Abstract
Modern agriculture has created a demand for plant biotechnology products that provide durable resistance to insect pests, tolerance of herbicide applications for weed control, and agronomic traits tailored for specific geographies. These transgenic trait products require a modular and sequential multigene stacking platform that is supported by precise genome engineering technology. Designed nucleases have emerged as potent tools for creating targeted DNA double strand breaks (DSBs). Exogenously supplied donor DNA can repair the targeted DSB by a process known as gene targeting (GT), resulting in a desired modification of the target genome. The potential of GT technology has not been fully realized for trait deployment in agriculture, mainly because of inefficient transformation and plant regeneration systems in a majority of crop plants and genotypes. This challenge of transgene stacking in plants could be overcome by Intra-Genomic Homologous Recombination (IGHR) that converts independently segregating unlinked donor and target transgenic loci into a genetically linked molecular stack. The method requires stable integration of the donor DNA into the plant genome followed by intra-genomic mobilization. IGHR complements conventional breeding with genetic transformation and designed nucleases to provide a flexible transgene stacking and trait deployment platform.
Similar content being viewed by others
Background
The Green Revolution in the 1960s combined advances in breeding and agricultural practice, and provided food security to millions of people [1]. Given an increasing global population, there is a projected need to increase world food production by 40 % in the next 20 years [2]. In addition to a growing population, climate change, degrading natural resources and changing food preferences have raised food and nutritional security to the level of the biggest challenge of the twenty-first century [3].
Genetically modified (GM) trait technology in the mid-1990s made a major impact in meeting the world food demand and there has been a rapid adoption of the technology. These first generation trait products involved simple herbicide and insect traits that required introduction of a single gene. Control of the broad range of insect pests and weeds desired today requires multiple insect and herbicide tolerance genes [4]. In addition, modern genomics and gene networking tools have revealed that many agronomic traits depend on different genes and complex interactions of proteins reacting to various external stimuli [1]. The next generation trait products, therefore, require integration of multiple transgenes and would also benefit from a flexible and modular trait stacking platform that would accommodate development of increasingly complex future products. Conventional breeding has been successfully employed for trait stacking, but this method requires substantial time and resources for sorting and deregulation of multiple unlinked transgenes [4, 5], and a limited number of independent loci can practically be stacked.
Designed nucleases have become a powerful gene targeting (GT) tool to create targeted DNA double strand breaks (DSBs) at specified genomic locations, which stimulate the cell’s DNA repair machinery leading to integration of exogenously supplied transgenes into a specified genomic site. While designed nuclease-mediated targeted mutagenesis is becoming routine in plants [6–9], site-directed transgene integration remains elusive, mainly due to low transformation and regeneration efficiencies in the majority of plant species and genotypes. A GT method that requires minimal transformation effort would be very attractive to address this challenge. This review focuses on conventional Intra-chromosomal somatic homologous recombination work in plants and its recent application using designed nucleases that can provide solutions to some of the challenges associated with the deployment of GT technology for transgene stacking in crop plants.
Gene targeting: a byproduct of genomic double-strand break
DSBs can arise spontaneously, may be induced by ionizing radiation and chemicals, or recently by designed nucleases (For review, see references [10–15]). Genomic DSBs could be negatively mutagenic or lethal to cells if not repaired efficiently. In plants, DSBs are repaired by homologous recombination (HR) or non-homologous end joining (NHEJ). HR and NHEJ mechanisms are conserved in eukaryotes; however, the efficiency of these pathways differs not only between species but also between cell types [16]. HR is a precise DSB repair pathway that requires sequences homologous (almost identical) to those flanking the DSB site [12, 13]. HR is the predominant DNA recombination pathway during meiosis in higher eukaryotes including plants [17]. NHEJ mainly involves ligation to unrelated sequences or to sequences with micro-homologies, resulting mostly in non-precise repair with small insertions or deletions at the DSB site. NHEJ is the primary DNA repair pathway in the somatic cells, while HR mainly occurs during S and G2 phases of the cell cycle [18].
Targeted DSB-induced NHEJ has been previously described for mutagenesis, deletions or imprecise insertions [6–9, 13, 19, 20]. In contrast, HR, a more precise mode of DNA repair, is preferred for GT [12, 13]. Gene targeting through HR requires simultaneous introduction of the nuclease to create targeted DSB at desired genomic location, and donor DNA containing flanking homologies, acting as a template for repair of the DSB [21].
Gene targeting challenges in plants
Targeted DSBs stimulate the cell’s DNA repair machinery making the DSB site accessible to a donor transgene for site-specific integration. The DSBs however do not preclude the ectopic integration of a donor transgene elsewhere in the genome. In addition, the GT process requires efficient delivery of the donor molecule to the DSB site and the ability to regenerate whole plants from the cells with a precisely repaired targeted genomic site. Random integration of the donor transgene and an inefficient transformation method for the donor delivery therefore are two major challenges for the routine deployment of GT technology in crop plants. Positive selection for GT such that precise insertion of the donor complements the non-functional selectable marker in the target locus has been used to avoid random integration of the donor [22, 23, 24] genes in the target locus. A positive–negative selection approach has also been used very successfully for GT in rice [25, 26]. A sequential GT method providing flexibility of incremental modifications of the target locus with new trait genes was recently developed [27]. That method exploited positive GT selection using an intron sequence homology between the donor and target that allowed sequential swapping of selection markers, providing a multi-generational GT method (Fig. 1) for trait product deployment [28].
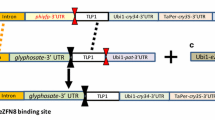
Basic design of constructs used for gene targeting via intra genomic homologous recombination. a Target construct contains a generation 1 (Gen 1) stack and a selection marker A flanked by unique homology sequences (HR1 and 2). A designed nuclease 1 binding site is inserted between selection marker A and HR2 sequence. b The donor construct contains selection marker B, downstream HR3 sequence, generation 2 (Gen 2) stack, and HR1 and 2 homology sequences matching the target. A designed nuclease 2 binding site is inserted between the selection marker and HR3 sequence for future targeting. The donor is flanked by a designed nuclease 1 binding site on each end. c The designed nuclease construct contains designed nuclease 1 coding sequence driven by appropriate promoter. d Target locus containing functional selection marker B gene and generation 2 (Gen 2) stack precisely inserted after gene targeting
The accessibility of the donor transgene to the DSB site is another key bottleneck for efficient GT process. Donor DNA is exogenously supplied either via direct DNA delivery [29], mostly using the microparticle bombardment, or via indirect DNA delivery, mainly mediated by Agrobacterium [30]. The production of a transgenic plant is the result of a sequence of events: a) transfer of exogenous DNA into the plant cell nucleus, b) integration of the foreign DNA in a transcriptionally active region of the host genome, and c) regeneration into a fully developed plant, either via organogenesis or somatic embryogenesis, of the original cell where the transgene integrated. Regardless of the gene transfer method used (direct or indirect) cell competence for foreign DNA integration and regeneration varies with cell type and developmental stage making the recovery of transgenic events a challenging task in most crop plants.
The nuclear targeting of the exogenous DNA is hindered by physical (e.g. cell wall), cellular (e.g. proteases, nucleases) and biological barriers (e.g. plant defense) and our understanding of how to best overcome these barriers is still limited [31, 32]. Actively dividing cells are the most amenable targets for DNA insertion and it has been shown that higher transformation is obtained in cells with nuclei at the S and G2 phases [33, 34] where chromatin remodeling takes place. A localized and temporary decondensation of the chromatin is believed to be necessary for efficient transgene integration in gene-rich euchromatic regions [35, 36].
Plant biology parameters associated with species, genotype, and explant type play an important role in the efficiency of transformation and regeneration. In rice, for example, between the two subspecies indica and japonica, indica is generally more recalcitrant to tissue culture and transformation [37]. Similarly for maize transformation and regeneration, the most responsive type of explant is the immature embryo where scutellum cells are induced to proliferate and undergo somatic embryogenesis [38], but this process is highly genotype-dependent and still limited mainly to crosses and derivatives of the maize inbred lines A188 [37]. Also in soybean, the ability to regenerate transgenic plants has been limited to a few soybean model genotypes (Jack and Williams 82) with some successful examples of competence for somatic embryogenesis transferred and combined in other cultivars via introgression [39].
Gene targeting via intra genomic homologous recombination
The challenges of inefficient transformation systems in crop plants could be overcome by intra-genomic homologous recombination (IGHR), which utilizes a cell’s recombinational machinery to replicate and supply donor DNA for IGHR mediated insertion of a donor within the target site. Intra-chromosomal HR in somatic cells of the whole plant was reported more than two decades ago (Reviewed in [40, 41, 42]). Two overlapping, non-functional pieces of a chimeric beta-glucuronidase (uidA) gene were used as recombination substrates, which upon HR led to a restoration of the functional uidA gene that was detected by histochemical staining of the encoded functional uidA protein. HR was reported in different organs and tissues during different stages of the plant development, including meristematic recombination events that revealed cell lineage patterns. The system was later used to demonstrate that an induced DSB in the target site resulted in twofold increase the HR frequency [22, 43]. The germline in plants is formed during later developmental stages, and any HR occurring during the life cycle of the plant could be germinally transmitted to the next generation. The demonstration of HR between linked overlapping DNA sequences within somatic cells of the whole plant was an important milestone in the GT field. The work paved the way for HR between unlinked DNA sequences in the genome of somatic cells, and regeneration of whole plants from these cells (see below).
The next significant development in the field was the application of designed nucleases for excision of the stably integrated transgene. In tobacco, a transgenic line containing a single copy of the codA gene flanked by cleavage sites specific to I-SceI nuclease was created. After induction of DSBs by transient expression of I-SceI, the codA gene was successfully removed from the calli, and plants lacking the codA gene were regenerated [44]. Similarly, tobacco plants containing a stably integrated uidA gene cassette flanked by designed nuclease sites were crossed with plants expressing the corresponding nuclease. The complete deletion of a 4.3 kb sequence comprising the uidA gene cassette was obtained in F1 progenies [45]. These reports were later followed by deletions of large endogenous genomic sequences in different plant species using designed nucleases [46–48].
Researchers in mammalian GT field were first to exploit cells’ recombination machinery to catalyze HR between a target locus and an in vivo-liberated donor [49]. In this system, the donor transgene is first inserted stably into the genome. The randomly inserted donor molecule is later released intragenomically within the genome of intact tissue. The IGHR based method was demonstrated using a site-specific recombinase (FLP) and a site-specific endonuclease (I-SceI) for the modification of the yellow locus in the Drosophila genome [49–51]. The method has been successfully applied for the modification of more than 20 loci in Drosophila [52].
A similar IGHR approach was also proposed for plant GT [53]; the first proof-of-principle in plants came several years later in Arabidopsis [54] using a single site-specific endonuclease (I-SceI). The GT system was designed using a non-functional truncated uidA target transgene containing cleavage sites for I-SceI nuclease, a donor transgene containing a complementary uidA GT cassette flanked by I-SceI sites, and a transgene containing a I-SceI expressing cassette that upon expression would generate in vivo release of linear donor after I-SceI expression. Single copy target and donor lines were crossed and lines homozygous for both the transgenes were obtained. The homozygous target/donor lines were then crossed with an I-SceI line and F1 progenies were screened for IGHR-mediated GT using uidA histochemical staining. Some F1 progenies revealed chimeric blue spots indicating GT in somatic cells during the plant development. The F1 lines were self-pollinated and F2 progenies were scored for the blue seedlings indicating germinal transmittance of GT. Targeted events were obtained up to one per 100 seeds. A similar approach was later attempted with some success in a native genomic target site in Arabidopsis using the CRISPR/Cas system [55].
After initial work on IGHR-mediated GT in a model system, the method was successfully demonstrated in maize by somatic ectopic recombination and tissue culture selection [56]. Similar to a previous effort in Arabidopsis, the target construct contained a non-functional partial neomycin phophotransferase II (nptII) gene and a cleavage site for I-SceI nuclease. The donor construct contained dexamethasone-inducible I-SceI, and an excisable nptII sequence complementing the partial sequence at the target locus such that GT would constitute the functional nptII gene. The target and donor plants were crossed and F1 progenies were selfed. No fully kanamycin-resistant plants were obtained from the dexamethasone-induced F2 progeny for target and donor. However kanamycin-resistant leaf sectors were observed indicating IGHR occurred in some somatic cells during plant development. The embryos isolated from immature kernels of F2 plants were subjected to callus induction on medium with and without dexamethasone. Kanamycin resistant GT events were recovered and the repair of the nptII gene was confirmed by molecular analyses. GT frequencies ranging from 0.13 to 0.55 % (per immature embryo treated) were obtained. The authors also made an interesting observation of GT at a cleaved target locus without excision of the donor molecule.
The demonstration of GT via IGHR in Arabidopsis and maize has created potential for the application of GT technology in transformation-inefficient crop plant species. Unlike direct transformation methods that limit donor molecules to a small number of treated cells, IGHR utilizes the plant system to replicate donor DNA in every cell throughout the life cycle. The extra-chromosomal donor molecule could be liberated and used by the target site as a template in plant tissues or stages that favor HR over NHEJ. The previous GT approaches relied on efficient transformation systems to produce a large number of events to obtain a few targeted plants. Since most economically important crop plants remain recalcitrant to transformation, GT technology has so far been practical in only a small number of crop plants. Additionally, IGHR releases only one to two copies of the donor, leading to high quality targeted events, in contrast to previous GT methods that require additional segregation work to remove randomly integrated undesired truncated donor molecules.
The IGHR method reviewed here generates tremendous opportunity for biotechnological application of GT in commercial transgenic trait deployment. This approach when combined with a sequential GT method (Fig. 1) [27] would provide the modular and flexible transgenic trait stacking platform (Fig. 2) currently needed for complex product needs in the agriculture industry. The strategy provides flexibility to stably integrate 1st generation or geography-specific traits in the target plant, while new traits are placed in the donor plant. Donor and target plants are crossed to create a breeding stack, which is then crossed with appropriate designed nuclease expressing plants. The F1 progenies are then subjected to tissue culture selection and targeted plant regeneration. Multiple donor lines containing different traits could strategically be made to keep modularity required for creating on-demand stacked transgenic traits. The additional tissue culture selection step restricts the use of this method to crop plants that are amenable to tissue culture techniques. Precise tissue-specific expression of designed nuclease in reproductive cells [57–59] can circumvent the need for a tissue culture regeneration process, providing broader application of this approach across different crops.
Crossing and targeted plant production strategy in maize using intra-genomic homologous recombination. Plants homozygous to donor (a) and target (b) are crossed and self-pollinated to obtain progenies that are homozygous to target and donor loci (c). The homozygous target donor plants are crossed with plants containing designed nuclease (DN) transgene (d) to obtain F1 progenies transgenic to target, donor and DN (e). The F1 immature embryos are treated on appropriate selection media (f) and targeted plants are regenerated on selection (g and h). Alternatively, the F1 plants could be selection sprayed (i) to obtain targeted plants (j)
Conclusions
Future biotech crops are projected to require multiple transgenes to confer resistance to a broad spectrum of insect pests and provide herbicide tolerance with different modes of action. Insects and weeds will eventually develop resistance, new target pests will emerge and new traits will inevitably be needed and desired, so designing those future products to be further modified and developing capabilities to accomplish the modifications are wise investments. It is clear that producing and modifying transgenic events through GT has many advantages over random integration, and technology continues to develop to make GT increasingly efficient and flexible. Intra genomic homologous recombination using designed nucleases has good potential to overcome limitations in plant transformation and breeding to achieve targeted and highly complex stacked trait crops.
Abbreviations
- DSBs:
-
double strand breaks
- GT:
-
gene targeting
- IGHR:
-
intra-genomic homologous recombination
- HR:
-
homologous recombination
- NHEJ:
-
non-homologous end joining
- uidA :
-
beta-glucuronidase
- nptII :
-
neomycin phophotransferase II
References
Dockter C, Hansson M. Improving barley culm robustness for secured crop yield in a changing climate. J Exp Bot. 2015;66(12):3499–509. doi:10.1093/jxb/eru521.
Parry MA, Hawkesford MJ. An integrated approach to crop genetic improvement. J Integr Plant Biol. 2012;54(4):250–9. doi:10.1111/j.1744-7909.2012.01109.x.
Bohra A, Sahrawat KL, Kumar S, Joshi R, Parihar AK, Singh U, et al. Genetics- and genomics-based interventions for nutritional enhancement of grain legume crops: status and outlook. J Appl Genet. 2015. doi:10.1007/s13353-014-0268-z.
Que Q, Chilton MD, de Fontes CM, He C, Nuccio M, Zhu T, et al. Trait stacking in transgenic crops: challenges and opportunities. GM Crops. 2010;1(4):220–9. doi:10.4161/gmcr.1.4.13439.
Siebert MW, Nolting SP, Hendrix W, Dhavala S, Craig C, Leonard BR, et al. Evaluation of corn hybrids expressing Cry1F, Cry1A.105, Cry2Ab2, Cry34Ab1/Cry35Ab1, and Cry3Bb1 against southern United States insect pests. J Econ Entomol. 2012;105(5):1825–34. doi:10.1603/Ec12155.
Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D, Christian M, et al. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc Natl Acad Sci U S A. 2010;107(26):12028–33. doi:10.1073/pnas.0914991107.
Liang Z, Zhang K, Chen K, Gao C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genomics. 2014;41(2):63–8. doi:10.1016/j.jgg.2013.12.001.
Gao J, Wang G, Ma S, Xie X, Wu X, Zhang X, et al. CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol Biol. 2015;87(1–2):99–110. doi:10.1007/s11103-014-0263-0.
Gao HR, Smith J, Yang MZ, Jones S, Djukanovic V, Nicholson MG, et al. Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 2010;61(1):176–87. doi:10.1111/j.1365-313X.2009.04041.x.
Camerini-Otero RD, Hsieh P. Homologous recombination proteins in prokaryotes and eukaryotes. Annu Rev Genet. 1995;29:509–52. doi:10.1146/annurev.ge.29.120195.002453.
Wood RD. DNA repair in eukaryotes. Annu Rev Biochem. 1996;65:135–67. doi:10.1146/annurev.bi.65.070196.001031.
Lieberman-Lazarovich M, Levy AA. Homologous recombination in plants: an antireview. Methods Mol Biol. 2011;701:51–65. doi:10.1007/978-1-61737-957-4_3.
Waterworth WM, Drury GE, Bray CM, West CE. Repairing breaks in the plant genome: the importance of keeping it together. New Phytol. 2011;192(4):805–22. doi:10.1111/j.1469-8137.2011.03926.x.
Puchta H, Fauser F. Synthetic nucleases for genome engineering in plants: prospects for a bright future. Plant J. 2014;78(5):727–41. doi:10.1111/tpj.12338.
Weeks DP, Spalding MH, Yang B. Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol J. 2015;. doi:10.1111/pbi.12448.
Puchta H, Fauser F. Synthetic nucleases for genome engineering in plants: prospects for a bright future. Plant J. 2013;. doi:10.1111/tpj.12338.
Osman K, Higgins JD, Sanchez-Moran E, Armstrong SJ, Franklin FC. Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol. 2011;190(3):523–44. doi:10.1111/j.1469-8137.2011.03665.x.
Puchta H. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot. 2005;56(409):1–14. doi:10.1093/jxb/eri025.
Weinthal DM, Taylor RA, Tzfira T. Nonhomologous end joining-mediated gene replacement in plant cells. Plant Physiol. 2013;162(1):390–400. doi:10.1104/pp.112.212910.
Chilton MD, Que Q. Targeted integration of T-DNA into the tobacco genome at double-stranded breaks: new insights on the mechanism of T-DNA integration. Plant Physiol. 2003;133(3):956–65. doi:10.1104/pp.103.026104.
D’Halluin K, Ruiter R. Directed genome engineering for genome optimization. Int J Dev Biol. 2013;57(6–8):621–7. doi:10.1387/ijdb.130217kd.
Puchta H, Dujon B, Hohn B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci U S A. 1996;93(10):5055–60.
Paszkowski J, Baur M, Bogucki A, Potrykus I. Gene targeting in plants. EMBO J. 1988;7(13):4021–6.
Cai CQ, Doyon Y, Ainley WM, Miller JC, Dekelver RC, Moehle EA, et al. Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol Biol. 2009;69(6):699–709. doi:10.1007/s11103-008-9449-7.
Nishizawa-Yokoi A, Nonaka S, Osakabe K, Saika H, Toki S. A universal positive-negative selection system for gene targeting in plants combining an antibiotic resistance gene and its antisense rna. Plant Physiol. 2015;169(1):362–70. doi:10.1104/pp.15.00638.
Shimatani Z, Nishizawa-Yokoi A, Endo M, Toki S, Terada R. Positive-negative-selection-mediated gene targeting in rice. Front Plant Sci. 2014;5:748. doi:10.3389/fpls.2014.00748.
Kumar S, AlAbed D, Worden A, Novak S, Wu H, Ausmus C, et al. A modular gene targeting system for sequential transgene stacking in plants. J Biotechnol. 2015;207:12–20. doi:10.1016/j.jbiotec.2015.04.006.
Petolino JF, Kumar S. Transgenic trait deployment using designed nucleases. Plant Biotechnol J. 2015;. doi:10.1111/pbi.12457.
Rivera AL, Gomez-Lim M, Fernandez F, Loske AM. Physical methods for genetic plant transformation. Phys Life Rev. 2012;9(3):308–45. doi:10.1016/j.plrev.2012.06.002.
Tzfira T, Citovsky V. Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr Opin Biotech. 2006;17(2):147–54. doi:10.1016/j.copbio.2006.01.009.
Taylor NJ, Fauquet CM. Microparticle bombardment as a tool in plant science and agricultural biotechnology. DNA Cell Biol. 2002;21(12):963–77. doi:10.1089/104454902762053891.
Pitzschke A. Agrobacterium infection and plant defense—transformation success hangs by a thread. Front Plant Sci. 2013;4:519. doi:10.3389/fpls.2013.00519.
Arias RS, Filichkin SA, Strauss SH. Divide and conquer: development and cell cycle genes in plant transformation. Trends Biotechnol. 2006;24(6):267–73. doi:10.1016/j.tibtech.2006.04.007.
Delporte F, Jacquemin JM, Masson P, Watillon B. Insights into the regenerative property of plant cells and their receptivity to transgenesis: wheat as a research case study. Plant Signal Behav. 2012;7(12):1608–20.
Szabados L, Kovács I, Oberschall A, Ábrahám E, Kerekes I, Zsigmond L, et al. Distribution of 1000 sequenced T-DNA tags in the Arabidopsis genome. Plant J. 2002;32(2):233–42. doi:10.1046/j.1365-313X.2002.01417.x.
Gelvin SB, Kim S-I. Effect of chromatin upon agrobacterium T-DNA integration and transgene expression. Biochimica et Biophysica Acta (BBA)—Gene Structure and Expression. 2007;1769(5–6):410–21. doi:http://dx.doi.org/10.1016/j.bbaexp.2007.04.005.
Hiei Y, Ishida Y, Komari T. Progress of cereal transformation technology mediated by Agrobacterium tumefaciens. Front Plant Sci. 2014;5:628. doi:10.3389/fpls.2014.00628.
Frame B, Main M, Schick R, Wang K. Genetic transformation using maize immature zygotic embryos. Methods Mol Biol. 2011;710:327–41. doi:10.1007/978-1-61737-988-8_22.
Kita Y, Nishizawa K, Takahashi M, Kitayama M, Ishimoto M. Genetic improvement of the somatic embryogenesis and regeneration in soybean and transformation of the improved breeding lines. Plant Cell Rep. 2007;26(4):439–47. doi:10.1007/s00299-006-0245-z.
Puchta H, Hohn B. In planta somatic homologous recombination assay revisited: a successful and versatile, but delicate tool. Plant Cell. 2012;24(11):4324–31. doi:10.1105/tpc.112.101824.
Swoboda P, Hohn B, Gal S. Somatic homologous recombination in planta—the recombination frequency is dependent on the allelic state of recombining sequences and may be influenced by genomic position effects. Mol Gen Genet. 1993;237(1–2):33–40.
Swoboda P, Gal S, Hohn B, Puchta H. Intrachromosomal homologous recombination in whole plants. EMBO J. 1994;13(2):484–9.
Chiurazzi M, Ray A, Viret JF, Perera R, Wang XH, Lloyd AM, et al. Enhancement of somatic intrachromosomal homologous recombination in Arabidopsis by the HO endonuclease. Plant Cell. 1996;8(11):2057–66. doi:10.1105/tpc.8.11.2057.
Siebert R, Puchta H. Efficient repair of genomic double-strand breaks by homologous recombination between directly repeated sequences in the plant genome. Plant Cell. 2002;14(5):1121–31.
Petolino JF, Worden A, Curlee K, Connell J, Strange Moynahan TL, Larsen C, et al. Zinc finger nuclease-mediated transgene deletion. Plant Mol Biol. 2010;73(6):617–28. doi:10.1007/s11103-010-9641-4.
Antunes MS, Smith JJ, Jantz D, Medford JI. Targeted DNA excision in Arabidopsis by a re-engineered homing endonuclease. BMC Biotechnol. 2012;12:86. doi:10.1186/1472-6750-12-86.
Qi Y, Li X, Zhang Y, Starker CG, Baltes NJ, Zhang F, et al. Targeted deletion and inversion of tandemly arrayed genes in Arabidopsis thaliana using zinc finger nucleases. G3 (Bethesda). 2013;3(10):1707–15. doi:10.1534/g3.113.006270.
Zhou H, Liu B, Weeks DP, Spalding MH, Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42(17):10903–14. doi:10.1093/nar/gku806.
Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288(5473):2013–8.
Rong YS, Golic KG. A targeted gene knockout in Drosophila. Genetics. 2001;157(3):1307–12.
Maggert KA, Gong WJ, Golic KG. Methods for homologous recombination in Drosophila. Methods Mol Biol. 2008;420:155–74. doi:10.1007/978-1-59745-583-1_9.
Bi X, Rong YS. Genome manipulation by homologous recombination in Drosophila. Brief Funct Genomic Proteomic. 2003;2(2):142–6.
Kumar S, Fladung M. Controlling transgene integration in plants. Trends Plant Sci. 2001;6(4):155–9. doi:10.1016/S1360-1385(01)01890-8.
Fauser F, Roth N, Pacher M, Ilg G, Sanchez-Fernandez R, Biesgen C, et al. In planta gene targeting. Proc Natl Acad Sci U S A. 2012;109(19):7535–40. doi:10.1073/pnas.1202191109.
Schiml S, Fauser F, Puchta H. The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 2014;80(6):1139–50. doi:10.1111/tpj.12704.
Ayar A, Wehrkamp-Richter S, Laffaire JB, Le Goff S, Levy J, Chaignon S, et al. Gene targeting in maize by somatic ectopic recombination. Plant Biotechnol J. 2013;11(3):305–14. doi:10.1111/Pbi.12014.
Even-Faitelson L, Samach A, Melamed-Bessudo C, Avivi-Ragolsky N, Levy AA. Localized egg-cell expression of effector proteins for targeted modification of the Arabidopsis genome. Plant J. 2011;68(5):929–37. doi:10.1111/j.1365-313X.2011.04741.x.
Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, et al. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015;16:144. doi:10.1186/s13059-015-0715-0.
Mao Y, Zhang Z, Feng Z, Wei P, Zhang H, Botella JR, et al. Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol J. 2015. doi:10.1111/pbi.12468.
Authors’ contributions
SK, PB, and MS drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We are grateful to Otto Folkerts, Lakshmi Sastry-Dent, and Rodrigo Sarria for reviewing the manuscript and providing valuable suggestions.
Competing interests
The authors in this publication certify that they are employees of Dow AgroSciences LLC, a subsidiary of The Dow Chemical Company. As employees, they authors may have some company stock (The Dow Chemical Company), but no other personal financial interest in the subject matter of the materials discussed in this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kumar, S., Barone, P. & Smith, M. Gene targeting and transgene stacking using intra genomic homologous recombination in plants. Plant Methods 12, 11 (2016). https://doi.org/10.1186/s13007-016-0111-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13007-016-0111-0