Abstract
Background
While the presence of SARS-CoV-2 in human breast milk is contentious, anti-SARS-CoV-2 antibodies have been consistently detected in human breast milk. However, it is uncertain when and how long the antibodies are present.
Methods
This was a prospective cohort study including all consecutive pregnant women with confirmed SARS-CoV-2 infection during pregnancy, recruited at six maternity units in Spain and Hong Kong from March 2020 to March 2021. Colostrum (day of birth until day 4 postpartum) and mature milk (day 7 postpartum until 6 weeks postpartum) were prospectively collected, and paired maternal blood samples were also collected. Colostrum samples were tested with rRT-PCR-SARS-CoV-2, and skimmed acellular milk and maternal sera were tested against SARS-CoV-2 specific immunoglobulin M, A, and G reactive to receptor binding domain of SARS-CoV-2 spike protein 1 to determine the presence of immunoglobulins. Then, we examined how each immunoglobulin type in the colostrum was related to the time of infection by logistic regression analysis, the concordance between these immunoglobulins in the colostrum, maternal serum, and mature milk by Cohen's kappa statistic, and the relationship between immunoglobulin levels in mature milk and colostrum with McNemar.
Results
One hundred eighty-seven pregnant women with confirmed SARS-CoV-2 infection during pregnancy or childbirth were recruited and donated the milk and blood samples. No SARS-CoV-2 was found in the human breast milk. Immunoglobulin A, G, and M were present in 129/162 (79·6%), 5/163 (3·1%), and 15/76 (19·7%) colostrum samples and in 17/62 (27·42%), 2/62 (3·23%) and 2/62 (3·23%) mature milk samples, respectively. Immunoglobulin A was the predominant immunoglobulin found in breast milk, and its levels were significantly higher in the colostrum than in the mature milk (p-value < 0.001). We did not find that the presence of immunoglobulins in the colostrum was associated with their presence in maternal, the severity of the disease, or the time when the infection had occurred.
Conclusions
Since anti-SARS-CoV-2 antibodies are found in the colostrum irrespective of the time of infection during pregnancy, but the virus itself is not detected in human breast milk, our study found no indications to withhold breastfeeding, taking contact precautions when there is active disease.
Similar content being viewed by others
Background
On 11 March 2020, the COVID-19 pandemic was declared by the World Health Organization (WHO) [1]. Since then, extensive efforts have focused on evaluating the effects of the new coronavirus on pregnancy. At the very beginning of the pandemic, newborns were separated from their mothers with confirmed SARS-CoV-2 infection to protect them against the virus. Breastfeeding was avoided because it was unknown if the virus could be transmitted via human breast milk. To date, some studies have reported the presence of SARS-CoV-2 in human breast milk [2,3,4,5,6,7] while others have not [8,9,10,11,12,13,14], but the sample size of these studies is small.
Currently, most healthcare systems and international organizations such as the Centers for Disease Control and Prevention (CDC) recommend breastfeeding for all mothers with active or past infection of SARS-CoV-2, as there appear to be more benefits of breastfeeding than the potential risk of transmission through human breast milk. One of the most important reasons to recommend breastfeeding is the possible passive immunization in newborns against SARS-CoV-2 [15]. In particular, IgA is important because it coats and seals the neonate's respiratory and intestinal tracts to prevent microorganisms from entering the body and bloodstream, constituting the first defense against the virus [16, 17]. Several studies have reported the presence of anti-SARS-CoV-2 antibodies [18,19,20,21,22,23,24,25] in human breast milk. Pace et al. have demonstrated that the specific IgG, IgM, and IgA anti-SARS-CoV-2 antibodies in human breast milk can effectively neutralize SARS-CoV-2 infectivity [11]. However, it is uncertain when the antibodies become present and how long they last in human breast milk.
The aims of this study were first, to determine the presence of anti-SARS-CoV-2 virus and antibodies in colostrum and mature human breast milk in women who had SARS-CoV-2 infection during pregnancy or at the time of childbirth; second, to investigate the association between the anti-SARS-CoV-2 antibodies in human milk with the levels of anti-SARS-CoV-2 antibodies in maternal blood, the severity of SARS-CoV-2 infection and the time interval from active illness; and third, to evaluate how each immunoglobulin type evolved from the colostrum to the mature milk.
Methods
Study population
This was a prospective cohort study aiming to include all consecutive pregnant women with laboratory-confirmed SARS-CoV-2 infection by deep throat saliva (DTS) or nasopharyngeal swab (NPS) real-time reverse-transcriptase-polymerase-chain-reaction (rRT-PCR) test or by rapid antigen-detection tests (Panbio™ COVID-19 Ag Rapid Test Device) [26], during pregnancy, labor or immediately after childbirth, who were able to provide consent to participate in the study, from six maternity units, five in Spain (Hospital Universitario de Torrejón and Hospital Universitario Príncipe de Asturias in Madrid, Hospital Universitario Vall d'Hebrón in Barcelona, Hospital Clínico Universitario San Cecilio in Granada and Hospital Clínico Universitario Virgen de la Arrixaca in Murcia) and one in Hong Kong SAR, China (The Chinese University of Hong Kong COVID-19 collaborative network), from March 2020 to March 2021. Eligibility criteria were: confirmed SARS-CoV2 infection, over 18 years old, and fluent in the investigator's language. Additionally, for suspected cases of COVID-19 where rRT-PCR was negative, if the symptoms had started within seven days of testing, the rRT-PCR was repeated 24 h after the first test. If the symptoms had started beyond seven days of testing, a serology test (ELISA) was performed [27] and women with positive results by either test were also offered participation.
Breast milk samples were collected from the six maternity units. All participants were unvaccinated against SARS-CoV-2, and it was their first SARS-CoV-2 infection.
The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement was used for reporting the results (Additional file. Table 1s).
Participants had one sample of colostrum (between the day of birth and day 4 postpartum) collected and stored at -80ºC. Maternal blood for serological analysis was also collected simultaneously; serum was separated and stored at -80ºC. One sample of fore mature milk (from day 7 to 6 weeks postpartum) was also collected and stored whenever possible.
Clinical data, including maternal age, body mass index (BMI) at the beginning of pregnancy, gestational age at the time of SARS-CoV-2 infection, and disease severity, were recorded for every participant, pseudo-anonymized, and entered into a secured common database. The COVID-19 severity was classified as asymptomatic, mild (when no hospitalization was required), and severe (when the diagnosis of pneumonia was established and hospitalization was needed) [28]. Gestational age was determined by first trimester sonographic assessment of fetal crown-rump length [29] or conception date in vitro fertilization pregnancy.
Biological sample collection and analysis
Breast milk (from 0.1 to 1.0 mL) was collected by manual expression with strict contact precautions to avoid contamination (facial mask and hand cleaning). Blood samples were collected in serum sep clot activator 8 mL tubes, centrifuged for five minutes at 3500 g, and then serum was collected. Both serum and breast milk samples were divided into 0.5 mL aliquots (when possible) in separate Eppendorf tubes, labeled with a unique patient identifier, and stored at -80ºC until subsequent analysis. Stored samples from Barcelona were analyzed locally at the end of the recruitment period. Samples from all other sites were sent without any further processing overnight on dry ice to Synlab Diagnósticos Globales Laboratory in Madrid every month from Spanish sites and in a single batch after rt-RT-PCR testing was performed locally at the end of the recruitment period from Hong Kong.
Breast milk samples were thawed at the laboratory and centrifuged at 800 g for 15 min. Fat was removed, and the supernatant was transferred to a new tube. Centrifugation was repeated twice to ensure the removal of all cells and fat [22]. Skimmed acellular milk was then tested against SARS-CoV-2 specific immunoglobulin M (IgM), immunoglobulin A (IgA), and immunoglobulin G (IgG) reactive to the receptor binding domain (RBD) of the SARS-CoV-2 spike protein 1 (protS1) [22]. As previously reported, serum samples were thawed and tested against SARS-CoV-2 specific antibodies. All equipment and reagents used for analyses are CE (Conformité Européenne) marked (Additional file. Table 2s).
Immuno-analyses
-
Determination of IgA and IgG antibodies was performed by the ELISA method (Enzyme-Linked Immunosorbent Assay), providing semiquantitative serology results against the S1 domain of the spike protein of SARS-CoV-2 in serum samples (Anti-SARS-CoV-2 ELISA IgG and Anti-SARS-CoV-2 ELISA IgA, Euroimmunn Medizinische Labordiagnostika AG, Lubeck, Germany) [30, 31]. Semiquantitative results were calculated as extinction of the control patient sample/extinction of calibrator (further details on this type of analysis are provided in Table 2). IgA and IgG were considered positive, indeterminate, and negative when results were > 1.1, 0.8 to 1.1 and < 0.8, respectively, as recommended by the manufacturer.
-
IgM determination was performed with chemiluminescence microparticle immunoassays, using spike protein-specific (Abbott test, SARS-CoV-2 IgM Abbott, Abbott Ireland Diagnostics Division Finisklin, Ireland) [32], providing semiquantitative (extinction of the control patient sample/extinction of calibrator). IgM was considered positive, indeterminate, and negative when results were > 1.1, 0.9 to 1.1 and < 0.9, respectively, as recommended by the manufacturer.
rRT-PCR-SARS-CoV-2 testing
Whenever available, a second colostrum aliquot was tested for SARS-CoV-2 by rRT-PCR to assess the presence of the virus in the sample. In the Spanish samples, viral RNA was extracted with Chemagic Viral DNA/RNA Kit using the Chemagic 360 with integrated dispense, which includes lyophilized Poly(A) RNA, lyophilized Proteinase K, and a lysis/binding buffer, and were analyzed with Euroinmune Kit (ORF1ab an N targets) and TaqMan™ 2019-nCov Assay kitv2 Thermofisher (s,ORF1ab and N targets). In the Hong Kong samples, viral RNA was extracted using RNeasy® Mini Kit (QIAGEN), and the detection of SARS-CoV-2 RNA was performed with the FDA-authorized CDC 2019-Novel Coronavirus (2019 nCoV) Real-Time RT-PCR Diagnostic Panel (EUA 200001). The N gene (N1 and N2) was assayed, with the human RNase P (RP) as an endogenous reference control. In all cases, samples containing organic or inorganic contaminants interfering with the PCR amplification process were considered inhibited (these samples contained organic or inorganic contaminants that interfered with the PCR amplification process).
For this study, we included all women with available colostrum; additional samples or analyses were not mandatory for inclusion. Given the limited volume of colostrum and serum collected, not all tests could be carried out in all cases. For some laboratory analyses that failed at the first attempt, repeat testing was not possible. Besides, many women did not return to the clinic after birth due to the lockdown. Therefore, we could not collect mature milk in these cases.
Statistical analysis
Descriptive data were expressed as median and interquartile range (IQR) and in proportions (absolute and relative frequencies). Cohen's Kappa was used to assess the colostrum and serum concordance. The kappa statistic was calculated without weighting; very good levels of agreement were considered when it is > 0.80, good 0.80–0.60, moderate 0.60–0.40, poor 0.40–0.20 and very poor < 0.20 [33]. Univariable logistic regression analysis was performed to assess if the presence of immunoglobulins in colostrum was associated with the presence of immunoglobulins in maternal serum, the severity of maternal symptoms, or the time passed from infection. Odds ratio (OR) and 95% confidence interval (CI) were calculated [34]. Lastly, the McNemar test was used to evaluate how each immunoglobulin type evolved in all paired colostrum-mature milk samples; this test reports p values based on the chi squared distribution with 1 degree of freedom. The level of significance was set at 0.05.
The statistical software R version 4.1.2 (Vienna, Austria) was used for all data analyses [35].
Results
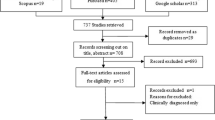
A total of 246 pregnant women with confirmed SARS-CoV2 infection during pregnancy or childbirth were eligible and were approached with information about the study. After exclusions, 191 women agreed to participate (4 were underaged, 7 were unable to provide consent, and 44 were not interested in participating). Among those, 187 had colostrum available for analysis (Figs. 1, 2). Of these, 38 (20.3%), 65 (34.8%), and 84 (44.9%) women acquired the infection in the first (< 14 weeks), second (14–28+6), and third trimester (> 28+6) of pregnancy, respectively. Among the cases with third-trimester infection, 29 (34.5%) had active SARS-CoV-2 infection at childbirth (rRT-PCR-SARS-CoV-2 positive at birth). Pregnancy and disease characteristics are shown in Table 1.
The colostrum and blood samples were collected between the day of birth and day 4 postpartum (median = 1; IQR 0 to 1). Mature milk samples were collected after day 7 postpartum (median = 39 days, IQR 25 to 44). Sample availability and serological status are displayed in Table 2.
Out of the 187 samples collected, only 162 yielded results for IgA, 163 for IgG, and 76 for IgM due to technical issues such as limited volume and assay failures (Table 3). IgA, IgG, and IgM were present in 129/162 (79.6%), 5/163 (3.1%), and 15/76 (19.7%) colostrum samples, respectively. All immunoglobulin-positive colostrum samples tested positive for IgA, except for one sample that only tested positive for IgG (IgA and IgM negative). Another tested positive for IgM and IgG, but there was insufficient sample for the detection of IgA. None of the samples had all 3 immunoglobulins detected.
Seventy-six colostrum samples were tested for rRT-PCR-SARS-CoV-2, including 29 with active disease at birth. 73 tested negative, and 3 were inhibited (these samples contained organic or inorganic contaminants that interfered with the PCR amplification process).
Association between colostrum and serum
One hundred eighteen women had at least one serology result. Association between the colostrum and serum measured with Cohen's Kappa was 0.09 (CI 95% -0.11 to 0.30) for IgA; 0.06 (CI 95% -0.01 to 0.12) for IgG, and 0.29 (CI 95% 0.03 to 0.54) for IgM (Table 4).
Factors related to colostrum positivity
There were no statistically significant differences between the immunoglobulin status in colostrum and the severity of the symptoms nor the time interval from the disease, either as a continuous variable or considering only active disease at birth vs. no active disease at birth (Table 5).
Antibody evolution from colostrum to mature milk
In mature milk samples, IgG was positive in 2/62 (3.23%) (two women with active disease at birth that tested negative in colostrum); IgA was positive in 17/62 (27.42%) (32 women that tested positive in colostrum but negative in mature milk; p-value for the difference between IgA in mature milk vs. IgA in colostrum < 0.001, McNemar's chi-squared statistic = 29.032); and IgM was positive in 0/51 (6 of 51 were positive in colostrum).
Discussion
Main findings
The study has demonstrated that, firstly, all human breast milk tested for rRT-PCR SARS-CoV-2 are negative; secondly, antibodies against SARS-CoV-2 present in the colostrum do not seem to vary significantly in relation to the time when the infection has occurred during pregnancy or with regard to their presence in the maternal blood; and thirdly, IgA is the predominant immunoglobulin found in human breast milk and its concentrations are significantly lower in the mature milk compared with colostrum.
Study strengths and limitations
To our knowledge, this is the largest series of colostrum samples from women with SARS-CoV-2 infection during pregnancy or at the time of birth (MEDLINE via Pubmed search (September 2023): ((Human breast milk[MeSH Terms]) AND ("COVID-19" [MeSH Terms])) AND ("antibodies" [MeSH Terms]), and where all three types of antibodies, as well as rRT-PCR-SARS-CoV-2, were tested. We also collected paired colostrum and mature milk samples and studied the serological status of the mother at the time of milk sampling, which allowed us to investigate the immunoglobulin association between the colostrum, mature milk, and maternal blood. Additionally, the protocol for collecting, handling, and storing samples was defined early and implemented in all centers [22]. Furthermore, we included 16 pregnant women with severe disease in the study, allowing us to investigate possible associations between the presence of immunoglobulins in colostrum and the severity of the disease.
The main limitations relate to the small sample size and the technical difficulties that further reduced the sample, which may have prevented us from recognizing other possible associations or significant findings. However, technical factors equally affect all samples, making it unlikely to be a source of bias. Besides, this study was conducted at the peak of the pandemic outbreak when vaccination was not a confounder, so the findings are still of great value. A second important limitation is that there is a wide range of gestational age at sampling, and the timing of colostrum and serum sample collection varied between days 0 to 4 postpartum, which may be responsible for physiological changes in immunoglobulin concentration. Nonetheless, we believe this also provides a better understanding of what happens during pregnancy and postpartum. Of note, there were fewer obese women and pregnancies ending in preterm birth than expected among infected COVID-19 pregnancies. However, this might be because most patients were recruited in non-tertiary referral centers, where the most severe cases were centralized.
Interpretation
It is well known that breastfeeding protects babies against gastrointestinal and respiratory infections [36,37,38,39]. IgA represents around 90% of all immunoglobulins in human milk, and its concentration is higher in the colostrum, decreasing during the first year of lactation [15]. Due to its low degradation and absorption rate in the infant's gastrointestinal system, IgA is the most important immunoglobulin in human milk since it protects the infant against infections at the mucosa level [16, 40]. Recently, it has been demonstrated that specific IgG, IgM, and IgA anti-SARS-CoV-2 antibodies in breast milk neutralize the virus in vitro [11, 41,42,43]. Therefore, anti-SARS-CoV-2 IgA in human breast milk could also protect the infant against the SARS-CoV-2 infection locally in their gastrointestinal mucosa, similar to what happens with other viral infections [44, 45].
In our study, most colostrum samples tested positive for IgA, irrespective of the time of SARS-CoV-2 infection. A significant reduction in IgA positivity was found when evaluating longitudinal changes in the colostrum and the mature milk. This is similar to what happens in other viral infections [15]. Importantly, IgA was present even in the colostrum of mothers with a negative serological status at childbirth, contrary to what happened with IgG, which was more likely to be detected when IgG in serum was also present. A possible explanation for this could be related to the fact that IgA is secreted from the maternal Gastrointestinal Antigen Linfoid Tissue (GALT) system and transported into the maternal mammary glands, where they are incorporated into the breast milk, while IgG is mostly filtered from the maternal plasma, albeit at a lower concentration [46]. When the infant nurses, they receive these antibodies along with essential nutrients from the maternal milk, providing passive immunity and protection against infections until their immune system matures [47, 48]. This system is responsible for secreting antibodies against common infections prevalent in maternal living area and, therefore, represent maternal memory [49]. This system also secretes IgM but at much lower concentrations.
In this study, 29 samples from women with active disease at childbirth were tested by rRT-PCR-SARS-CoV-2, and all were negative. Evidence suggesting the presence of SARS-CoV-2 in breast milk is conflicting [2,3,4,5, 8,9,10, 20], and it is possible that cross-contamination was responsible for the positive results [11]. Goad et al. investigated the presence of cell-specific expression of angiotensin-converting enzyme 2 (ACE2), proteases TMPRSS2, and cathepsins CTSB and CTSL in breast epithelium, and they did not find co-expression of ACE2/TMPRSS2 or ACE2/CTSB/L, which is essential for the entry of the virus into the cell. Therefore, they concluded that there was no risk of vertical transmission of SARS-CoV-2 in neonates through breastfeeding [50].
Clinical implications
This study confirms that SARS-CoV-2 is not detected in breast milk, even when active infection occurs at birth. Therefore, the possibility of vertical transmission while breastfeeding is extremely low. Furthermore, since antibodies are found in the colostrum irrespective of the time of infection, all women should be encouraged to breastfeed their infants, regardless of the time when the condition has occurred during the pregnancy, undertaking contact precautions when there is active disease. Nevertheless, since IgA concentrations drop significantly from the colostrum to mature milk, we could speculate that they might be even lower beyond six weeks postpartum, so public health measures should still be maintained to reduce the risk of the babies acquiring the infection.
Conclusions
Our study has provided further evidence that breastfeeding is safe during maternal SARS-CoV-2 infection as the virus has not been detected in human breast milk, and protective antibodies have been found instead. However, larger studies with longer follow-ups are still needed.
Availability of data and materials
The data presented in this study are available on request from the corresponding author and conditioned to approval from the relevant Research Ethics Committees due to data protection regulations.
Abbreviations
- ACE2:
-
Angiotensin-converting enzyme 2
- BMI:
-
Body mass index
- CI:
-
Confidence interval
- ELISA:
-
Enzyme-linked immunosorbent assay
- GALT:
-
Gastrointestinal antigen Linfoide tissue
- IgA:
-
Immunoglobulin A
- IgG:
-
Immunoglobulin G
- IgM:
-
Immunoglobulin M
- IQR:
-
Interquartile range
- LMP:
-
Last menstrual period
- protS1:
-
SARS-CoV-2 spike protein 1
- RBD:
-
Receptor binding domain
- rRT-PCR:
-
Real-time reverse-transcriptase-polymerase-chain-reaction
References
WHO’s COVID-19 response. 2021. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/interactive-timeline.
Groß R, Conzelmann C, Müller JA, Stenger S, Steinhart K, Kirchhoff F, et al. Detection of SARS-CoV-2 in human breastmilk. Lancet. 2020;395(10239):1757–8.
Buonsenso D, Costa S, Sanguinetti M, Cattani P, Posteraro B, Marchetti S, et al. Neonatal late onset Infection with Severe Acute Respiratory Syndrome Coronavirus 2. Am J Perinatol. 2020;37(8):869–72.
Kirtsman M, Diambomba Y, Poutanen SM, Malinowski AK, Vlachodimitropoulou E, Parks WT, et al. Probable congenital SARS-CoV-2 infection in a neonate born to a woman with active SARS-CoV-2 infection. CMAJ. 2020;192(24):E647–50.
Tam PCK, Ly KM, Kernich ML, Spurrier N, Lawrence D, Gordon DL, et al. Detectable Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in human breast milk of a mildly symptomatic patient with Coronavirus disease 2019 (COVID-19). Clin Infect Dis. 2021;72(1):128–30.
Kilic T, Kilic S, Berber NK, Gunduz A, Ersoy Y. Investigation of SARS-CoV-2 RNA in milk produced by women with COVID-19 and follow-up of their infants: A preliminary study. Int J Clin Pract. 2021;75(7):e14175.
Kumar J, Meena J, Yadav A, Kumar P. SARS-CoV-2 detection in human milk: a systematic review. J Matern Fetal Neonatal Med. 2022;35(25):5456–63.
Chen H, Guo J, Wang C, Luo F, Yu X, Zhang W, et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395(10226):809–15.
Lackey KA, Pace RM, Williams JE, Bode L, Donovan SM, Järvinen KM, et al. SARS-CoV-2 and human milk: what is the evidence? Matern Child Nutr. 2020;16(4):e13032.
Rajewska A, Mikołajek-Bedner W, Lebdowicz-Knul J, Sokołowska M, Kwiatkowski S, Torbé A. COVID-19 and pregnancy - where are we now? A review J Perinat Med. 2020;48(5):428–34.
Pace RM, Williams JE, Järvinen KM, Belfort MB, Pace CDW, Lackey KA, et al. Characterization of SARS-CoV-2 RNA, Antibodies, and Neutralizing Capacity in Milk Produced by Women with COVID-19. mBio. 2021;12(1):e03192–20.
Prasad A, N Y, Kumar P, Chaudhary BK, Pati BK, Anant M, et al. Excretion of SARS-CoV-2 in breast milk: a single-centre observational study. BMJ Paediatr Open. 2021;5(1):e001087.
Krogstad P, Contreras D, Ng H, Tobin N, Chambers CD, Bertrand K, et al. No infectious SARS-CoV-2 in breast milk from a cohort of 110 lactating women. Pediatr Res. 2022;92(4):1140–5.
Vigil-Vázquez S, Carrasco-García I, Hernanz-Lobo A, Manzanares Á, Pérez-Pérez A, Toledano-Revenga J, et al. Impact of Gestational COVID-19 on Neonatal Outcomes: Is Vertical Infection Possible? Pediatr Infect Dis J. 2022;41(6):466–72.
Goldman AS, Garza C, Nichols BL, Goldblum RM. Immunologic factors in human milk during the first year of lactation. J Pediatr. 1982;100(4):563–7.
Mestecky J. Antibody-dependent passive protection of mucosal surfaces. Hum Vaccines Immunother. 2022;18(2):1899549.
Fagarasan S, Kawamoto S, Kanagawa O, Suzuki K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu Rev Immunol. 2010;28:243–73.
Dong Y, Chi X, Hai H, Sun L, Zhang M, Xie WF, et al. Antibodies in the breast milk of a maternal woman with COVID-19. Emerg Microbes Infect. 2020;9(1):1467–9.
Demers-Mathieu V, Do DM, Mathijssen GB, Sela DA, Seppo A, Järvinen KM, et al. Difference in levels of SARS-CoV-2 S1 and S2 subunits- and nucleocapsid protein-reactive SIgM/IgM, IgG and SIgA/IgA antibodies in human milk. J Perinatol. 2020;41(4):850–9. https://doi.org/10.1038/s41372-020-00805-w.
Lebrão CW, Cruz MN, da Silva MH, Dutra LV, Cristiani C, Affonso Fonseca FL, et al. Early Identification of IgA Anti-SARSCoV-2 in milk of mother with COVID-19 Infection. J Hum Lact. 2020;36(4):609–13.
Gao X, Wang S, Zeng W, Chen S, Wu J, Lin X, et al. Clinical and immunologic features among COVID-19-affected mother-infant pairs: antibodies to SARS-CoV-2 detected in breast milk. New Microbes New Infect. 2020;37: 100752.
Fox A, Marino J, Amanat F, Krammer F, Hahn-Holbrook J, Zolla-Pazner S, et al. Robust and Specific Secretory IgA Against SARS-CoV-2 Detected in Human Milk. iScience. 2020;23(11):101735.
Fox A, Marino J, Amanat F, Oguntuyo KY, Hahn-Holbrook J, Lee B, et al. The IgA in milk induced by SARS-CoV-2 infection is comprised of mainly secretory antibody that is neutralizing and highly durable over time. PLoS ONE. 2022;17(3): e0249723.
Szczygioł P, Łukianowski B, Kościelska-Kasprzak K, Jakuszko K, Bartoszek D, Krajewska M, et al. Antibodies in the breastmilk of COVID-19 recovered women. BMC Pregnancy Childbirth. 2022;22(1):635.
Bode L, Bertrand K, Najera JA, Furst A, Honerkamp-Smith G, Shandling AD, et al. Characterization of SARS-CoV-2 antibodies in human milk from 21 women with confirmed COVID-19 infection. Pediatr Res. 2023;93(6):1626–33.
Merino P, Guinea J, Muñoz-Gallego I, González-Donapetry P, Galán JC, Antona N, et al. Multicenter evaluation of the PanbioTM COVID-19 rapid antigen-detection test for the diagnosis of SARS-CoV-2 infection. Clin Microbiol Infect. 2021;27(5):758–61. https://doi.org/10.1016/j.cmi.2021.02.001.
Lamouroux A, Attie-Bitach T, Martinovic J, Leruez-Ville M, Ville Y. Evidence for and against vertical transmission for severe acute respiratory syndrome coronavirus 2. Am J Obstet Gynecol. 2020;223(1):91.e1–91.e4.
Wu Z, McGoogan JM. Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–42.
Robinson HP, Fleming JE. A critical evaluation of sonar ‘crown-rump length’ measurements. Br J Obstet Gynaecol. 1975;82(9):702–10.
Beavis KG, Matushek SM, Abeleda APF, Bethel C, Hunt C, Gillen S, et al. Evaluation of the EUROIMMUN Anti-SARS-CoV-2 ELISA Assay for detection of IgA and IgG antibodies. J Clin Virol. 2020;129: 104468.
Taylor SC, Hurst B, Martiszus I, Hausman MS, Sarwat S, Schapiro JM, et al. Semi-quantitative, high throughput analysis of SARS-CoV-2 neutralizing antibodies: Measuring the level and duration of immune response antibodies post infection/vaccination. Vaccine. 2021;39(39):5688–98.
Nicol T, Lefeuvre C, Serri O, Pivert A, Joubaud F, Dubée V, et al. Assessment of SARS-CoV-2 serological tests for the diagnosis of COVID-19 through the evaluation of three immunoassays: Two automated immunoassays (Euroimmun and Abbott) and one rapid lateral flow immunoassay (NG Biotech). J Clin Virol. 2020;129: 104511.
Lantz CA, Nebenzahl E. Behavior and interpretation of the kappa statistic: resolution of the two paradoxes. J Clin Epidemiol. 1996;49(4):431–4.
Cleveland WS, Grosse E, Shyu WM. Local Regression Models. In: Statistical Models in S. Routledge; 1992.
Core Team. R: A language and environment for statistical computing. 2020. https://www.R-project.org/.
Jason JM, Nieburg P, Marks JS. Mortality and infectious disease associated with infant-feeding practices in developing countries. Pediatrics. 1984;74(4 Pt 2):702–27.
Hanson LA. Human milk and host defence: immediate and long-term effects. Acta Paediatr. 1999;88(430):42–6.
Kovar MG, Serdula MK, Marks JS, Fraser DW. Review of the epidemiologic evidence for an association between infant feeding and infant health. Pediatrics. 1984;74(4 Pt 2):615–38.
Castilla J, Pintado B, Sola I, Sánchez-Morgado JM, Enjuanes L. Engineering passive immunity in transgenic mice secreting virus-neutralizing antibodies in milk. Nat Biotechnol. 1998;16(4):349–54.
Pullen KM, Atyeo C, Collier ARY, Gray KJ, Belfort MB, Lauffenburger DA, et al. Selective functional antibody transfer into the breastmilk after SARS-CoV-2 infection. Cell Rep. 2021;37(6): 109959.
Low JM, Low YW, Zhong Y, Lee CYC, Chan M, Ng NBH, et al. Titres and neutralising capacity of SARS-CoV-2-specific antibodies in human milk: a systematic review. Arch Dis Child Fetal Neonatal Ed. 2022;107(2):174–80.
Pace RM, Williams JE, Järvinen KM, Belfort MB, Pace CD, Lackey KA, et al. COVID-19 and human milk: SARS-CoV-2, antibodies, and neutralizing capacity. MedRxiv. 2020;2020.09.16.20196071.
Morniroli D, Signorini L, Dolci M, Vizzari G, Ronchi A, Pietrasanta C, et al. Breastmilk from COVID-19 negative lactating mothers shows neutralizing activity against SARS-COV-2. Sci Rep. 2023;13(1):15521.
Jawhara S. Can Drinking Microfiltered Raw Immune Milk From Cows Immunized Against SARS-CoV-2 Provide Short-Term Protection Against COVID-19? Front Immunol. 2020;11:1888.
Aknouch I, Sridhar A, Freeze E, Giugliano FP, van Keulen BJ, Romijn M, et al. Human milk inhibits some enveloped virus infections, including SARS-CoV-2, in an intestinal model. Life Sci Alliance. 2022;5(12): e202201432.
Larson BL, Heary HL, Devery JE. Immunoglobulin production and transport by the mammary gland. J Dairy Sci. 1980;63(4):665–71.
Brandtzaeg P, Farstad IN, Haraldsen G, Jahnsen FL. Cellular and molecular mechanisms for induction of mucosal immunity. Dev Biol Stand. 1998;92:93–108.
Mestecky J. The common mucosal immune system and current strategies for induction of immune responses in external secretions. J Clin Immunol. 1987;7(4):265–76.
Nathavitharana KA, Catty D, McNeish AS. IgA antibodies in human milk: epidemiological markers of previous infections? Arch Dis Child Fetal Neonatal Ed. 1994;71(3):F192–197.
Goad J, Rudolph J, Rajkovic A. Female reproductive tract has low concentration of SARS-CoV2 receptors. PLoS ONE. 2020;15(12):e0243959.
Acknowledgements
The authors are grateful to all participants and their attending obstetricians, nurses, midwives, and laboratory technicians at all participating hospitals for facilitating the performance of this study. We are also grateful to Dr. Santiago Valor, CMO at Synlab International GmbH, for his contribution to the study execution and valuable comments, iMaterna Foundation (registry number: 2148, Spain) for supporting this study by grant, and Synlab Diagnósticos Globales and Perkin Elmer for providing the reagents, devices, and human resources to perform the analyses of samples. This study is part of the Ph.D. thesis of Dr. Rayo. To all collaborators: Anna Suy, Department of Obstetrics, Hospital Universitari Vall d’Hebron, Barcelona. Maria de la Calle, Department of Obstetrics, Hospital Universitario La Paz, Madrid. Juan Luis Delgado, department of Obstetrics, Hospital Virgen de la Arrixaca, Murcia. Leire Rodriguez, department of Obstetrics, Hospital Universitario Cruces, Barakaldo. Sara Ruiz-Martínez, department of Obstetrics, Hospital Clínico Universitario Lozano Blesa, Zaragoza. Francisca Sonia Molina, department of Obstetrics, San Cecilio University Hospital, Instituto de Investigación Biosanitaria (IBS), Granada. Vicente Diago, Department of Obstetrics, Hospital Universitario La Fe, Valencia. Adriana Aquise, department of Obstetrics, Hospital Universitario de Torrejón, Madrid. On behalf of the Gesta- COVID-19 Collaboration Group. Mónica García Alvarez, department of clinical microbiology at Synlab Madrid. Miriam Sanchez Tudela, department of andrology laboratory, Hospital Universitario de Torrejón. T. Ma, F. N. Yu, Queen Elizabeth Hospital, Hong Kong SAR, China. C. W. Kong, United Christian Hospital, Hong Kong SAR, China. T. K. Lo, Princess Margaret Hospital, Hong Kong SAR, China. P. L. So, Tuen Mun Hospital, Hong Kong SAR, China, W. C. Leung, Kwong Wah Hospital, Hong Kong SAR, China. W. Shu, Pamela Youde Nethersole Eastern Hospital, Hong Kong SAR, China; K. W. Cheung, Queen Mary Hospital, Hong Kong SAR, China. S. Moungmaithong, The Chinese University of Hong Kong, Hong Kong SAR, China. On behalf of the Chinese University of Hong Kong COVID-19 collaborative network.
This study is part of the Ph.D. Thesis of Dr. Nieves Rayo, for Universidad Francisco de Vitoria, Madrid.
Funding
This study was supported by grants from Instituto de Salud Carlos III 208 (ISCIII) COV20/00188 and iMaterna Foundation (Fundación para la Investigación y el Desarrollo de la Medicina Materno-Fetal y Neonatal, Registry No: 2148). Perkin Elmer provided the reagents for laboratory testing, and Synlab Diagnósticos Globales provided the instruments and human resources to analyze the samples. The funders had no role in study conception and execution, data collection, analyses of the results, nor in the writing of the manuscript or decision to submit for publication.
Author information
Authors and Affiliations
Contributions
Conceptualization: IFB, LCP, MMG. Data curation: IFB, NR, JCS, BS, OOH, BWL, JLD, DSN, SV, LM, AS, RPT, VR, BS, MMG, LCP. Formal analysis: IFB, NR, VR, MMG. Investigation: IFB, NR, JCS, BS, OOH, BWL, JLD, DSN, SV, LM, AS, RPT, VR, BS, MMG, LCP. Methodology: IFB, MMG, LCP. Project administration: MMG, LCP. Supervision: MMG, LCP. Validation: IFB, NR, JCS, BS, OOH, BWL, JLD, DSN, SV, LM, AS, RPT, VR, BS, MMG, LCP. Writing – original draft: IFB, NR, MMG, LCP. Writing – review & editing: IFB, NR, JCS, BS, OOH, BWL, JLD, DSN, SV, LM, AS, RPT, VR, BS, MMG, LCP. Statistics: VR.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Vall d'Hebron University Ethics Committee (PR(AMI)181/2020) and Hong Kong (no number provided) on March 27, 2020, and subsequently validated by each Local Research Ethics Committee at the participating centers. Verbal and written information about the study was provided to the woman by a medical team member during pregnancy, labor, or immediately after childbirth. Written informed consent was obtained from every participant.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Table S1. STROBE Statement-Checklist of items that should be included in reports of cohort studies.
Additional file 2: Table S2.
Conformité Européene (CE) registration number of the reagents used for sample analyses.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Fernández-Buhigas, I., Rayo, N., Silos, J.C. et al. Anti-SARS-CoV-2-specific antibodies in human breast milk following SARS-CoV-2 infection during pregnancy: a prospective cohort study. Int Breastfeed J 19, 5 (2024). https://doi.org/10.1186/s13006-023-00605-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13006-023-00605-w