Abstract
Co-infections have a key role in virus transmission in wild reservoir hosts. We investigated the simultaneous presence of astroviruses, coronaviruses, and paramyxoviruses in bats from Madagascar, Mayotte, Mozambique, and Reunion Island. A total of 871 samples from 28 bat species representing 8 families were tested by polymerase chain reactions (PCRs) targeting the RNA-dependent RNA-polymerase genes. Overall, 2.4% of bats tested positive for the presence of at least two viruses, only on Madagascar and in Mozambique. Significant variation in the proportion of co-infections was detected among bat species, and some combinations of co-infection were more common than others. Our findings support that co-infections of the three targeted viruses occur in bats in the western Indian Ocean region, although further studies are needed to assess their epidemiological consequences.
Similar content being viewed by others
Co-infection (sometimes written as coinfection) can be defined as the simultaneous infection by at least two genetically different infectious agents in the same host [1,2,3,4,5]. It can affect both host fitness and disease transmission dynamics, therefore playing a critical role in the epidemiology of infectious agents [4,5,6,7]. In bats, although many studies have focused on the detection of emerging viruses such as astroviruses (AstVs), coronaviruses (CoVs), and paramyxoviruses (PMVs), limited data is available regarding co-infection patterns and its potential effect on host fitness and disease transmission dynamics [8,9,10,11,12,13].
In previous studies, we have investigated the presence of either AstVs, CoVs or PMVs in bats in the western Indian Ocean (WIO) region [14,15,16,17,18,19,20,21,22,23]. Here, we conducted a meta-analysis of published data [19,20,21, 23] and performed additional molecular screening in order to obtain a final dataset of 871 bat samples tested for AstVs, CoVs, and PMVs from 28 species representing 8 bat families (see Additional file 1 and Additional file 2).
Biological material was collected on Madagascar, in Mozambique, on Mayotte, and on Reunion Island as part of previous investigations on infectious agents circulation in bats (details relating to the collection of biological material are available in [23]). The list of samples included in this study (e.g. bat species, location, date, type of samples) is provided in the Additional file 1. All samples were previously tested for the presence of CoV [23]; some of them were also tested for the presence of AstV (516 samples; [20, 21]) and PMV (167 samples; [19]). Additional assays were thus performed for the detection of the AstV (355 samples) and PMV (704 samples) RNA-dependent RNA-polymerase (RdRp) genes. Molecular detection was performed using semi-nested polymerase chain reactions (PCRs), as previously described [16, 17, 19,20,21, 24, 25]. PCR products were visualized on 2% agarose gels stained with 2% Gelred (Biotium, Hayward, CA, USA). Pearson Chi square tests were conducted to examine the effect of the roost sites (i.e. cave, building, tree), host species, sex, and sampling location (i.e. country or island), on virus detection, and to investigate potential associations between AstV, CoV, and PMV. Analyses were conducted with R, version 4.0.5 [26].
PCR products of expected size were submitted for direct Sanger sequencing (Genoscreen, Lille, France). Nucleotide sequences were aligned to generate consensus sequences, and were edited manually using ChromasLite 2.6.5 (Technelysium Pty, South Brisbane, Australia). The 33 partial AstV sequences and 13 partial PMV sequences generated in this study were deposited in GenBank respectively under the accession numbers MZ614404 to MZ614436 and MZ614437 to MZ614449. Genetic diversity was explored with pairwise distance values obtained from phangorn package in R, version 2.6.3 [27]. Sequences were compared to reference sequences in NCBI GenBank using the Basic Local Alignment Search Tool (BLAST) with the standard nucleotide BLAST (BLASTn) algorithm (BLAST was performed on August 18th, 2021) [28, 29]. Then, AstV and PMV sequences generated in this study were respectively aligned with 105 and 74 reference partial nucleotide sequences, using CLC Sequence Viewer version 7.6.1 (CLC Bio, Aarhus, Denmark). Phylogenetic trees were generated by maximum-likelihood using PhyML software 3.1 [30], with a GTR evolutionary model, and 1000 bootstrap replicates.
One hundred and forty-two samples tested positive for AstV (mean detection rate ± 95% confidence interval: 16.3% ± 2.5%) (Additional file 2). Positive samples were detected only in Mozambique (20.2% ± 3.6%) and on Madagascar (18.6% ± 3.5%), without significant variation between these two locations (χ2 = 0.3, df = 1, P > 0.5). Significant differences in AstV detection were observed between roost types (χ2 = 6.2, df = 1, P < 0.05), between species (χ2 = 311.8, df = 24, P < 0.001) and between males (20.9% ± 3.8%) and females (11.6% ± 3.0%) (χ2 = 5.9, df = 1, P < 0.05). AstV prevalence was significantly higher in bats species roosting in caves (27.5% ± 3.9%) than in buildings (2.5% ± 2.0%). On Madagascar, a high detection rate was found in species of the genus Miniopterus as compared to the other taxa (χ2 = 162.4, df = 1, P < 0.05), and especially in Miniopterus manavi (88.9% ± 14.5%), Miniopterus gleni (86.7% ± 17.2%), and Miniopterus sororculus (71.4% ± 33.5%). The highest AstV detection rate in Mozambique was observed in Triaenops afer (68.6% ± 12.7%).
A total of 32 samples tested positive for PMV (3.7% ± 1.3%) (Additional file 2). Positive samples were detected only on Madagascar (5.2% ± 1.3%) and in Mozambique (2.7% ± 1.1%), without significant difference between locations (χ2 = 2.5, df = 1, P > 0.5). Significant differences were detected between males (5.4% ± 2.1%) and females (1.9% ± 1.3%) (χ2 = 7.6, df = 1, P < 0.05), and between roost sites (χ2 = 7.3, df = 2, P < 0.01). Bats living in caves were more frequently positive (5.7% ± 2.0%) than those living in buildings (1.7% ± 1.6%). The proportion of positive bats was also significantly different between species on Madagascar (χ2 = 91.9, df = 27, P < 0.001), particularly in Triaenops menamena (42.4% ± 16.9%). In Mozambique, differences between species were not statistically significant (χ2 = 1.3, df = 3, P > 0.05).
The detection of CoV was conducted as part of a previous study [23]. Briefly, 82 of the 871 samples were positive for CoV (9.4% ± 1.9%), with a higher prevalence in Mozambique (20.5% ± 2.7%) (χ2 = 50.4, df = 3, P < 0.001) and no difference between males and females (χ2 = 2.7, df = 1, P > 0.05) (Additional file 2). Positive samples were detected only in bats roosting in caves (10.7% ± 2.6%) and buildings (8.0% ± 3.1%), without statistical difference between these two roost sites (χ2 = 1.7, df = 1, P > 0.05). However, a significant variation was detected among bat species (χ2 = 125.7, df = 27, P < 0.001). In Mozambique, the highest prevalence was detected in the cave roosting Rhinolophus lobatus (66.7% ± 30.8%). No difference was observed among the species that tested positive on Madagascar (χ2 = 2.3, df = 3, P > 0.05).
The overall proportion of positive bats detected for either AstVs or PMVs was consistent with previous studies performed in the WIO region, and in other tropical regions [31,32,33,34]. Interestingly, higher detection rates were found for both viruses in bats using caves as day-roost sites, suggesting that cave-roosting behavior maybe favorable for horizontal transmission between bats [35]. Differences between locations, sex, and bat species may be explained by a range of factors. For example, seasonality has been identified as a major driver of the infection dynamics of many pathogens, affecting both host susceptibility and transmission [17, 36,37,38,39]. Important seasonal variation in the prevalence of infected animals can depend on the period the samples were collected, and, in turn, can lead to misrepresentative conclusions regarding the level of bat exposure to viruses, in particular in cross-sectional studies. Longitudinal studies in wild animals are thus important to precisely assess prevalence of infected animals and its temporal variation [17, 37, 40].
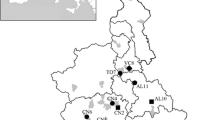
Twenty-one of the 871 samples tested positive for more than one virus (2.4% ± 1.0%). These co-infections were detected only in Mozambique (5.0% ± 2.7%) and on Madagascar (1.7% ± 1.1%), with significant variation among these locations (χ2 = 7.0, df = 1, P < 0.01) (Fig. 1, Additional file 3). In both location, co-infections were detected only in bats roosting in cave (Madagascar: χ2 = 6.4, df = 1, P < 0.05; Mozambique: χ2 = 1.9, df = 1, P > 0.05), and no significant variation was observed between males and females (Madagascar: χ2 = 3.6, df = 1, P > 0.05; Mozambique: χ2 = 3.3, df = 1, P > 0.05). Globally, co-infections involving AstV-CoV (1.4% ± 0.8%) and AstV-PMV (1.1% ± 0.7%) were more frequently detected than CoV-PMV (0.1% ± 0.3%) and AstV-CoV-PMV (0.3% ± 0.4%) (χ2 = 11.3, df = 3, P < 0.05) (Fig. 1). The presence of AstVs was strongly correlated to the presence of PMVs (χ2 = 4.6, df = 1, P < 0.05) suggesting a potential positive association between these two viruses. Co-infections involving AstVs, CoVs or PMVs were reported in other tropical bat species. For example, a study of Hipposideros cervinus on Borneo reported 4.1% of AstV-CoV coinfected bats with a positive association between these two viruses [11].
On Madagascar, co-infections were detected in three of the 18 tested species: Triaenops menamena (15.2% ± 24.7%), Miniopterus gleni (13.3% ± 14.8%), and Paratriaenops furculus (3.2% ± 0.2%), without significant difference between species (χ2 = 2.7, df = 2, P > 0.05) (Fig. 2). In Mozambique, co-infections were detected in four of the eight tested species: Triaenops afer (15.7% ± 10.0%), Nycteris thebaica (7.1% ± 13.5%), Hipposideros caffer (5.1% ± 5.6%), and Miniopterus mossambicus (4.8% ± 9.1%) (χ2 = 4.4, df = 3, P > 0.05), also without significant difference between species (Fig. 3). Interestingly, co-infections were detected in bats of the family Rhinonycteridae both in Mozambique (Triaenops) and on Madagascar (Triaenops and Paratriaenops). In Mozambique, the proportion of co-infections was significantly higher in Triaenops afer than in all other tested species (χ2 = 4.4, df = 1, P < 0.05). Also, as compared to other species that tested positive for co-infection, different combinations of co-infections (i.e. different associations between viruses) were detected in species of the genus Triaenops. Indeed, Triaenops menamena on Madagascar presented two types of co-infections: AstV-PMV and CoV-PMV, whereas Triaenops afer in Mozambique harbored three types: AstV-CoV, AstV-PMV, and AstV-CoV-PMV; while other species, including Paratriaenops furculus, only presented one co-infection type (Figs. 2 and 3 and Additional file 3). Altogether, these results highlight multiple infections in WIO region bats, and give rise to additional questions concerning variation among species, as well as their consequences on viral infection dynamics.
High genetic diversity was detected for AstV, with pairwise differences up to 46% between sequences, without support for host family or species association (Fig. 4). Based on BLASTn comparisons, we found that our sequences had a high level of identity (between 80 and 92%) with AstV previously described in bats of the WIO region [20, 21], as well as with AstVs detected in bats from continental Africa (e.g. Gabon, Democratic Republic of Congo), and in other regions in the world (e.g. China, Thailand) (Additional file 4). However, one sequence obtained from a Triaenops menamena, a species endemic to Madagascar, showed 92% identity to an AstV sequence detected in a mouse from China (Additional file 4). Another example, even more unexpected, one AstV sequence obtained from Chaerephon leucogaster on Madagascar had 96% identity with an AstV sequence from a bird of the order Passeriformes (Additional file 4). These findings were consistent with phylogenetic results and were statistically supported (Fig. 4). A recent study also reported AstVs related to avastrovirus in environmental samples collected in a colony of Mormopterus francoismoutoui, a member of the family Molossidae endemic to Reunion Island [41]. These findings may suggest introduction of AstVs on the island by non-native rodents, and could also support environmental transmission of AstVs between species of different taxa, as previously suggested [42]. Nevertheless, studies investigating the circulation of AstVs in terrestrial small mammals in the WIO region are required to assess these potential host-shifts.
Maximum likelihood consensus tree derived from 138 astrovirus (AstV) RNA-dependent RNA-polymerase partial nucleotide sequences (397 bp). Black dots indicate nodes with bootstrap values higher or equal than 60. Sequence names colored in red indicate bat AstVs detected in this study. Sequence accession numbers are indicated in parentheses
Genetic diversity was less important for PMV sequences, with pairwise differences up to 29%. All our sequences were genetically related to PMVs previously described on Madagascar [16, 19] or in continental Africa (e.g. Ghana, Kenya), with sequence identity ranging from 78 to 99% (Additional file 5 and Fig. 5). Phylogenetic analyses highlighted some degree of host-specificity, as previously described for PMV in the western Indian Ocean (Fig. 5) [16]. For instance, most sequences clustered either with PMV sequences detected in bats on the same genus captured in the region or elsewhere (e.g. Hipposideros caffer from Mozambique), or with sequences obtained in bats from the same family (e.g. Mops condylurus sequence from Mozambique, and Chaerephon leucogaster sequence from Madagascar clustered with a sequence detected in Mops leucostigma on Madagascar). However, some sequences were included in more diversified groups including different bat families (e.g. two sequences obtained from Paratriaenops furculus on Madagascar that clustered with sequences detected in Miniopterus griveaudi and Chaerephon leucogaster on Madagascar).
Maximum likelihood consensus tree derived from 87 paramyxovirus (PMV) RNA-dependent RNA-polymerase partial nucleotide sequences (439 bp). Black dots indicate nodes with bootstrap values higher or equal than 60. Sequence names colored in red indicate bat PMVs detected in this study. Sequence accession numbers are indicated in parentheses
We report co-infections in bats on Madagascar and in Mozambique, ranging from 3.2% to 15.7% of the positive samples, and depending on the tested bat species. Although our cross-sectional sampling precludes detailed interpretation of the biological drivers of such variation, our results nevertheless highlight that interactions between infectious agents in bats may exist with potential consequences on their epidemiology. AstVs, CoVs, and PMVs are emerging viruses that represent a major challenge for human and animal health. Further knowledge on virus interaction in wildlife, based on long-term longitudinal sampling is needed to fully assess the epidemiological consequences of co-infections [5].
Availability of data and materials
Data are available in the supplementary material (detailed list of samples and results of molecular screening) and in GenBank (sequence accession numbers of the sequences generated in this study: MZ614404 to MZ614449).
Abbreviations
- AstV:
-
Astrovirus
- BLAST:
-
Basic local alignment search tool
- BLASTn:
-
Standard nucleotide BLAST
- CoV:
-
Coronavirus
- PCR:
-
Polymerase chain reaction
- PMV:
-
Paramyxovirus
- RdRp:
-
RNA-dependent RNA-polymerase
- WIO:
-
Western Indian Ocean
References
Petney TN, Andrews RH. Multiparasite communities in animals and humans: frequency, structure and pathogenic significance. Int J Parasitol. 1998;28:377–93.
Cox FEG. Concomitant infections, parasites and immune responses. Parasitology. 2001;122:S23-38.
Rigaud T, Perrot-Minnot M-J, Brown MJF. Parasite and host assemblages: embracing the reality will improve our knowledge of parasite transmission and virulence. Proc R Soc B Biol Sci. 2010;277:3693–702.
Vaumourin E, Vourc’h G, Gasqui P, Vayssier-Taussat M. The importance of multiparasitism: examining the consequences of co-infections for human and animal health. Parasites Vectors. 2015;8:545.
Hoarau AOG, Mavingui P, Lebarbenchon C. Coinfections in wildlife: focus on a neglected aspect of infectious disease epidemiology. PLOS Pathogens. 2020;16:e1008790.
Pedersen AB, Fenton A. Emphasizing the ecology in parasite community ecology. Trends Ecol Evol. 2007;22:133–9.
Hellard E, Fouchet D, Vavre F, Pontier D. Parasite–parasite interactions in the wild: How to detect them? Trends Parasitol. 2015;31:640–52.
Peel AJ, Wells K, Giles J, Boyd V, Burroughs A, Edson D, et al. Synchronous shedding of multiple bat paramyxoviruses coincides with peak periods of Hendra virus spillover. Emerg Microbes Infect. 2019;8:1314–23.
Kaufman E. Two (or more) viruses in one bat: a systematic quantitative literature review of viral coinfection in bats. Independent Study Project (ISP); 2020 p. 3345. https://digitalcollections.sit.edu/isp_collection/3345/.
Rizzo F, Edenborough KM, Toffoli R, Culasso P, Zoppi S, Dondo A, et al. Coronavirus and paramyxovirus in bats from northwest Italy. BMC Vet Res. 2017;13(1):396.
Seltmann A, Corman VM, Rasche A, Drosten C, Czirják GÁ, Bernard H, et al. Seasonal fluctuations of astrovirus, but not coronavirus shedding in bats inhabiting human-modified tropical forests. EcoHealth. 2017;14:272–84.
Luis AD, Hayman DTS, O’Shea TJ, Cryan PM, Gilbert AT, Pulliam JRC, et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: Are bats special? Proc R Soc B Biol Sci. 2013;280:20122753.
Anthony SJ, Epstein JH, Murray KA, Navarrete-Macias I, Zambrana-Torrelio CM, Solovyov A, et al. A strategy to estimate unknown viral diversity in mammals. mBio. 2013;4(5):e00598-13.
Iehlé C, Razafitrimo G, Razainirina J, Andriaholinirina N, Goodman SM, Faure C, et al. Henipavirus and Tioman virus antibodies in pteropodid bats. Madagascar Emerg Infect Dis. 2007;13:159–61.
Wilkinson DA, Temmam S, Lebarbenchon C, Lagadec E, Chotte J, Guillebaud J, et al. Identification of novel paramyxoviruses in insectivorous bats of the Southwest Indian Ocean. Virus Res. 2012;170:159–63.
Wilkinson DA, Melade J, Dietrich M, Ramasindrazana B, Soarimalala V, Lagadec E, et al. Highly diverse Morbillivirus-related paramyxoviruses in wild fauna of the southwestern Indian Ocean islands: evidence of exchange between introduced and endemic small mammals. J Virol. 2014;88:8268–77.
Dietrich M, Wilkinson DA, Benlali A, Lagadec E, Ramasindrazana B, Dellagi K, et al. Leptospira and paramyxovirus infection dynamics in a bat maternity enlightens pathogen maintenance in wildlife: dual infection dynamics in a bat maternity. Environ Microbiol. 2015;17:4280–9.
Razanajatovo NH, Nomenjanahary LA, Wilkinson DA, Razafimanahaka JH, Goodman SM, Jenkins RK, et al. Detection of new genetic variants of Betacoronaviruses in endemic frugivorous bats of Madagascar. Virol J. 2015;12:42.
Mélade J, Wieseke N, Ramasindrazana B, Flores O, Lagadec E, Gomard Y, et al. An eco-epidemiological study of Morbilli-related paramyxovirus infection in Madagascar bats reveals host-switching as the dominant macro-evolutionary mechanism. Sci Rep. 2016;6:23752.
Lebarbenchon C, Ramasindrazana B, Joffrin L, Bos S, Lagadec E, Le Minter G, et al. Astroviruses in bats, Madagascar. Emerg Microbes Infect. 2017;6:e58.
Hoarau F, Le Minter G, Joffrin L, Schoeman CM, Lagadec E, Ramasindrazana B, et al. Bat astrovirus in Mozambique. Virol J. 2018;15:104.
Brook CE, Ranaivoson HC, Broder CC, Cunningham AA, Héraud J, Peel AJ, et al. Disentangling serology to elucidate henipa‐ and filovirus transmission in Madagascar fruit bats. J Anim Ecol. 2019;88:1001–16.
Joffrin L, Goodman SM, Wilkinson DA, Ramasindrazana B, Lagadec E, Gomard Y, et al. Bat coronavirus phylogeography in the western Indian Ocean. Sci Rep. 2020;10(1):6873.
Chu DKW, Poon LLM, Guan Y, Peiris JSM. Novel Astroviruses in insectivorous bats. J Virol. 2008;82:9107–14.
Tong S, Chern S-WW, Li Y, Pallansch MA, Anderson LJ. Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J Clin Microbiol. 2008;46:2652–8.
R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing Vienna, Austria; 2015
Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–3.
Altschul S. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402.
Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–14.
Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 30. Syst Biol. 2010;59:307–21.
Drexler JF, Corman VM, Müller MA, Maganga GD, Vallo P, Binger T, et al. Bats host major mammalian paramyxoviruses. Nat Commun. 2012;3:796.
Fischer K, Pinho dos Reis V, Balkema-Buschmann A. Bat astroviruses: towards understanding the transmission dynamics of a neglected virus family. Viruses. 2017;9:34.
Ithete NL, Stoffberg S, Corman VM, Cottontail VM, Richards LR, Schoeman MC, et al. Close relative of human Middle East respiratory syndrome coronavirus in bat. S Afr Emerg Infect Dis. 2013;19:1697–9.
Smith CS, de Jong CE, Meers J, Henning J, Wang L-F, Field HE. Coronavirus infection and diversity in bats in the Australasian region. EcoHealth. 2016;13:72–82.
Willoughby A, Phelps K, PREDICT Consortium, Olival K. A comparative analysis of viral richness and viral sharing in cave-roosting bats. Diversity. 2017;9:35.
Altizer S, Dobson A, Hosseini P, Hudson P, Pascual M, Rohani P. Seasonality and the dynamics of infectious diseases. Ecol Lett. 2006;9:467–84.
Drexler JF, Corman VM, Wegner T, Tateno AF, Zerbinati RM, Gloza-Rausch F, et al. Amplification of emerging viruses in a bat colony. Emerg Infect Dis. 2011;17:449–56.
Mühldorfer K, Speck S, Kurth A, Lesnik R, Freuling C, Müller T, et al. Diseases and causes of death in European bats: dynamics in disease susceptibility and infection rates. PLoS ONE. 2011;6:e29773.
George DB, Webb CT, Farnsworth ML, O’Shea TJ, Bowen RA, Smith DL, et al. Host and viral ecology determine bat rabies seasonality and maintenance. Proc Natl Acad Sci. 2011;108:10208–13.
Wacharapluesadee S, Boongird K, Wanghongsa S, Ratanasetyuth N, Supavonwong P, Saengsen D, et al. A longitudinal study of the prevalence of Nipah virus in Pteropus lylei bats in Thailand: evidence for seasonal preference in disease transmission. Vector-Borne Zoonot Dis. 2010;10:183–90.
Joffrin L, Hoarau AOG, Lagadec E, Köster M, Ramanantsalama RV, Mavingui P, et al. Astrovirus in Reunion free-tailed bat (Mormopterus francoismoutoui). Viruses. 2021;13:1524.
Mendenhall IH, Smith GJD, Vijaykrishna D. Ecological drivers of virus evolution: astrovirus as a case study. J Virol. 2015;89:6978–81.
Acknowledgements
We thank L. Joffrin, J. Mélade, D.A. Wilkinson, and their collaborating co-authors of the three studies we retrieved data from. We are grateful to all involved in the field for the collection of biological material: M. Dietrich, C. Dionisio, L. Domergues, Y. Gomard, L. Joffrin, T. Mbohoahy, T. Nekena, M. Pannequin, J. Rakotoarivelo, M. Rakotomanga, C. F. Rakotondramanana, A. Randrenjarison, P. Tortosa, and D. A. Wilkinson. We also thank K. Dellagi and H. Pascalis for the development and the management of the “partenariat Mozambique-Réunion dans la recherche en santé : pour une approche intégrée d’étude des maladies infectieuses à risque épidémique (MoZaR)” research program.
Funding
This work was funded by the “Pathogènes associés à la Faune Sauvage océan Indien (FS-OI)”, the “Leptospirose océan Indien (LeptOI)”, the “Paramyxovirus océan Indien (ParamyxOI)”, the “Partenariat Mozambique-Réunion dans la recherche en santé: pour une approche intégrée d’étude des maladies infectieuses à risque épidémique (MoZaR)”, and by the VIROPTERE (INTERREG V océan Indien) programs. A.H. was supported by a “Ministère de l’Enseignement supérieur, de la Recherche et de l’Innovation” PhD fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
A.H., C.L., and P.M. conceived and designed the study. S.M.G., E.L., G.L.M., B.R., A.D.S., and M.C.S. collected biological material. D.A.H., A.H., M.K, and C.L. performed the molecular analyses. D.A.H. and A.H. analyzed the data. E.S.G. (Mozambique), S.M.G. (Madagascar), and P.M. (France) contributed to the project management for the respective country institutions. A.H. was the principal author of the paper. All authors edited, read, and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study is based on archived samples collected as part of previous investigations of infectious agents circulation in bats on islands in the western Indian Ocean and Mozambique.Procedures were evaluated and approved by an ethic committee (Comité d’éthique du CYROI number 114; Cyclotron Reunion Océan Indien, Sainte Clotilde, Reunion Island), and authorized by the French Ministry of Higher Education, of Research and Innovation. On Madagascar, research permits were delivered by “Direction du Système des Aires Protégées and Direction Générale de l’Environnement et des Forêts” (350/10/MEF/SG/DGF/DCB.SAP/SCB, 032/12/MEF/SG/DGF/ DCB.SAP/SCBSE, 067/12/MEF/SG/DGF/DCB.SAP/SCBSE, 194/12/ MEF/SG/DGF/DCB.SAP/SCB, N◦283/11/MEF/SG/DGF/DCB.SAP/SCB, N◦077/12/MEF/SG/DGF/DCB.SAP/SCBSE, 238/14/MEEF/SG/ DGF/DCB.SAP/SCB and 268/14/MEEF/SG/DGF/DCB.SAP/SCB) and samples were exported to Reunion Island thanks to a permit delivered by the Malagasy national authority (243C-EA06/MG12). On Mayotte, research permits were issued by the ‘Direction de l’Environnement, de l’Aménagement et du Logement’ (Arreté n°158/DEAL/SEPR/2014). In Mozambique, research permits were issued by the Museum of Natural History (Ref. 01/MHN/E.27/2015) and the Ministry of Health (N°S/N/SDI/0233/15). Finally, on Reunion Island, research permits were delivered by the Préfecture de La Réunion (Arrêté préfectoral du 11 Février 2013 and Arrêté préfectoral du 11 Septembre 2014 N°2014-07).
Consent for publication
Not applicable.
Competing interests
The authors have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. List and origin of samples, day roost sites, and result of the molecular screening.
Additional file 2
. Number of tested and positive samples, per location, bat family, species, samples types, and collection year.
Additional file 3
. Number of tested and positive samples for co-infections, per country, bat family, bat species, sample type, and collection year.
Additional file 4
. Astrovirus nucleotide sequence similarity obtained with BLASTn.
Additional file 5
. Paramyxovirus nucleotide sequence similarity obtained with BLASTn
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Hoarau, A.O.G., Goodman, S.M., Al Halabi, D. et al. Investigation of astrovirus, coronavirus and paramyxovirus co-infections in bats in the western Indian Ocean. Virol J 18, 205 (2021). https://doi.org/10.1186/s12985-021-01673-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-021-01673-2