Abstract
Introduction
Comprehensive information about the genome analysis and its prognostic values of NSCLC patients in Chinese population are still needed.
Patients
A total of 117 Chinese patients with NSCLC were enrolled in this study. Tumor tissues or blood were collected and sequenced by targeted next-generation sequencing of 556 cancer related genes. The associations between clinical outcomes and clinical characteristics, TMB, mutated genes, treatment therapies were analyzed using Kaplan-Meier methods and further evaluated using multivariable Cox proportional hazards regression model.
Results
A total of 899 mutations were identified by targeted NGS. The most frequently mutations included EGFR (47%), TP53 (46%), KRAS (18%), LRP1B (12%) and SPTA1 (10%). Patients with mutant TP53, PREX2, ARID1A, PTPRT and PIK3CG had lower median overall survival (OS) than those patients with wild-type (P = 0.0056, P < 0.001, P < 0.0001, P < 0.0001 and P = 0.036, respectively). Using a multivariate Cox regression model, PREX2 (P < 0.001), ARID1A (P < 0.001) and PIK3CG (P = 0.04) were independent prognostic factors in NSCLC. In the patients received chemotherapy, squamous patients had a significantly longer median OS than adenocarcinoma patients (P = 0.011). In the patients received targeted therapy, adenocarcinoma patients had a significantly longer survival period than squamous patients (P = 0.01).
Conclusions
Our study provided comprehensive genomic alterations in a cohort of Chinese NSCLC. We also identified new prognostic biomarkers, which could provide potential clues for targeted therapies.
Similar content being viewed by others
Introduction
Lung cancer is one of the leading causes of cancer related mortality worldwide, as well as in China [1]. According to 2015 statistics, there were approximately 730,000 new cases of lung cancer in China and more than 430,000 people died from this disease [2, 3]. Lung cancer is divided into non-small-cell lung carcinoma (NSCLC) and small-cell lung carcinoma, with NSCLC accounting for the most of all of cases (approximately 85%) [4]. Despite more and more researches on different treatment strategies, the 5-year overall survival rate of patients with NSCLC is less than 18% [5], which suggests that there is still a need for new targeted therapies in NSCLC. Previous studies have shown differences in the frequency of driver genes among lung cancer patients in different countries, which affect the efficacy of targeted drugs [6,7,8,9]. Thus, it is necessary to study the genomic profiles in Chinese NSCLC patients, which can help identify specific predictive-biomarkers to promote the development of precision medicine for lung cancer treatment and prevention.
Over the last decade, next-generation sequencing (NGS) technology has increasingly used for clinical diagnosis and therapies. Lots of evidences have shown the capability in accurately capturing and identifying multiple genetic alterations of NGS, including single nucleotide variant (SNV), insertions and deletions, copy number variations (CNV) and structure variations (SV), which can significantly reduce sequencing costs, improve accuracy of detection and achieve real-time monitoring progression of tumors, with high sensitivity for detecting extremely low levels of mutation frequency [10, 11]. Many studies have used NGS to analyze variations in genes and tumor mutation burden (TMB) in solid tumors [12,13,14,15]. As a result, several important genes in lung cancer have been identified, such as EGFR, ALK and ROS1. EGFR tyrosine kinase inhibitors (TKIs) erlotinib and gefitinib were the first class of molecularly targeted agents approved by the U.S, Food and Drug Administration (FDA) in 2004, up to today, erlotinib, gefitinib and other EGFR TKIs such as afatinib and osimertinib are all used for the treatment of metastatic NSCLC patients whose tumors have EGFR exon 19 deletions or exon 21 L858R mutations [16,17,18,19,20]. Subsequently, ALK inhibitor crizotinib was the first FDA-approved targeted therapy for the treatment of ALK-positive advanced NSCLC patients in 2011, the second generation ALK TKIs ceritinib, alectinib and ensartinib were all approved for metastatic ALK + NSCLC [21,22,23,24,25,26]. And crizotinib was approved for metastatic ROS1 positive NSCLC in 2016 [24, 27,28,29]. These discoveries have revolutionized treatment of patients whose tumor harbor these genes. Recent years, many studies have explored the prognostic values of the mutated genes which have been considered as potential important therapeutic target in NSCLC, such as MET [30], KMT2D [31] and PIK3CA [32], which has greatly facilitated the discovery of gene-based tumor biomarkers [33, 34]. However, until now, the knowledge of genetic variations and molecular biomarkers in NSCLC are still lacking in the Chinese population. Therefore, it is necessary to explore the genetic mutational landscape and identify prognostic biomarkers of Chinese patients to deeply understand the clinical outcomes and find new treatment options for lung cancer.
In the present study, we established a panel comprised 556 genes to detect somatic mutations in 117 samples from Chinese NSCLC patients. Furthermore, we explored the prognostic values of tumor burden mutation (TMB), clinical characteristics, gene mutations and treatment therapies. Our study aimed to provide a comprehensive genomic profiling of 117 Chinese patients with NSCLC, and provide new prognostic biomarkers to help find new therapeutic targets.
Patients and methods
Patient cohort and DNA extraction
Tumor tissues or blood were obtained from 117 Chinese patients with NSCLC at Shanxi Cancer Hospital between 2019 and 2022. The diagnosis of all the samples in the cohort was performed on the morphology of hematoxylin & eosin staining (HE) by two experienced molecular pathologists and the content of tumor cell (tumor purity) was higher than 50%. 8–10 of 5–10 μm tumor slices of formalin-fixed paraffin-embedded (FFPE) of tumor tissues and 8–10 mL plasma samples from patients were collected for further use [35]. Genomic DNA was isolated from tissues or blood using the DNeasy Blood and Tissue Kit (69,504, QIAGEN, Venlo, Netherlands). DNA content was determined by Agilent 2100 Bioanalyzer (USA). Libraries were constructed if the gDNA amount from the tumor tissue/plasma samples ≥ 200 ng [35]. All patients signed an informed consent before joining the study. Demographic and clinical characteristics were collected from patients. This study was approved by the Ethics Committee of Shanxi Cancer Hospital and performed in accordance with the World Medical Association Declaration of Helsinki (as revised in 2013).
Next-generation sequencing
Next-generation sequencing (NGS) performed on ZhenXinan ctDNA NGS Panel (Tongshu BioTech, Shanghai, China). Sequencing libraries of different components were prepared using the KAPA Hyper Prep kit (KAPA Biosystems) with an optimized manufacturer’s protocol. Enriched libraries were amplified and detected on Illumina Novaseq 6000 platform (Illumina) in accordance with manufacturer’s instructions. The average sequencing depth in tissues is ≥ 1000×; the average sequencing depth in plasma cfDNA is ≥ 7000×. The variant allele frequency (VAF) is ≥ 1% for tissue DNA and ≥ 0.1% for cfDNA from plasma [35]. The sequencing data in the FASTQ format were mapped to the human genome (hg38) using BWA aligner 0.7.10. Local alignment optimization, variant calling and annotation were performed using GATK 3.2 (https://gatk.broadinstitute.org/), MuTect [36] and VarScan [37] respectively. Somatic mutations existing in at least 2 of the results of the 3 software were selected as high confident mutations and to be involved in the further bioinformatics and biostatistical analysis.
Statistical analysis
Oncoplot mutations were plotted by using the MAfTools R package. Mutations in cancer-related driver genes were also analyzed. The clusterProfiler R package was used to visualize the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment results of mutated genes in all the samples [38]. Fisher’s exact test was used to analyze the associations between two groups. Overall survival (OS) was defined as the time from the date of diagnosis to the date of death or the last follow-up visit for patients. The definition of progression-free survival (PFS) was the time from start of treatment to the clinical or radiographic progression, or the end of follow-up. Median OS and PFS were calculated using Kaplan–Meier method and survival curves were compared with log-rank tests. The variables putatively associated with patient survival were analyzed with the log-rank test and Cox proportional hazards model. The statistical software package SPSS 22.0 was used for statistical analysis. All tests were bilateral, with P values < 0.05 indicating significant statistical difference.
Results
Clinicopathological characteristics of NSCLC patients
A total of 117 patients were included in this study. The clinical and pathological information of all patients in the cohort are summarized in Table 1. A total of 72 (61.5%) patients are males, and 60 patients (51.3%) presented a smoking history. The median age of the patients was 61 years (range, 28–78 years). Most of the patients showed metastases to the lungs and/or other sites (78/117, 66.7%). Regarding histological subtypes, lung adenocarcinoma was the most common subtype (96/117, 82.1%), followed by lung squamous cell (15/117, 12.8%). 8 patients (6.8%) were classified as stage I and II, and 109 patients (93.2%) as stage III and IV. Males were especially prevalent in late-stage (III-IV) group. In all patients, 44 (37.6%) received targeted therapy. A total of 55 patients (47.0%) received chemotherapy; of these, 36 (65.5%) underwent chemotherapy alone, 14 (25.5%) underwent chemotherapy and immunotherapy, while the remaining patients received chemotherapy and antivascular therapy.
Somatic mutation landscape in NSCLC patients
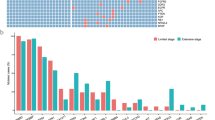
NGS results showed that different somatic mutations occur in all genes, including amplification and fusion, chromosomal structural variation, insertion and deletion, and point mutation. In the 117 samples tested, a total of 899 mutations were identified, and the dominant mutation type was missense mutation (693/899, 77.1%; Fig. 1A). Variant spectrum showed that C > T had the highest mutation percentage in all patients (Fig. 1B). The distribution of top 30 mutations is shown in Fig. 1C. The top 10 most frequently mutated genes in all the samples were EGFR (55/117, 47%), TP53 (54/117, 46%), KRAS (21/117, 18%), LRP1B (14/117, 12%), SPTA1 (12/117, 10%), KEAP1 (11/117, 9%), KMT2D (11/117, 9%), KMT2C (10/117, 9%), GNAS (9/117, 8%) and PIK3CA (9/117, 8%). The number of patients with EGFR and TP53 mutations was basically the same (47% and 46%, respectively), of these patients, 26 had EGFR and TP53 co-mutations (47.3% of patients with EGFR and 48.1% of patients with TP53). Then, we further subdivided top 10 gene mutations into exons, the results are shown in supplementary Fig. 1.
CNV analysis showed that 394 genes had copy number amplification (supplementary Fig. 2A). Among these, EVI2A (82/117, 70.09%), EVI2B (82/117, 70.09%), NF1 (82/117, 70.09%), OMG (82/117, 70.09%) and BRAC1 (72/117, 61.54%) were the genes with highest amplification. A total of 830 genes had copy number deletion (supplementary Fig. 2B). The genes with the highest copy number deletions were KAT6A (76/117, 64.96%) and RASA1 (72/117, 61.54%).
GO and KEGG enrichment analysis
Next, we performed GO and KEGG enrichment analysis on top 30 mutated genes. GO enrichment analysis is divided into molecular function analysis (MF), biological process analysis (BP) and cellular component analysis (CC). As shown in supplementary Fig. 3A, the results showed that the top five biological process are response to radiation, response to light stimulus, phosphatidylinositol 3-kinase signaling, somitogenesis and cellular response to heat. At the molecular function level, these genes are significantly enriched in RNA polymerase II-specific DNA-binding transcription factor binding, DNA-binding transcription factor binding, transcription coactivator activity, transcription coregulator activity and p53 binding (supplementary Fig. 3B). The top five cellular components are extrinsic component of membrane, cytoplasmic side of plasma membrane, extrinsic component of plasma membrane, npBAF complex and Wnt signalosome (supplementary Fig. 3C). KEGG analysis revealed that these genes are mainly concentrated in human papillomavirus infection, microRNAs in cancer, hepatocellular carcinoma, phospholipase D signaling pathway, colorectal cancer and other signaling pathways (supplementary Fig. 3D).
Relationships between TMB and clinicopathological characteristics
Tumor mutation burden (TMB) has been regarded as a biomarker to predict immunotherapy response in clinical oncology, including NSCLC. We performed comparative analysis of the clinical characteristics to explore the association between TMB and Chinese NSCLC patients. Somatic mutations were obtained after removing splicing mutations, and the valid TMB values were obtained after dividing the number of somatic mutations by the size of the panel. Wilcoxon Signed Rank Test was used to analyze the differences of TMB in clinical characteristics. There were significant differences in TMB between smokers and nonsmokers (median, 4.65 Mutations/Mb and 2.9 Mutations/Mb, respectively; P = 0.014; Fig. 2A). The median TMB of squamous is 2.97 times that of adenocarcinoma (median, 8.6 Mutations/Mb and 2.9 Mutations/Mb, respectively). Significant difference was observed between these two tumor types (P = 0.0018; Fig. 2B). There were no significant differences in TMB between younger patients and elder patients (range, 0.7–84.9 Mutations/Mb; median, 3.6 Mutations/Mb; P = 0.30), males and females (median, 4.3 Mutations/Mb and 2.9 Mutations/Mb, respectively; P = 0.10), patients with metastasis and without metastasis (2.9 Mutations/Mb and 5.0 Mutations/Mb, respectively; P = 0.21), and early-stage and late-stage patients (median, 1.75 Mutations/Mb and 3.6 Mutations/Mb, respectively; P = 0.065).
Relationships between mutations and clinicopathological features
We compared clinicopathological characteristics of the cohort of patients with the top 30 mutated genes (supplementary Table 1). The results showed that the EGFR mutation rate was significantly higher in females (30/45, 66.7%) than in males (25/72, 34.7%; P = 0.0011), in never smokers (35/57, 61.4%) than in smokers (20/60, 33.3%; P = 0.0030), in patients with metastasis (44/78, 56.4%) than in those without metastasis (11/39, 28.2%; P = 0.0057), and in patients with adenocarcinoma (51/96, 53.1%) than in those with squamous (3/15, 20%; P = 0.025). In contrast, the KRAS mutation rate was higher in males (17/72, 23.6%) than in females (4/45, 8.9%; P = 0.050). In addition, the KRAS mutation rate was higher in smokers (14/60, 23.3%) than in never smokers (7/57, 12.3%; P = 0.15), in patients with squamous (3/15, 20%) than in those with adenocarcinoma (18/96, 18.8%; P = 1), and in patients with early-stage (2/8, 25%) than in those in late-stage (19/109, 17.4%; P = 0.63). However, there were no significant differences between KRAS mutations with histological subtypes, smoking status and the stage. KEAP1 mutations were only occurred in males (11/72, 15.3%), which is significantly more frequent than in females (0%; P = 0.0064); and the KEAP1 mutation rate was significantly higher in smokers (10/60, 16.7%) than in never smokers (1/57, 1.8%; P = 0.0085), and in more prevalent among patients in early stage (3/8, 37.5%) than in those in late stage (8/109, 7.3%; P = 0.027). Besides, PREX2 mutations were only occurred in males (7/72, 9.7%), which is significantly more frequent than in females (0%; P = 0.042); PREX2 mutations were only occurred in smokers (7/60, 12%), which was significantly higher in never smokers (0%; P = 0.013), and PREX2 mutations were significantly higher in patients with squamous (4/15, 26.7%) than those with adenocarcinoma (2/96, 2.1%; P = 0.0029). In addition, the NSD2 mutation rate was significantly higher in elder patients (3/17, 17.6%) than in younger patients (2/100, 2%; P = 0.022). The KMT2D, GNAS, EP300, GRM3, and PIK3CG mutations were all significantly higher in patients with squamous than those with adenocarcinoma (P = 0.0066, P = 0.019, P = 0.0075, P = 0.017, and P = 0.0075, respectively). The KMT2C mutation rate was significantly more frequent in patients without metastasis (7/39, 17.9%) than those with metastasis (3/78, 3.8%; P = 0.015). No association was found between TP53, LRP1B, SPTA1 and clinicopathological characteristics.
Survival analysis
The relationship between TMB and prognosis
Nonsynonymous TMB was calculated and analysis was performed to assess the effect of TMB on prognosis. We used the median TMB (3.6 Mutations/Mb) as the cut-off value to divide TMB into two groups, TMB ≤ 3.6 Mutations/Mb as TMB-1 group and TMB > 3.6 Mutations/Mb as TMB-2 group. Univariate analysis showed that OS was significantly longer in patients with lower TMB than those patients with higher TMB (64 months vs. 36 months, P = 0.028) (Fig. 3). However, multivariate analysis showed that TMB was not an independent prognostic factor (P = 0.49; supplementary Table 1).
Effects of clinicopathological features on overall survival
The average follow-up was for 25.7 months, and the median OS was 18 months (range, 1-127 months). As shown in supplementary Fig. 4, patients with squamous has a significantly longer survival period than those patients with adenocarcinoma and other lung cancer types (median survival time, 62 months vs. 39 months vs. 11 months; P < 0.001, by log-rank test), and there were no significantly correlations between other clinicopathological features and OS (supplementary Table 1).
Effects of genomic alterations on overall survival
Patients with TP53 (median survival time, 27 months vs. 62 months; P = 0.0056), PREX2 (median survival time, 8 months vs. 42 months; P < 0.001), ARID1A (median survival time, 10 months vs. 44 months; P < 0.0001), PTPRT (median survival time, 8 months vs. 44 months; P < 0.0001) and PIK3CG (median survival time, 24 months vs. 42 months; P = 0.036) mutations survived for a significantly shorter period than those without the mutations, suggesting that these mutations predict positive factors for NSCLC prognosis (Fig. 4A-E). In addition, we further validated these five prognosis-related genes in the cohort of lung adenocarcinoma and lung squamous from the Cancer Genome Atlas (TCGA) data set. In the squamous patients from the TCGA database, the TP53 mutation was significantly associated with prognosis (Fig. 4F). Other four genes had no significantly associations with prognosis in lung adenocarcinoma or/and lung squamous. In contrast, patients with KRAS mutations survived longer than those without mutations (median survival time, 64 months vs. 39 months), whereas there was no significant difference (P = 0.45).
Overall survival analysis of the mutated Genes. (A) Kaplan-Meier curve of TP53 mutant. (B) Kaplan-Meier curve of PREX2 mutant. (C) Kaplan-Meier curve of ARID1A mutant. (D) Kaplan-Meier curve of PTPRT mutant. (E) Kaplan-Meier curve of PIK3CG mutant. (F) TCGA validation between TP53 mutant and overall survival in lung squamous cohort
Multivariate Cox proportional hazard analyses of clinicopathological features showed that smoking (P = 0.02) and the histological subtype (P < 0.001) were independent prognostic factors for overall survival (Fig. 5). In addition, the results demonstrated that the mutated gene PREX2 (P < 0.001), ARID1A (P < 0.001) and PIK3CG (P = 0.04) were independent prognostic factors (Fig. 5). However, TP53 and PTPRT were not independent prognostic factors (Fig. 5; supplementary Table 1).
Effects of treatments on progression-free survival
Univariate analysis revealed that the patients underwent targeted therapy (PFS group 1) had a significantly prolonged PFS as compared to those with chemotherapy (PFS group 2) (15 months vs. 6 months; P = 0.0031; Fig. 6A). In the patients who underwent chemotherapies, squamous patients had a significantly longer PFS than adenocarcinoma patients (12 months vs. 7 months; P = 0.011; Fig. 6B). In the patients who underwent targeted therapies, adenocarcinoma patients had a significantly longer PFS than squamous patients (15 months vs. 3 months; P = 0.01; Fig. 6C).
Progression-free survival analysis of treatments. (A) Kaplan-Meier curve of the patients received chemotherapies and targeted therapies. (B) Kaplan-Meier curve of different pathological types of the patients who received chemotherapies. (C) Kaplan-Meier curve of different pathological types of the patients who received targeted therapies
Discussion
In this study, we used a targeted NGS panel to perform comprehensive genomic profiling on tumor specimens from 117 Chinese NSCLC patients. We identified 899 somatic mutations by 556-genes panel. The most frequently mutated genes were obtained. Notably, EGFR and TP53 were co-mutated in most sample. 394 genes and 830 genes showed copy number amplification and copy number deletion, respectively. The top 30 mutated genes were specifically enriched in the cancer-associated pathways. Then, we analyzed the association between TMB and clinical characteristics. There were significant differences in TMB between the smoking status and pathological subtypes. Similar results were shown in previous studies. In addition, we explored the relationship between clinical characteristics and driver alterations. Finally, we performed survival analysis, including the relationships between TMB and overall survival, mutated genes and overall survival, treatments and progression-free survival. We have identified genes whose mutations are significantly associated with survival in NSCLC and further confirmed these mutated genes in the TCGA database.
Based on the targeted NGS, potentially genetic alterations were detected in 87.18% of Chinese NSCLC patients in the present cohort, which was similar to the previously study [39]. We determined the most frequently mutated genes in the NSCLC patients, including EGFR, TP53, KRAS and LRP1B. These mutated genes have been reported in NSCLC previously [40,41,42]. The distribution of genetic alterations in the Chinese patients showed differences with those in the Caucasian patients. Compared with a study on the American population, their results showed that the most frequently mutated gene is KRAS, followed by EGFR [43]. Interestingly, in a study of Chinese NSCLC, the highest mutated genes were TP53, EGFR and CREBBP [3]. However, our results showed certainly consistency with those in some studies. In a study on the Chinese patients, the most frequently mutated genes were EGFR, TP53, KRAS and ALK [15]. In a study on the East Asian patients, the most frequently mutated genes were EGFR, TP53, ALK and KRAS [44]. EGFR was the most frequently mutated gene in both this study and our study, accounting for about 47% in all the samples.
Asian population have unique clinical features and tumor histology, exhibiting different prevalence of oncogenic mutations [45]. Association between driver genes and clinical features were consistent with prior reports. For example, EGFR mutations were more common in females, never smokers and in patients with adenocarcinoma [41, 46]. In addition, it was more common in early-stage patients than in those with late-stage, which was not found in the study of Liu’s et al [44]. The KRAS mutations were also more common in males than females [47]. Besides, we found other two driver alterations that associated with clinical characteristics. KEAP1 mutations were only occurred in males, enriched in smokers than never smokers, and in early-stage patients than in late-stage patients. According to the previous studies, approximately 20–30% of lung adenocarcinoma harbor the KEAP1 mutations which correlate with poor prognosis [42, 48, 49]. Romero et al’ s study provided the evidence for stratification of patients with KEAP1 mutation as possible responders to targeted SLC33A1 inhibition [50]. Saleh et al’ s study showed that KEAP1 mutations accounted for about 17% of NSCLC patients, and KEAP1 mutation was significantly associated with higher age, male sex, adenocarcinoma differentiation and advanced stage, and also represented an independently negative prognostic biomarker [51]. PREX2 mutations only occurred in males and smokers, and PREX2 mutations were significantly higher in patients with squamous than those with adenocarcinoma. In Wang et al’ s study, PREX2 mutations were found to be highly frequent in patients with multiple primary lung adenocarcinoma than those with single primary lung adenocarcinoma (P = 0.0456).
In terms of clinical outcomes, patients with low TMB and adenocarcinoma were associated with better overall survival. Patients with TP53, PREX2, ARID1A, PTPRT and PIK3CG had worse prognosis. TP53 was commonly considered as a prognostic factor with poor prognosis of lung cancer in many studies [52, 53]. However, opposite or neural results were also reported [54, 55]. We found that patients with TP53 mutations had better survival than those with wild type in TCGA cohort of lung squamous patients, which is contrast with our results. Since most of the patients are lung adenocarcinoma in our study, which could explain these results. PREX2 mutations were frequently occurred in melanoma [56] and were considered as new candidate drivers of pancreatic carcinogenesis [57]. Previous studies have found that ARID1A mutations are likely to be related to the higher immune infiltrates in endometrial cancer, stomach cancer and colon cancer [58]. The study of Zhu et al. suggested that ARID1A mutations were related to the good prognosis of immune checkpoint inhibitors (ICI) therapy based on the pan-cancer population [59]. The study of Chen et al. suggested that PTPRT mutation is associated with poor progression-free survival in pan-cancer and NSCLC [60]. Contrary to our results, the study of Zhang et al. showed that melanoma patients with PTPRT mutations harbored a significantly elevated ICI response rate and a prolonged survival outcome, and in the NSCLC cohort, the favorable response and immunotherapy survival were also observed in PTPRT-mutated patients [61]. The study of the association between PIK3CG and prognosis is still lacked. In a study of Wu et al., PIK3CG considered as favorable prognostic factor [62]. In multivariate analysis accounting for gender, smoking status, pathology, stage, TMB and the mutated genes with potential prognostic values, ARID1A, PREX2 and PIK3CG retained the significant independent prognostic factors. However, for TP53 and PTPRT, the survival impact of mutations as independent variations did not remain significant. There were also no significant differences in overall survival between the patients with EGFR, KRAS mutations and those without these genes in this study. However, recent advances of studying KRAS biology have led the discovery of KRAS p.G12C-specific inhibitors which show the great promising clinical results [63]. In a phase I trial (NCT03600883), sotorasib showed anticancer activity in advanced NSCLC patients who harbored KRAS p.G12C mutation with a median follow-up of 11.7 months [64]. Furthermore, in a phase II trial (NCT03785249), adagrasib also showed clinical efficacy without new safety signals in NSCLC patients with advanced or metastatic tumors harboring KRAS p.G12C mutation [65]. As the KRAS p.G12C mutation is the most frequent variation among all KRAS mutations in NSCLC, presenting in 10–13% of patients with lung adenocarcinomas, the potentially effective therapies of KRAS p.G12C mutant NSCLC may help to optimize clinical treatment outcomes in this important and common subtype of lung cancers [63, 66].
Targeted therapy has led an important impact in lung cancer management and clinical outcomes over the past two decades. In the present cohort, patients received targeted therapy had a better PFS than those received chemotherapies. In addition, we found that in the patients that received chemotherapies, squamous patients had a better PFS than adenocarcinoma patients, and in the patients that received targeted therapies, adenocarcinoma patients had a better PFS than squamous patients. Adenocarcinoma represents 50–60% of total NSCLC cases, whereas squamous cell carcinoma represents 20–30% [67]. Most prevalent actionable mutations in adenocarcinoma include KRAS, EGFR, ALK, RET, ROS1, BRAF, HER2, MET [42, 68,69,70]. However, most of these mutations are rare in squamous NSCLC, so there were much more adenocarcinoma patients who could receive targeted therapies than squamous patients, which could explain the above results.
Conclusion
In this study, we assessed the mutation profiles, copy number variations and tumor mutation burden of 117 NSCLC patients. There were significant differences in TMB between smokers and nonsmokers, and between patients with adenocarcinoma and those with squamous. However, TMB was not associated with other clinical characteristics. Patients with low TMB had significantly longer survival period than those with high TMB. Our discovery suggested that mutant TP53, PREX2, ARID1A, PTPRT and PIK3CG considered as prognostic factors with poor prognosis of NSCLC. Adenocarcinoma patients had a significantly longer survival period than squamous patients who received targeted therapies. Our findings indicated that targeted NSG panel is a good tool for tumor molecular characterization. In addition, our results were expected to provide implications for cancer translational research and management of NSCLC.
References
Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7–33.
Zhang S, Sun K, Zheng R, et al. Cancer incidence and mortality in China, 2015. Journal of the National Cancer Center 2021;1:2–11.
Zheng S, Wang X, Fu Y, et al. Targeted next-generation sequencing for cancer-associated gene mutation and copy number detection in 206 patients with non-small-cell lung cancer. Bioengineered 2021;12:791–802.
Wang L, Zhao D, Qin K, et al. Effect and biomarker of Nivolumab for non-small-cell lung cancer. Biomed Pharmacother 2019;117:109199.
Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res 2016;5:288–300.
Blons H, Garinet S, Laurent-Puig P, et al. Molecular markers and prediction of response to immunotherapy in non-small cell lung cancer, an update. J Thorac Dis 2019;11:S25-s36.
Cao J, Chen L, Li H, et al. An accurate and comprehensive clinical sequencing assay for Cancer targeted and Immunotherapies. Oncologist 2019;24:e1294-e1302.
Seo JS, Ju YS, Lee WC, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res 2012;22:2109–19.
Teixeira VH, Pipinikas CP, Pennycuick A, et al. Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat Med 2019;25:517–525.
Nikiforova MN, Wald AI, Roy S, et al. Targeted next-generation sequencing panel (ThyroSeq) for detection of mutations in thyroid cancer. J Clin Endocrinol Metab 2013;98:E1852-60.
Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics 2019;13:34.
Han SW, Kim HP, Shin JY, et al. Targeted sequencing of cancer-related genes in colorectal cancer using next-generation sequencing. PLoS One 2013;8:e64271.
Cai H, Jing C, Chang X, et al. Mutational landscape of gastric cancer and clinical application of genomic profiling based on target next-generation sequencing. J Transl Med 2019;17:189.
Chang YS, Tu SJ, Chen YC, et al. Mutation profile of non-small cell lung cancer revealed by next generation sequencing. Respir Res 2021;22:3.
Zhao S, Zhang Z, Zhan J, et al. Utility of comprehensive genomic profiling in directing treatment and improving patient outcomes in advanced non-small cell lung cancer. BMC Med 2021;19:223.
Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129–39.
Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497–500.
Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306–11.
Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 2005;353:123–32.
Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall survival with Osimertinib in untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med 2020;382:41–50.
Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693–703.
Shaw AT, Kim DW, Mehra R, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med 2014;370:1189–97.
Seto T, Kiura K, Nishio M, et al. CH5424802 (RO5424802) for patients with ALK-rearranged advanced non-small-cell lung cancer (AF-001JP study): a single-arm, open-label, phase 1–2 study. Lancet Oncol 2013;14:590–8.
Chen R, Manochakian R, James L, et al. Emerging therapeutic agents for advanced non-small cell lung cancer. J Hematol Oncol 2020;13:58.
Shaw AT, Kim TM, Crinò L, et al. Ceritinib versus chemotherapy in patients with ALK-rearranged non-small-cell lung cancer previously given chemotherapy and crizotinib (ASCEND-5): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol 2017;18:874–886.
Horn L, Infante JR, Reckamp KL, et al. Ensartinib (X-396) in ALK-Positive Non-Small Cell Lung Cancer: results from a first-in-human phase I/II, Multicenter Study. Clin Cancer Res 2018;24:2771–2779.
Davies KD, Le AT, Theodoro MF, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res 2012;18:4570–9.
Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963–71.
Shaw AT, Riely GJ, Bang YJ, et al. Crizotinib in ROS1-rearranged advanced non-small-cell lung cancer (NSCLC): updated results, including overall survival, from PROFILE 1001. Ann Oncol 2019;30:1121–1126.
Awad MM, Oxnard GR, Jackman DM, et al. MET exon 14 mutations in Non-Small-Cell Lung Cancer are Associated with Advanced Age and Stage-Dependent MET genomic amplification and c-Met overexpression. J Clin Oncol 2016;34:721–30.
Ardeshir-Larijani F, Bhateja P, Lipka MB, et al. KMT2D mutation is Associated with Poor Prognosis in Non-Small-Cell Lung Cancer. Clin Lung Cancer 2018;19:e489-e501.
McGowan M, Hoven AS, Lund-Iversen M, et al. PIK3CA mutations as prognostic factor in squamous cell lung carcinoma. Lung Cancer 2017;103:52–57.
Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med 2011;17:297–303.
Ansorge WJ. Next-generation DNA sequencing techniques. N Biotechnol 2009;25:195–203.
Xie J, Yao W, Chen L, et al. Plasma ctDNA increases tissue NGS-based detection of therapeutically targetable mutations in lung cancers. BMC Cancer 2023;23:294.
McKenna A, Hanna M, Banks E, et al. The genome analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303.
Koboldt DC, Chen K, Wylie T, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 2009;25:2283–5.
Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7.
Jordan EJ, Kim HR, Arcila ME, et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov 2017;7:596–609.
Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet 2013;382:720–31.
Singal G, Miller PG, Agarwala V, et al. Association of patient characteristics and Tumor Genomics with Clinical Outcomes among patients with Non-Small Cell Lung Cancer using a clinicogenomic database. Jama 2019;321:1391–1399.
Comprehensive molecular Profiling of lung adenocarcinoma. Nature 2014;511:543–50.
Hagemann IS, Devarakonda S, Lockwood CM, et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer 2015;121:631–9.
Liu L, Liu J, Shao D, et al. Comprehensive genomic profiling of lung cancer using a validated panel to explore therapeutic targets in east asian patients. Cancer Sci 2017;108:2487–2494.
Toh CK, Wong EH, Lim WT, et al. The impact of smoking status on the behavior and survival outcome of patients with advanced non-small cell lung cancer: a retrospective analysis. Chest 2004;126:1750–6.
La Fleur L, Falk-Sörqvist E, Smeds P, et al. Mutation patterns in a population-based non-small cell lung cancer cohort and prognostic impact of concomitant mutations in KRAS and TP53 or STK11. Lung Cancer 2019;130:50–58.
Kosaka T, Yatabe Y, Onozato R, et al. Prognostic implication of EGFR, KRAS, and TP53 gene mutations in a large cohort of japanese patients with surgically treated lung adenocarcinoma. J Thorac Oncol 2009;4:22–9.
Shen R, Martin A, Ni A, et al. Harnessing clinical sequencing data for survival stratification of patients with metastatic lung adenocarcinomas. JCO Precis Oncol 2019;3.
Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev 2013;27:2179–91.
Romero R, Sanchez-Rivera FJ, Westcott PMK, et al. Keap1 mutation renders lung adenocarcinomas dependent on Slc33a1. Nat Cancer 2020;1:589–602.
Saleh MM, Scheffler M, Merkelbach-Bruse S, et al. Comprehensive analysis of TP53 and KEAP1 mutations and their impact on Survival in localized- and Advanced-Stage NSCLC. Journal of Thoracic Oncology 2022;17:76–88.
Jiao XD, Qin BD, You P, et al. The prognostic value of TP53 and its correlation with EGFR mutation in advanced non-small cell lung cancer, an analysis based on cBioPortal data base. Lung Cancer 2018;123:70–75.
Molina-Vila MA, Bertran-Alamillo J, Gascó A, et al. Nondisruptive p53 mutations are associated with shorter survival in patients with advanced non-small cell lung cancer. Clin Cancer Res 2014;20:4647–59.
Labbé C, Cabanero M, Korpanty GJ, et al. Prognostic and predictive effects of TP53 co-mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC). Lung Cancer 2017;111:23–29.
Nishio M, Koshikawa T, Kuroishi T, et al. Prognostic significance of abnormal p53 accumulation in primary, resected non-small-cell lung cancers. J Clin Oncol 1996;14:497–502.
Berger MF, Hodis E, Heffernan TP, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 2012;485:502–6.
Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495–501.
Jiang T, Chen X, Su C, et al. Pan-cancer analysis of ARID1A alterations as biomarkers for Immunotherapy Outcomes. J Cancer 2020;11:776–780.
Zhu Y, Yan C, Wang X, et al. Pan-cancer analysis of ARID family members as novel biomarkers for immune checkpoint inhibitor therapy. Cancer Biol Ther 2022;23:104–111.
Chen C, Liu H, Xu Q, et al. Association of PTPRT mutations with Cancer metastasis in multiple Cancer types. Biomed Res Int 2022;2022:9386477.
Zhang W, Shi F, Kong Y, et al. Association of PTPRT mutations with immune checkpoint inhibitors response and outcome in melanoma and non-small cell lung cancer. Cancer Med 2022;11:676–691.
Wu W, Jia L, Zhang Y, et al. Exploration of the prognostic signature reflecting tumor microenvironment of lung adenocarcinoma based on immunologically relevant genes. Bioengineered 2021;12:7417–7431.
Veluswamy R, Mack PC, Houldsworth J, et al. KRAS G12C-Mutant Non-Small Cell Lung Cancer: Biology, Developmental therapeutics, and Molecular Testing. J Mol Diagn 2021;23:507–520.
Hong DS, Fakih MG, Strickler JH, et al. KRAS(G12C) inhibition with Sotorasib in Advanced Solid Tumors. N Engl J Med 2020;383:1207–1217.
Janne PA, Riely GJ, Gadgeel SM, et al. Adagrasib in Non-Small-Cell Lung Cancer harboring a KRAS(G12C) mutation. N Engl J Med 2022;387:120–131.
Arbour KC, Jordan E, Kim HR, et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-Mutant Non-Small Cell Lung Cancer. Clin Cancer Res 2018;24:334–340.
Wang M, Herbst RS, Boshoff C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat Med 2021;27:1345–1356.
Bar-Sagi D, Knelson EH, Sequist LV. A bright future for KRAS inhibitors. Nat Cancer 2020;1:25–27.
Li T, Kung HJ, Mack PC, et al. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Oncol 2013;31:1039–49.
Drilon A, Cappuzzo F, Ou SI, et al. Targeting MET in Lung Cancer: will expectations finally be MET? J Thorac Oncol 2017;12:15–26.
Acknowledgements
We thank Tongshu Biotechnology Co., Ltd. for technical support.
Author information
Authors and Affiliations
Contributions
Fangfang Shen conceived and designed the study. Fangfang Shen, Wei Guo and Xia Song collected data. Fangfang Shen, Wei Guo, Xia Song and Bei Wang analyzed and interpreted data. All authors participated in manuscript writing and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All the methods involved in the study were in accordance with the Helsinki Declaration and were approved by the ethics committees of Affiliated Cancer Hospital of Shanxi Medical, and informed consent has been obtained from all individuals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
13000_2023_1349_MOESM2_ESM.jpg
Supplementary Fig. 2: Copy number variations of the top 30 mutated genes. (A) Distribution of the copy number amplification. (B) Distribution of the copy number deletion.
13000_2023_1349_MOESM3_ESM.jpg
Supplementary Fig. 3: Go and KEGG enrichment analysis of the top 30 mutated genes. (A) Biological process analysis. (B) Molecular function analysis. (C) Cellular component analysis. (D) KEGG enrichment analysis.
13000_2023_1349_MOESM5_ESM.doc
Supplementary Table 1: Multivariate Cox proportional hazard analyses of clinicopathological factors for OS in NSCLC cohort.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Shen, F., Guo, W., Song, X. et al. Molecular profiling and prognostic biomarkers in chinese non-small cell lung cancer cohort. Diagn Pathol 18, 71 (2023). https://doi.org/10.1186/s13000-023-01349-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13000-023-01349-1