, number of patients with high or low circRNA expression; (3) clinicopathologic data: age, gender, tumor site, tumor size, clinical stage, histologic classification, differentiation, metastasis; and (4) prognostic information: overall survival (OS), disease-free survival (DFS) or progression-free survival (PFS), hazard ratio (HR) and its 95% confidence interval (CI) for prognostic outcome, analysis method, data availability, follow-up duration. Any disagreement was resolved by discussion or help from a third reviewer.
If the studies have reported prognostic information in the article, we documented the data directly; otherwise, we extracted available data from the Kaplan-Meier curve (K-M curve) via an open-source Engauge Digitizer software version 12.1 [27]. The Engauge Digitizer digitizes image files containing graphs by placing points along axes and curves and recovers the data points from those graphs. Then, we reconstructed the necessary data through several established practical methods for meta-analysis [28] (Supplementary Note 2). The corresponding authors were contacted to request the data, if the articles did not report sufficient data or impossible to reconstruct based on reported data. When there was no response, the article was only qualitatively analyzed.
Quality assessment
Two reviewers independently assessed the quality of included studies conducting the Newcastle-Ottawa Quality Assessment Scale (NOS) [29, 30]. NOS used a star system to judge the study on three broad perspectives: the selection of the study groups; the comparability of the groups; and the ascertainment of either the exposure or outcome of interest for case-control or cohort studies, respectively. In our review, studies with prognostic outcomes were treated as cohort studies, while those only reported cross-sectional clinicopathologic features were considered as case-control studies. A modified version of NOS was used in our review (Supplementary Table 1). If there were disagreements between the two reviewers, they would be resolved through discussion or consultation with a third reviewer.
Data synthesis and analysis
The meta-analysis was conducted with Stata software version 15.1 (Stata Corp., College Station, TX, USA) using relevant packages (Supplementary Note 3). A p value < 0.05 suggested statistical significance, unless otherwise specified. To merge the outcomes of up- and downregulated circRNAs, we translated the HRs and 95%CI into a form that HRs > 1 suggested poor prognosis and was considered statistically significant if the 95%CI did not contain 1. The heterogeneity was assessed through the Higgins I-square statistic and chi-square Q test. A random-effect model was applied with the existence of marked heterogeneity as I-square > 50% and chi-square Q p value < 0.10; otherwise, a fixed-effect model was used. The publication bias was objectively evaluated by funnel plots and Begg’s funnel plots. Begg’s and Egger’s tests were quantitatively conducted to detect underlying publication bias. A p value > 0.1 was considered as low publication bias. By omitting the included studies one by one, the reliability of the pooled effect size was assessed. A trim and fill method was also used to assess the reliability of results. Subgroup analyses were performed to explore potential sources of heterogeneity, according to (1) regulation pattern: upregulated, or downregulated; (2) sample size: < 53 samples (median), or ≥ 53 samples; (3) data availability: reported or K-M curve; (4) cutoff value: median, average, or others; and (5) NOS: score < 5.5 stars (median), score ≥ 5.5 stars. An additional analysis was performed to investigate the relation between circ_0002052 and osteosarcoma, since the data from multiple studies allowed a more convictive conclusion.
Results
Literature search
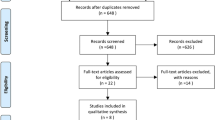
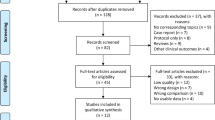
As the flow diagram shows (Fig. 1), our systematic review identified 968 records from electronic databases. We screened 305 titles and abstracts after the exclusion of 663 duplicates. Sixty articles were considered to be potentially eligible. We further identified 115 records from gray literature sources; however, no additional eligible article was found. Full-text assessment included 60 articles, and hand search did not identify additional relevant articles. Finally, 52 articles were included in the qualitative analysis [31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82]. Thirty-eight articles on clinicopathology and 36 articles on prognosis were included in the quantitative analysis after the exclusion of articles with incomplete data.
Study characteristics
Table 1 summarizes the characteristics of included studies. Fifty-two studies with 2934 osteosarcoma patients were included. All the studies were conducted in China. Forty-eight and 4 articles were published in English and Chinese, respectively. Forty-three dysregulated circRNAs were detected, in which 7 were downregulated and 36 were upregulated in osteosarcoma patients. Fifty-one studies measured circRNA expression in tissue samples from osteosarcoma patients by qRT-PCR, while one study used serum as a test sample.
Quality assessment
The sum of the NOS score is present in Table 1 and Fig. 2. The sum of the NOS score ranged from 3 to 8 stars, with a median of 5.5 stars, indicating the moderate quality of selected studies. The risk of bias was found mainly related to unclear patient inclusion criteria, inadequate treatment procedure, unreported cutoff value of circRNAs, and various cutoff values of clinicopathologic features, as well as unclear follow-up plan and high loss rate. Detailed quality assessment results are presented in Supplementary Table 2.
CircRNAs and clinicopathologic features of osteosarcoma
Table 2 and Fig. 3 show the correlations between circRNAs and clinicopathologic features in 38 selected studies with 2284 osteosarcoma patients. Original data of included studies on clinicopathogical features is summarized in Supplementary Table 3. Dysregulated circRNAs were related to advanced clinicopathologic features, including larger tumor size (OR 2.122, 95%CI 1.418–3.175), advanced clinical stage (OR 2.847, 95%CI 2.059–3.935), and present of metastasis (OR 2.630, 95%CI 1.583–4.371). For chemotherapy, dysregulated circRNAs suggested a better response (OR 0.443, 95%CI 0.231–0.849), but a higher probability of resistance (OR 9.343, 95%CI 5.352–16.309). The heterogeneity of studies on tumor size, clinical stage, metastasis, and chemotherapy response was high. Begg’s and Egger’s tests indicated that studies on tumor size and metastasis have potential high publication bias. The sensitivity analysis showed that the pooled results were stable except for studies on tumor size. The cutoff values of age, tumor size, and clinical stage varied, and corresponding forest plots are presented in Supplementary Fig. 1.
Forest plots evaluated the association between circRNA dysregulation and clinicopathological features of osteosarcoma: (A) age, (B) gender, (C) tumor site, (D) tumor size, (E) clinical stage, (F) metastasis, (G) histologic classification, (H) histologic pattern, (I) differentiation grade, (J) chemotherapy response (K) chemotherapy resistance, and (L) alkaline phosphatase
CircRNAs and prognosis of osteosarcoma
Table 3 shows the studies on circRNAs and survival prognosis in 44 selected studies, in which 36 studies with 2213 osteosarcoma patients were included in quantitative analysis. Original data of included studies on prognosis is summarized in Supplementary Table 4. Figure 4 and Table 4 present that circRNAs were significantly correlated with OS (HR 2.437, 95%CI 2.224–2.670) with low heterogeneity and reliability. On the other hand, circRNAs were significantly correlated with DFS (HR 2.125, 95%CI 1.621–2.786) with high heterogeneity. Figure 5 reveals the leave-one-out analysis of pooled DFS, indicating that one included study had a significant effect. The funnel plot with Begg’s test and Egger’s test suggested that the likelihood of publication bias was low.
Funnel plots and Begg’s funnel plots judged publication bias of (A, C) overall survival and (B, D) disease-free survival in osteosarcoma. Leave-one-out analysis and trim and fill analysis showed the relationship between circRNA dysregulation and prognosis (E, G) overall survival and (F, H) disease-free survival of osteosarcoma patients
Subgroup analysis
Subgroup analysis results of OS can be found in Table 5. All of the subgroups showed a significant correlation between circRNAs and OS of the patients. The results did not show differences among subgroups according to the regulation pattern, sample size, data availability, cutoff value, or NOS. The corresponding forest plots of OS are presented in Supplementary Figure 2.
Circ_0002052 and osteosarcoma
There were 4 studies repeatably investigated circ_0002052 in osteosarcoma. Table 6 summarizes the 3 available studies with 140 patients and showed that a higher expression of circ_0002052 has a relation with poorer OS (HR 3.197, 95%CI 2.054–4.976). The sensitivity and publication bias analyses have limited significance, since only three studies were included. The corresponding forest plots are presented in Supplementary Figure 3.
Discussions
Dysregulated circRNA expression has been demonstrated to be important in cancer initiation, development, and immigration [7,8,9], and has potential as diagnostic and prognostic biomarkers in various tumors [10,11,12]. Our systematic review conducted a structural literature review and included 52 studies investigating 43 dysregulated circRNAs in 2934 patients with osteosarcoma. We revealed that abnormal circRNA expression was related to tumor size, clinical stage, metastasis, and chemotherapy response and resistance. Further, dysregulated circRNAs were also prognostic biomarkers for OS and DFS. Additionally, dysregulated circ_0002052 was repeatably studied and showed a relation with poorer OS.
Two previous systematic reviews have performed meta-analyses on the clinicopathologic significance and prognostic value of circRNAs in osteosarcoma [21, 22]. The latest review included 31 studies, including 22 on clinicopathologic features and 23 on survival prognosis [22]. Thus, the pooled results may be underpowered due to insufficient data. The review summarized the relation between dysregulated circRNAs and age, gender, tumor size, clinical stage, and metastasis, while our review conducted more analyses on the influence of circRNAs on 12 features with 38 studies. Especially, our analysis on treatment response and resistance provided more practicable insight on treatment decision-making. Moreover, our analysis on survival prognosis included 36 studies to reach more convincing results with increased statistical power. The sensitivity analysis showed the reliability of results that dysregulated circRNAs were promising prognostic biomarkers for osteosarcoma patients. Additionally, our study summarized for the first time that circ_0002052 was significantly correlated with poorer OS with multiple datasets to confirm the efficacy.
Our sensitivity analysis showed that the correlations between dysregulated circRNAs and tumor size and DFS were not reliable, indicating that future studies might change the current results. The publication bias was detected in the analysis of dysregulated circRNAs on tumor size and metastasis, which encouraged more studies on this clinically relevant topic. Subgroup analyses were performed to explore the influence of study characteristics on the pooled results and found that the results remained stable regardless of regulation pattern, sample size, data availability, cutoff value, or study quality, suggesting a potential application in clinical practice.
The quality of included studies was assessed according to the NOS tool, although the overall quality of studies showed a moderate score with a median of 5.5 stars. There were several concerns releveled during our assessment. Most of the included studies put an emphasis on the function of circRNAs in osteosarcoma cells instead of their clinical significance. Therefore, the patient inclusion criteria, treatment procedure, and follow-up were usually unclearly described, which might hinder the clinical translation of circRNAs. The cutoff values were unreported in half of the included studies. Thus, further validation might be impossible. On the other hand, the various cutoff values of clinicopathologic features might introduce a risk of bias into our analysis, including age, tumor size, and clinical stage. To confirm circRNAs as clinically practicable biomarkers, more well-designed and high-quality studies were needed.
The summary of all available circRNAs indicated that circRNAs were significantly correlated with both OS and DFS, while circ_0002052 was the only circRNA that had been studied repeatedly in osteosarcoma patients [41, 58, 65, 72]. The meta-analysis showed that higher expression of circ_0002052 has a relation with poorer OS, but its relation with DFS was not available. Since efficacy confirmed in multiple datasets tends to be more convictive [83], more repeatable and reproducible studies are encouraged to provide more robust evidence for circRNAs as biomarkers for osteosarcoma, to allow translation of circRNAs into clinical practice.
Except for circRNAs, microRNAs and long non-coding RNAs have also shown potential diagnostic, prognostic, and therapeutic values in musculoskeletal malignancies [16,17,18,19,20,21,22, 84,85,86]. On the other hand, evidence is being produced on non-coding RNAs being of importance in benign musculoskeletal diseases [87,88,89,90]. These non-coding RNAs could be useful for diagnostic or management purposes in musculoskeletal conditions. However, before they can be applied in clinical practice, the issue of delivery of RNAs needs to be overcome [87, 88].
Our review has several limitations. Firstly, the number of included studies on several clinicopathologic features was comparatively small. Although up to four studies showed that dysregulated circRNA expression has a relation with chemotherapy response and resistance, more studies were encouraged. Secondly, two-thirds of HRs with 95% CIs of OS were indirectly extracted. However, the subgroup analysis demonstrated that there was no significant difference between pooled results according to extraction methods. Thirdly, data from eight studies were impossible to reconstruct, and not available through contraction to the author, which might generate possible bias. Fourthly, the subgroup analysis of DFS was not performed since the number of included studies was limited to draw any stable results. Moreover, we also failed to perform subgroup analyses according to the clinicopathological features of patients, due to varying cutoffs. A more in-depth analysis is encouraged if more future studies provide further details. Fifthly, all of the studies were performed in China, which might lead to biased results due to ethics groups. The role of circRNAs in osteosarcoma among different populations can be evaluated, if investigations in other ethnic groups are available. Finally, only one study obtained circRNA expression data from serum. It is still unclear whether the serum was suitable for circRNA detection in osteosarcoma patients. It might be more practicable and less invasive if the expression detected from serum or plasma had comparable efficiency to those from tissue samples.
Conclusions
In conclusion, our study showed that there is a significant correlation between the dysregulated expression of circRNAs and advanced clinicopathologic features, and it did affect the survival prognosis of osteosarcoma patients. CircRNAs might play an important role in the occurrence and development of osteosarcoma and showed potential as prognostic biomarkers for osteosarcoma. Our review also pointed out the quality insufficiency in current studies and emphasized the need for prospective high-quality studies with multiple datasets to promote clinical translation.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ALP:
-
Alkaline phosphatase
- CI:
-
Confidence interval
- circRNA:
-
Circular ribonucleic acid
- HR:
-
Hazard ratio
- LDH:
-
Lactate dehydrogenase
- OR:
-
Odds ratio
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
- PLR:
-
Positive likelihood ratio
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- NOS:
-
Newcastle-Ottawa Scale
- K-M curve:
-
Kaplan-Meier curve
References
The WHO Classification of Tumours Editorial Board. World Health Organization classification of tumors: WHO classification of tumours of soft tissue and bone, Chapter 3 Bone tumours. 5th ed. Lyon: IARC Press; 2020. p. 400–21.
Whelan JS, Davis LE. Osteosarcoma, Chondrosarcoma, and Chordoma. J Clin Oncol. 2018;36(2):188–93. https://doi.org/10.1200/JCO.2017.75.1743.
Casali PG, Bielack S, Abecassis N, Aro HT, Bauer S, Biagini R, et al. ESMO Guidelines Committee, PaedCan and ERN EURACAN. Bone sarcomas: ESMO-PaedCan-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29(Suppl 4):iv79–95. https://doi.org/10.1093/annonc/mdy310.
National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology. Bone Cancer. 2021;v1 2020-11-20. Accessed via https://www.nccn.org/professionals/physician_gls/pdf/bone.pdf on 2021-02-10.
Chou AJ, Geller DS, Gorlick R. Therapy for osteosarcoma: where do we go from here? Paediatr Drugs. 2008;10(5):315–27. https://doi.org/10.2165/00148581-200810050-00005.
Xin S, Wei G. Prognostic factors in osteosarcoma: A study level meta-analysis and systematic review of current practice. J Bone Oncol. 2020;21:100281. https://doi.org/10.1016/j.jbo.2020.100281.
Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21(1):22–36. https://doi.org/10.1038/s41568-020-00306-0.
Anfossi S, Babayan A, Pantel K, Calin GA. Clinical utility of circulating non-coding RNAs - an update. Nat Rev Clin Oncol. 2018;15(9):541–63. https://doi.org/10.1038/s41571-018-0035-x.
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–91. https://doi.org/10.1038/s41576-019-0158-7.
Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–65. https://doi.org/10.1038/onc.2017.361.
Weng W, Wei Q, Toden S, et al. Circular RNA ciRS-7 - a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23(14):3918–28. https://doi.org/10.1158/1078-0432.CCR-16-2541.
Smid M, Wilting SM, Uhr K, et al. The circular RNome of primary breast cancer. Genome Res. 2019;29(3):356–66. https://doi.org/10.1101/gr.238121.118.
Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–4. https://doi.org/10.1038/cr.2015.82.
Bahn JH, Zhang Q, Li F, et al. The landscape of microRNA, piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61(1):221–30. https://doi.org/10.1373/clinchem.2014.230433.
Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015;10(10):e0141214. https://doi.org/10.1371/journal.pone.0141214.
Soghli N, Qujeq D, Yousefi T, Soghli N. The regulatory functions of circular RNAs in osteosarcoma. Genomics. 2020;112(4):2845–56. https://doi.org/10.1016/j.ygeno.2020.03.024.
Wan B, Hu H, Wang R, Liu W, Chen D. Therapeutic potential of circular RNAs in osteosarcoma. Front Oncol. 2020;10:370. https://doi.org/10.3389/fonc.2020.00370.
Liu J, Yang L, Fu Q, Liu S. Emerging roles and potential biological value of circRNA in osteosarcoma. Front Oncol. 2020;10:552236. https://doi.org/10.3389/fonc.2020.552236.
Tu C, He J, Qi L, et al. Emerging landscape of circular RNAs as biomarkers and pivotal regulators in osteosarcoma. J Cell Physiol. 2020;235(12):9037–58. https://doi.org/10.1002/jcp.29754.
Li Z, Li X, Xu D, et al. An update on the roles of circular RNAs in osteosarcoma. Cell Prolif. 2021;54(1):e12936. https://doi.org/10.1111/cpr.12936.
Huang X, Yang W, Zhang Z, Shao Z. Dysregulated circRNAs serve as prognostic and diagnostic markers in osteosarcoma by sponging microRNA to regulate the downstream signaling pathway. J Cell Biochem. 2020;121(2):1834–41.
Zhang C, He J, Qi L, et al. Diagnostic and prognostic significance of dysregulated expression of circular RNAs in osteosarcoma. Expert Rev Mol Diagn. 2021;21(2):235–44. https://doi.org/10.1080/14737159.2021.1874922.
Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. https://doi.org/10.1136/bmj.n71.
International prospective register of systematic reviews (PROSPERO). Accessed via https://www.crd.york.ac.uk/prospero/ 2021.
McGowan J, Sampson M, Salzwedel DM, Cogo E, Foerster V, Lefebvre C. PRESS peer review of electronic search strategies: 2015 guideline statement. J Clin Epidemiol. 2016;75:40–6. https://doi.org/10.1016/j.jclinepi.2016.01.021.
Bramer WM, Giustini D, de Jonge GB, Holland L, Bekhuis T. De-duplication of database search results for systematic reviews in EndNote. J Med Libr Assoc. 2016;104(3):240–3. https://doi.org/10.3163/1536-5050.104.3.014.
Mitchell M, Muftakhidinov B, Winchen T, et al. Engauge Digitizer Software. Accessed via http://markummitchell.github.io/engauge-digitizer on 2021-02-10. https://doi.org/10.5281/zenodo.3941227.
Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. https://doi.org/10.1186/1745-6215-8-16.
Ma LL, Wang YY, Yang ZH, Huang D, Weng H, Zeng XT. Methodological quality (risk of bias) assessment tools for primary and secondary medical studies: what are they and which is better? Mil Med Res. 2020;7(1):7. https://doi.org/10.1186/s40779-020-00238-8.
Wells G, Shea B, O'Connell D, Peterson J, Welch V, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Accessed via http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp on 2021-02-14.
Chen Y, Zhang S, Bai C, Guan Z, Chen W. Circ_0000885 Enhances Osteosarcoma Progression by Increasing FGFR1 Expression via Sponging MiR-1294. Cancer Manag Res. 2020;12:6441–52. https://doi.org/10.2147/CMAR.S244382.
Ding S, Zhang G, Gao Y, Chen S, Cao C. Circular RNA hsa_circ_0005909 modulates osteosarcoma progression via the miR-936/HMGB1 axis. Cancer Cell Int. 2020;20:305. https://doi.org/10.1186/s12935-020-01399-1.
Gao Y, Ma H, Gao Y, Tao K, Fu L, Ren R, et al. CircRNA Circ_0001721 promotes the progression of osteosarcoma through miR-372-3p/MAPK7 axis. Cancer Manag Res. 2020;12:8287–302. https://doi.org/10.2147/CMAR.S244527.
Hu Y, Gu J, Shen H, Shao T, Li S, Wang W, et al. Circular RNA LARP4 correlates with decreased Enneking stage, better histological response, and prolonged survival profiles, and it elevates chemosensitivity to cisplatin and doxorubicin via sponging microRNA-424 in osteosarcoma. J Clin Lab Anal. 2020;34(2):e23045. https://doi.org/10.1002/jcla.23045.
Huang L, Chen M, Pan J, Yu W. Circular RNA circNASP modulates the malignant behaviors in osteosarcoma via miR-1253/FOXF1 pathway. Biochem Biophys Res Commun. 2018;500(2):511–7. https://doi.org/10.1016/j.bbrc.2018.04.131.
Ji X, Shan L, Shen P, He M. Circular RNA circ_001621 promotes osteosarcoma cells proliferation and migration by sponging miR-578 and regulating VEGF expression. Cell Death Dis. 2020;11(1):18. https://doi.org/10.1038/s41419-019-2204-y.
Jiang Y, Hou J, Zhang X, Xu G, Wang Y, Shen L, et al. Circ-XPO1 upregulates XPO1 expression by sponging multiple miRNAs to facilitate osteosarcoma cell progression. Exp Mol Pathol. 2020;117:104553. https://doi.org/10.1016/j.yexmp.2020.104553.
Jiang X, Chen D. Circular RNA hsa_circ_0000658 inhibits osteosarcoma cell proliferation and migration via the miR-1227/IRF2 axis. J Cell Mol Med. 2021;25(1):510–20. https://doi.org/10.1111/jcmm.16105.
Jin Y, Li L, Zhu T, Liu G. Circular RNA circ_0102049 promotes cell progression as ceRNA to target MDM2 via sponging miR-1304-5p in osteosarcoma. Pathol Res Pract. 2019;215(12):152688. https://doi.org/10.1016/j.prp.2019.152688.
Jin J, Chen A, Qiu W, Chen Y, Li Q, Zhou X, et al. Dysregulated circRNA_100876 suppresses proliferation of osteosarcoma cancer cells by targeting microRNA-136. J Cell Biochem. 2019;120(9):15678–87. https://doi.org/10.1002/jcb.28837.
Jin Z, Cui J, Zeng Y, Zhou Q, Liu J. Expression of circular RNA circ_0002052 in osteosarcoma and its clinical significance. J Chin Prac Diag Ther. 2019;33(1):32–4. https://doi.org/10.13507/j.isn.1674-3474.2019.01.010 Article in Chinese.
Lei S, Xiang L. Up-regulation of circRNA hsa_circ_0003074 expression is a reliable diagnostic and prognostic biomarker in patients with osteosarcoma. Cancer Manag Res. 2020 Sep 29;12:9315–25. https://doi.org/10.2147/CMAR.S262093.
Li B, Li X. Overexpression of hsa_circ_0007534 predicts unfavorable prognosis for osteosarcoma and regulates cell growth and apoptosis by affecting AKT/GSK-3β signaling pathway. Biomed Pharmacother. 2018;107:860–6. https://doi.org/10.1016/j.biopha.2018.08.086.
Li L, Guo L, Yin G, Yu G, Zhao Y, Pan Y. Upregulation of circular RNA circ_0001721 predicts unfavorable prognosis in osteosarcoma and facilitates cell progression via sponging miR-569 and miR-599. Biomed Pharmacother. 2019;109:226–32. https://doi.org/10.1016/j.biopha.2018.10.072.
Li X, Liu Y, Zhang X, Shen J, Xu R, Liu Y, et al. Circular RNA hsa_circ_0000073 contributes to osteosarcoma cell proliferation, migration, invasion and methotrexate resistance by sponging miR-145-5p and miR-151-3p and upregulating NRAS. Aging. 2020;12(14):14157–73. https://doi.org/10.18632/aging.103423.
Li S, Pei Y, Wang W, Liu F, Zheng K, Zhang X. Extracellular nanovesicles-transmitted circular RNA has_circ_0000190 suppresses osteosarcoma progression. J Cell Mol Med. 2020;24(3):2202–14. https://doi.org/10.1111/jcmm.14877.
Li L, Kong XA, Zang M, Dong J, Feng Y, Gui B, et al. Hsa_circ_0003732 promotes osteosarcoma cells proliferation via miR-545/CCNA2 axis. Biosci Rep. 2020;40(6):BSR20191552. https://doi.org/10.1042/BSR20191552.
Liu J, Li X, Yue L, Lv H. Circ_0105346 knockdown inhibits osteosarcoma development via regulating miR-1182/WNT7B axis. Cancer Manag Res. 2021;13:521–35. https://doi.org/10.2147/CMAR.S281430.
Liu YD, Liu LP. Circ100284 promotes invasion and migration of osteosarcoma cells by down-regulating PTEN and EMP1. Eur Rev Med Pharmacol Sci. 2020;24(12):6540–50. https://doi.org/10.26355/eurrev_202006_21638.
Liu DY, Li Z, Zhang K, Jiao N, Lu DG, Zhou DW, et al. Circular RNA CircMTO1 suppressed proliferation and metastasis of osteosarcoma through miR-630/KLF6 axis. Eur Rev Med Pharmacol Sci. 2021;25(1):86–93. https://doi.org/10.26355/eurrev_202101_24352.
Ma XL, Zhu KP, Zhang CL. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J Cancer. 2018;9(10):1856–62. https://doi.org/10.7150/jca.24619.
Mao X, Guo S, Gao L, Li G. Circ-XPR1 promotes osteosarcoma proliferation through regulating the miR-214-5p/DDX5 axis. Hum Cell. 2021;34(1):122–31. https://doi.org/10.1007/s13577-020-00412-z.
Nie WB, Zhao LM, Guo R, Wang MX, Ye FG. Circular RNA circ-NT5C2 acts as a potential novel biomarker for prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22(19):6239–44. https://doi.org/10.26355/eurrev_201810_16030.
Pan G, Hu T, Chen X, Zhang C. Upregulation of circMMP9 promotes osteosarcoma progression via targeting miR-1265/CHI3L1 axis. Cancer Manag Res. 2019;11:9225–31. https://doi.org/10.2147/CMAR.S226264.
Pan Y, Lin Y, Mi C. Cisplatin-resistant osteosarcoma cell-derived exosomes confer cisplatin resistance to recipient cells in an exosomal circ_103801-dependent manner. Cell Biol Int. 2020. https://doi.org/10.1002/cbin.11532.
Qi H, Sun Y, Jiang Y, Li X. Upregulation of circular RNA circ_0000502 predicts unfavorable prognosis in osteosarcoma and facilitates cell progression via sponging miR-1238. J Cell Biochem. 2018. https://doi.org/10.1002/jcb.28134.
Wang L, Du ZG, Huang H, Li FS, Li GS, Xu SN. Circ-0003998 promotes cell proliferative ability and invasiveness by binding to miR-197-3p in osteosarcoma. Eur Rev Med Pharmacol Sci. 2019;23(24):10638–46. https://doi.org/10.26355/eurrev_201912_19761.
Wang C, Feng XB, Qu JH. Expression of circ-0002052 in osteosarcoma and its correlation with clinicopathological features and prognosis. J Clin Exp Med. 2019;18(23):2521–4. https://doi.org/10.3969/j.issn.1671-4695.2019.23.017 Article in Chinese.
Wang L, Zhang GC, Kang FB, Zhang L, Zhang YZ. hsa_circ0021347 as a potential target regulated by B7-H3 in modulating the malignant characteristics of osteosarcoma. Biomed Res Int. 2019, 2019:9301989. https://doi.org/10.1155/2019/9301989.
Wang JH, Wu XJ, Duan YZ, Li F. Circular RNA_CNST promotes the tumorigenesis of osteosarcoma cells by sponging miR-421. Cell Transplant. 2020;29:963689720926147. https://doi.org/10.1177/0963689720926147.
Wang Y, Shi S, Zhang Q, Dong H, Zhang J. MicroRNA-206 upregulation relieves circTCF25-induced osteosarcoma cell proliferation and migration. J Cell Physiol. 2020. https://doi.org/10.1002/jcp.29570.
Wang L, Wang P, Su X, Zhao B. Circ_0001658 promotes the proliferation and metastasis of osteosarcoma cells via regulating miR-382-5p/YB-1 axis. Cell Biochem Funct. 2020;38(1):77–86. https://doi.org/10.1002/cbf.3452.
Wei W, Ji L, Duan W, Zhu J. Circular RNA circ_0081001 knockdown enhances methotrexate sensitivity in osteosarcoma cells by regulating miR-494-3p/TGM2 axis. J Orthop Surg Res. 2021;16(1):50. https://doi.org/10.1186/s13018-020-02169-5.
Wen Y, Li B, He M, Teng S, Sun Y, Wang G. circHIPK3 promotes proliferation and migration and invasion via regulation of miR-637/HDAC4 signaling in osteosarcoma cells. Oncol Rep. 2021;45(1):169–79. https://doi.org/10.3892/or.2020.7833.
Wu Z, Shi W, Jiang C. Overexpressing circular RNA hsa_circ_0002052 impairs osteosarcoma progression via inhibiting Wnt/β-catenin pathway by regulating miR-1205/APC2 axis. Biochem Biophys Res Commun. 2018;502(4):465–71. https://doi.org/10.1016/j.bbrc.2018.05.184.
Xiang L, Zhang W. Expression level of circular RNA hsa_circ_0005721 in tissues and serum of patients with osteosarcoma and its clinical significance. J Clin Pathol Res. 2020;40(5):1136–43. https://doi.org/10.3978/j.issn.2095-6959.2020.05.010 Article in Chinese.
Yan M, Gao H, Lv Z, Liu Y, Zhao S, Gong W, et al. Circular RNA PVT1 promotes metastasis via regulating of miR-526b/FOXC2 signals in OS cells. J Cell Mol Med. 2020;24(10):5593–604. https://doi.org/10.1111/jcmm.15215.
Yang J, Han Q, Li C, Yang H, Chen X, Wang X. Circular RNA circ_0001105 inhibits progression and metastasis of osteosarcoma by sponging miR-766 and activating YTHDF2 expression. Onco Targets Ther. 2020;13:1723–36. https://doi.org/10.2147/OTT.S234668.
Zhang H, Wang G, Ding C, Liu P, Wang R, Ding W, et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. 2017;8(37):61687–97. https://doi.org/10.18632/oncotarget.18671.
Zhang H, Yan J, Lang X, Zhuang Y. Expression of circ_001569 is upregulated in osteosarcoma and promotes cell proliferation and cisplatin resistance by activating the Wnt/β-catenin signaling pathway. Oncol Lett. 2018;16(5):5856–62. https://doi.org/10.3892/ol.2018.9410.
Zhang Z, Zhao M, Wang G. Hsa_circ_0051079 functions as an oncogene by regulating miR-26a-5p/TGF-β1 in osteosarcoma. Cell Biosci. 2019;9:94. https://doi.org/10.1186/s13578-019-0355-2.
Zhang PR, Ren J, Wan JS, Sun R, Li Y. Circular RNA hsa_circ_0002052 promotes osteosarcoma via modulating miR-382/STX6 axis. Hum Cell. 2020;33(3):810–8. https://doi.org/10.1007/s13577-020-00335-9.
Zhang C, Zhou H, Yuan K, Xie R, Chen C. Overexpression of hsa_circ_0136666 predicts poor prognosis and initiates osteosarcoma tumorigenesis through miR-593-3p/ZEB2 pathway. Aging. 2020;12(11):10488–96. https://doi.org/10.18632/aging.103273.
Zhang C, Na N, Liu L, Qiu Y. CircRNA hsa_circ_0005909 promotes cell proliferation of osteosarcoma cells by targeting miR-338-3p/HMGA1 axis. Cancer Manag Res. 2021;13:795–803. https://doi.org/10.2147/CMAR.S285118.
Zhang G, Wang X, Zhu L. Highly expressed circular RNA hsa_circ_0017247 promotes osteosarcoma cell invasion and migration. Chinese Journal of Cellular and Molecular Immunology. 2021;36(7):634–9. https://doi.org/10.13423/j.cnki.cjcmi.009037 Article in Chinese.
Zhao Y, Zhang J. CircSAMD4A accelerates cell proliferation of osteosarcoma by sponging miR-1244 and regulating MDM2 mRNA expression. Biochem Biophys Res Commun. 2019;516(1):102–11. https://doi.org/10.1016/j.bbrc.2019.05.182.
Zheng S, Qian Z, Jiang F, Ge D, Tang J, Chen H, et al. CircRNA LRP6 promotes the development of osteosarcoma via negatively regulating KLF2 and APC levels. Am J Transl Res. 2019;11(7):4126–38.
Zhou X, Natino D, Qin Z, Wang D, Tian Z, Cai X, et al. Identification and functional characterization of circRNA-0008717 as an oncogene in osteosarcoma through sponging miR-203. Oncotarget. 2017;9(32):22288–300. https://doi.org/10.18632/oncotarget.23466.
Zhu KP, Ma XL, Zhang CL. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int J Biol Sci. 2018;14(3):321–30. https://doi.org/10.7150/ijbs.24360.
Zhu KP, Zhang CL, Hu JP, Zhang L. A novel circulating hsa_circ_0081001 act as a potential biomarker for diagnosis and prognosis of osteosarcoma. Int J Biol Sci. 2018;14(11):1513–20. https://doi.org/10.7150/ijbs.27523.
Zhu KP, Ma XL, Zhang L, Zhang CL, Hu JP, Cai T, et al. Screening circular RNA related to chemotherapeutic resistance in osteosarcoma by RNA sequencing. Epigenomics. 2018;10(10):1327–46. https://doi.org/10.2217/epi-2018-0023.
Zhu K, Niu L, Wang J, Wang Y, Zhou J, Wang F, et al. Circular RNA hsa_circ_0000885 levels are increased in tissue and serum samples from patients with osteosarcoma. Med Sci Monit. 2019;25:1499-1505. doi: 10.12659/MSM.914899.
Zhang W, Ren X, Qi L, Zhang C, Tu C, Li Z. The value of lncRNAs as prognostic biomarkers on clinical outcomes in osteosarcoma: a meta-analysis. BMC Cancer. 2021;21(1):202. https://doi.org/10.1186/s12885-021-07882-w.
Chen D, Wang H, Zhang M, Jiang S, Zhou C, Fang B, et al. Abnormally expressed long non-coding RNAs in prognosis of Osteosarcoma: A systematic review and meta-analysis. J Bone Oncol. 2018;13:76–90. https://doi.org/10.1016/j.jbo.2018.09.005.
Gao SS, Wang YJ, Zhang GX, Zhang WT. Potential diagnostic value of miRNAs in peripheral blood for osteosarcoma: A meta-analysis. J Bone Oncol. 2020;23:100307. https://doi.org/10.1016/j.jbo.2020.100307.
Gao SS, Wang YJ, Zhang GX, Zhang WT. Potential diagnostic value of circulating miRNA for multiple myeloma: A meta-analysis. J Bone Oncol. 2020;25:100327. https://doi.org/10.1016/j.jbo.2020.100327.
Oliviero A, Della Porta G, Peretti GM, Maffulli N. MicroRNA in osteoarthritis: physiopathology, diagnosis and therapeutic challenge. Br Med Bull. 2019;130(1):137–47. https://doi.org/10.1093/bmb/ldz015.
Giordano L, Della Porta G, Peretti GM, Maffulli N. Therapeutic potential of microRNA in tendon injuries. Br Med Bull. 2020;133(1):79–94. https://doi.org/10.1093/bmb/ldaa002.
Mao X, Cao Y, Guo Z, Wang L, Xiang C. Biological roles and therapeutic potential of circular RNAs in osteoarthritis. Mol Ther Nucleic Acids. 2021;24:856–67. https://doi.org/10.1016/j.omtn.2021.04.006.
Sikora M, Marycz K, Smieszek A. Small and Long Non-coding RNAs as Functional Regulators of Bone Homeostasis, Acting Alone or Cooperatively. Mol Ther Nucleic Acids. 2020;21:792–803. https://doi.org/10.1016/j.omtn.2020.07.017.
Acknowledgements
The authors would like to thank Prof. Huan Zhang for her kindness and enlightening comments on this study, and Dr. Shiqi Mao for sharing his knowledge in cancer genetics. The authors would like to express their gratitude to the editors for thier help and support in article publication, and reviewers for their careful review and constructive suggestions on this manuscript.
Funding
This study was supported by the National Natural Science Funds of China (81771790) and the Medicine and Engineering Combination Project of Shanghai Jiao Tong University (YG2019ZDB09). They played no role in the study design, data collection or analysis, decision to publish, or manuscript preparation.
Author information
Authors and Affiliations
Contributions
Jingyu Zhong: conceptualization, methodology, investigation, formal analysis, validation, software, visualization, writing—original draft, writing—review and editing
Guangcheng Zhang: conceptualization, methodology, investigation, formal analysis, validation, writing—review and editing
Weiwu Yao: conceptualization, writing—review and editing, supervision, funding acquisition
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhong, J., Zhang, G. & Yao, W. Clinicopathologic significance and prognostic value of circRNAs in osteosarcoma: a systematic review and meta-analysis. J Orthop Surg Res 16, 578 (2021). https://doi.org/10.1186/s13018-021-02568-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13018-021-02568-2