Abstract
Background
The persistence of severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) RNA in the body fluids of patients with the novel coronavirus disease 2019 (COVID-19) may increase the potential risk of viral transmission. There is still uncertainty on whether the recommended quarantine duration is sufficient to reduce the risk of transmission. This study aimed to investigate the persistence of SARS-CoV-2 RNA in the nasopharyngeal, blood, urine, and stool samples of patients with COVID-19.
Methods
In this hospital-based longitudinal study, 100 confirmed cases of COVID-19 were recruited between March 2020 and August 2020 in Guilan Province, north of Iran. Nasopharyngeal, blood, urine, and stool samples were obtained from each participant at the time of hospital admission, upon discharge, 1 week after discharge, and every 2 weeks until all samples were negative for SARS-CoV-2 RNA by reverse transcription-polymerase chain reaction (RT-PCR) assay. A survival analysis was also performed to identify the duration of viral persistence.
Results
The median duration of viral RNA persistence in the nasopharyngeal samples was 8 days from the first positive RT-PCR result upon admission (95% CI 6.91–9.09); the maximum duration of viral shedding was 25 days from admission. Positive blood, urine, and stool RT-PCR results were detected in 24%, 7%, and 6% of the patients, respectively. The median duration of viral persistence in the blood, urine, and stool samples was 7 days (95% CI 6.07–7.93), 6 days (95% CI 4.16–8.41), and 13 days (95% CI 6.96–19.4), respectively. Also, the maximum duration of viral persistence in the blood, urine, and stool samples was 17, 11, and 42 days from admission, respectively.
Conclusion
According to the present results, immediately after the hospitalized patients were discharged, no evidence of viral genetic materials was found. Therefore, appropriate treatments were selected for the patients at this hospital. However, we recommend further investigations on a larger sample size in multi-center and prospective randomized controlled trials (RCTs) to evaluate the effects of different drugs on the shedding of the virus through body secretions.
Similar content being viewed by others
Introduction
Coronavirus disease 2019 (COVID-19) was declared as a global pandemic by the World Health Organization (WHO) in December 2020 [1]. By November 22, 2020, this virus infected almost 57 million people around the world and caused more than 1,300,000 deaths [1]. Iran is ranked the eighth country in terms of COVID-19 mortality [2]. There were 1,189,203 confirmed cases of COVID-19 and 54,440 deaths in Iran from February 15, 2019, until December 25, 2020 [3].
The main route of COVID-19 transmission seems to be direct or indirect exposure to respiratory droplets [4, 5]. Although other routes of transmission, such as mother-to-fetus, fecal–oral, and airborne transmission, are currently controversial and subject to future investigations [5,6,7,8], epidemiological studies have shown that COVID-19 patients have had close contact with an infected individual or have been in close proximity to a patient [9].
Initially, severe acute respiratory syndrome-coronavirus 2 (SARS-Cov-2) was isolated and identified in respiratory samples by real-time reverse transcription-polymerase chain reaction (RT-PCR) assay [10]. However, in recent studies, viral nucleic acids have been also detected in urine, stool, and gastric mucosa samples [10, 11]. In a previous study, ten children with COVID-19, despite having negative nasopharyngeal RT-PCR results, still showed positive RT-PCR of throat swabs [12]. Based on the Centers for Disease Control and Prevention (CDC) guidelines, all patients with a positive respiratory RT-PCR result must be isolated for at least 10 days after symptom onset and after resolution of fever for at least 24 h [13].
Nevertheless, there are several case reports on the persistence of positive RT-PCR in patients with COVID-19, indicating the possibility of positive results after the symptoms have resolved [14, 15]. Therefore, the persistence of the virus in body fluids can increase the potential risk of viral transmission in asymptomatic or recovered patients [15]. It also remains uncertain whether the quarantine duration, recommended by the CDC, is sufficient to reduce viral transmission [16]. Since the frequency and detection time of SARS-CoV-2 RNA in body, fluids are not well understood, in this longitudinal study, we aimed to determine the persistence of SARS-CoV-2 RNA in the nasopharyngeal, blood, urine, and stool samples of patients with COVID-19, which were collected every 2 weeks by sequential sampling.
Methods
Study population and design
This hospital-based longitudinal study was performed between March 2020 and August 2020 during 6 months. The participants were selected by convenience sampling among hospitalized patients with a confirmed diagnosis of COVID-19 in the only referral hospital of Rasht, Guilan Province, in north of Iran. A positive case of COVID-19 was defined as a patient with a positive quantitative real-time RT-PCR of nasopharyngeal samples [17]. For inclusion in this study, a confirmed diagnosis of COVID-19, defined as a positive PCR result, was required.
The sample size was estimated to be 100 participants at a confidence level of 95% and test power of 80%. If any of the participants refused to give a sample, he/she was excluded from the study. This study was approved by the local ethics committee of Guilan University of Medical Sciences, Rasht, Iran (code: IR.GUMS.REC.1399.013). Written informed consent was obtained from each participant.
Measurements
Nasopharyngeal, blood, urine, and stool samples were obtained from each participant at the time of admission, upon discharge, 1 week after discharge, and every 2 weeks until all samples were negative for SARS-CoV-2 RNA on RT-PCR. Upon admission, all four samples (nasopharyngeal, blood, urine, and stool) were collected from each participant and analyzed for SARS-CoV-2 RNA by PCR. Also, upon discharge, nasopharyngeal and stool samples were collected from each participant; blood and urine samples were collected if they were positive for SARS-CoV-2 RNA at the time of admission. Besides, in each follow-up visit, nasopharyngeal, blood, urine, and stool samples were collected if the patient was positive for SARS-CoV-2 RNA in the previous visit.
The nasopharyngeal and stool samples were obtained by sterile Dacron swabs. Also, 5-mL samples of whole blood and urine were taken for SARS-CoV-2-specific real-time RT-PCR. The samples were processed by a trained laboratory technician immediately after sampling. The coronavirus genome was isolated using the RNJia Virus Kit (ROJETechnologies, Yazd, Iran) for RNA extraction, and real-time PCR was carried out using the COVID-19 One-Step RT-PCR Kit (Pishtazteb, Iran), according to the manufacturer’s instructions. The results of quantitative RT-PCR are shown as cycle threshold (Ct) values. A positive control and a negative control were also included in each run to generate valid results. A Ct value < 40 was defined as a positive test result. Besides, the viral load was categorized as high (< 20), medium (20–29.9), and low (30–39.9) [18].
Moreover, the clinical and demographic characteristics of the participants were collected in this study. The sociodemographic characteristics of the patients included age, gender, marital status, job, education, type of residence, and socioeconomic status. Besides, the clinical manifestations of COVID-19 included fever, cough, sore throat, dyspnea, weakness, muscular pain, headache, diarrhea, nausea and vomiting, and chill. Besides, information on underlying diseases (e.g., diabetes, hypertension, cardiovascular disease, immunodeficiency, cancer, and respiratory disease) and inflammatory markers (e.g., white blood cell count, erythrocyte sedimentation rate, and C-reactive protein) were collected.
In this study, the hospital treatment plans were categorized into four groups, including hydroxychloroquine, antiviral treatment (e.g., lopinavir, Remdesivir, and Sovodak consisting of 400 mg sofosbuvir and 60 mg daclatasvir), interferon β1 (at five subcutaneous doses of 44 µg daily, 3 days a week), and local treatment (e.g., diphenhydramine, acetaminophen, zinc, vitamin C, and famotidine). All patients received corticosteroids (8 mg dexamethasone) daily.
Data analysis
Comparison of qualitative variables (clinical and demographic characteristics) between the groups, categorized based on the duration of SARS-CoV-2 RNA persistence in the nasopharynx, was performed using Chi-square or Fisher’s exact test. A survival analysis was also performed to identify the median and 95th percentile of SARS-CoV-2 persistence. To find clinical and demographic characteristics that may be associated with the persistence of SARS-CoV-2 RNA, a Cox regression analysis was performed. Pearson’s correlation coefficient was also calculated to determine the relationship between the Ct value and the length of hospital stay. Data analysis was performed in SPSS version 17.0 (SPSS Inc., Chicago, IL, USA).
Results
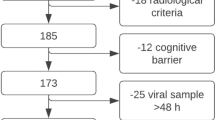
During the study, 106 hospitalized patients with positive nasopharyngeal RT-PCR of SARS-CoV-2 RNA were enrolled, six of whom expired (5.66%). The mean age of the participants was 53.30 ± 13.03 years (range 29–86 years). The majority of the patients were male (60%), married (84%), urban residents (85%), and employed (68%). Also, the majority of the participants had less than high school diploma (63%) and a low socioeconomic status (82%). The results of SARS-CoV-2 RNA RT-PCR in different samples over time are shown in Fig. 1.
Only 24% of blood RT-PCR samples were positive upon admission; one sample remained positive upon discharge and became negative after 1 week. Also, the RT-PCR of urine samples was positive in 6% of the patients upon admission, while all samples became negative at discharge. Moreover, the RT-PCR result was positive in the stool samples of four patients upon admission, and two samples remained positive at discharge. Besides, two new stool samples were positive at discharge. Three of these samples were negative after 1 week, and one was negative 5 weeks after discharge. Upon discharge, 30% of nasopharyngeal RT-PCR samples were positive, and all were cleared after one weak.
There was no significant difference in terms of the demographic characteristics among the three groups. The participants’ clinical and demographic characteristics, based on the duration of viral persistence in the nasopharynx, are shown in Table 1. Almost 11% of patients showed viral persistence in the nasopharynx longer than 14 days from the first positive PCR. The demographic characteristics of the patients were not significantly different among the three groups.
Similarly, there was no significant association between the COVID-19 symptoms and the duration of viral persistence in the nasopharynx. Our findings revealed that the type of hospital treatment plan did not contribute to the viral persistence in the nasopharynx. In this study, only one out of 100 patients (1%) had a high viral load, 47% had a moderate viral load, and 52% had a low viral load. There was no significant relationship between the Ct value and the length of hospital stay (r = 0.030, P = 0.766). Also, no significant difference was detected between the duration of SARS-CoV-2 RNA persistence in the nasopharynx and the nasopharyngeal viral load upon admission (r = 0.030, P = 0.766).
The median duration of SARS-CoV-2 RNA persistence in the nasopharynx was 8 days (95% CI 6.91–9.09) from the first positive RT-PCR upon admission, and the maximum duration of viral persistence was 25 days from admission (Fig. 2). The median duration of SARS-CoV-2 RNA persistence in the blood was 7 days (95% CI 6.07–7.93) from the first positive RT-PCR at admission, and the maximum duration of viral persistence was 17 days from admission (Fig. 2). The median duration of SARS-CoV-2 RNA persistence in the stool samples was 13 days (95% CI 6.96–19.4) from the first positive RT-PCR, and the maximum duration of viral persistence was 42 days from admission (Fig. 2). Besides, the median duration of SARS-CoV-2 RNA persistence in the urine was 6 days (95% CI 4.16–8.41) from the first positive RT-PCR upon admission, and the maximum duration of viral persistence was 11 days from admission (Fig. 2).
In the Cox regression analysis of factors possibly associated with the duration of SARS-CoV-2 RNA persistence, no significant associations were found (data not presented). The frequency of some COVID-19 gastrointestinal (GI) symptoms, according to the stool RT-PCR, is presented in Fig. 3. Abdominal pain and diarrhea were significantly more common in patients with positive stool RT-PCR samples as compared to those with negative stool RT-PCR. Moreover, in the analysis of clinical and demographic characteristics that could be associated with positive blood and urine RT-PCR, no significant relationships were detected (data not presented).
Discussion
In this longitudinal study, the maximum duration of nasopharyngeal (25 days) and fecal (42 days) SARS-CoV-2 RNA shedding was longer than the recommended quarantine duration (at least 10 days after symptom onset and after resolution of fever for at least 24 h) [13]. Nearly one-third of hospitalized COVID-19 patients had positive nasopharyngeal RT-PCR results upon discharge, and about one-tenth of them showed viral persistence in the nasopharynx longer than 14 days from the first positive RT-PCR. The maximum duration of nasopharyngeal shedding was 25 days from admission.
The mentioned findings are compatible with the results of some studies, which revealed that some patients continued to have positive upper respiratory tract RT-PCR results after discharge from the hospital for the next few days [19, 20]. Although we did not identify any determinants of viral persistence, a study from China demonstrated that the prolonged presence of the virus in the upper respiratory tract was associated with disease severity [21]. On the other hand, another study from Portugal revealed that viral RNA persistence was not associated with disease severity, and a stronger immune response was a determinant of viral RNA clearance [22].
Positive blood SARS-CoV-2 RNA RT-PCR results were reported in about one-third of our patients, although most of them became negative at the time of discharge. Although studies on SARS-CoV-2 RNA detection in the blood are limited, a study from China revealed that SARS-CoV-2 RNA was detected in the blood of six out of 57 Chinese patients; all six patients with positive blood RT-PCR had a severe clinical condition [23]. The higher positive rate of blood RT-PCR in our study was probably attributed to disease severity in our patients and the hospital-based design of this study.
The present findings revealed that positive results of urine SARS-CoV-2 RNA RT-PCR were less common (found in 7/100 patients) in our study population, all of which became negative at the time of discharge. This result is consistent with previous studies from Turkey and China, which demonstrated that nearly 5–7% of COVID-19 patients had positive urine RT-PCR results [24, 25].
Positive stool SARS-CoV-2 RNA RT-PCR was detected in only six out of 100 patients in our study, while the time required for the positive stool RT-PCR to become negative was longer than the nasopharyngeal, urine, and blood RT-PCR tests, with a maximum fecal shedding duration of 42 days. Therefore, positive stool RT-PCR results in hospitalized patients represent the need to take precautions and use protective equipment in interventional procedures involving the gastrointestinal tract in a hospital environment. Similarly, in another study from China, an asymptomatic case remained positive in the stool RT-PCR for a period of 42 days. Also, in almost two-thirds of the patients, fecal shedding took longer than nasopharyngeal shedding [20]. Consistent with these findings, longer periods were reported in previous studies for detecting and shedding other coronaviruses in the gastrointestinal tract [26, 27].
According to our findings, the viral load of SARS-CoV-2, measured by the Ct method upon admission, is not a valuable parameter for predicting the duration of SARS-CoV-2 RNA clearance from the nasopharynx. The Ct value was inversely associated with the length of hospital stay and the time until viral clearance; in other words, a higher Ct value was indicative of a faster viral clearance [28, 29].
Although our findings demonstrated that the maximum duration of nasopharyngeal and fecal SARS-CoV-2 RNA shedding was longer than the recommended quarantine duration by the CDC [13], it is unclear whether individuals with persistent positive RNA PCR results have an infection risk. Therefore, further studies are needed to determine whether PCR positivity is associated with infective or non-infective nucleic acid fragments. In the present study, as soon as the patients were discharged from the hospital, there was no viral genetic material in the blood, urine, or nasopharyngeal samples, except in two out of 100 patients with a persistent PCR. Therefore, patients treated for SARS-CoV-2 and COVID-19 did not show infectivity once discharged from the hospital; this finding provides further evidence for applying successful treatments [30,31,32].
The strengths of the present study include the long-term follow-up and detection of viral RNA in respiratory and extra-respiratory sites. On the other hand, the hospital-based design of this study is one of its limitations, leading to a more strict patient enrollment that limited the generalizability of our findings. Another limitation of this study is that the day of symptom onset was not determined, and all calculations and analyses were based on the first nasopharyngeal RT-PCR at admission; therefore, the duration of viral shedding was underestimated. Finally, since SARS-CoV-2 RNA was found upon discharge in a reduced number of samples (especially in the blood and stool samples), further large-scale studies are recommended.
Conclusion
The present findings revealed that in hospitalized COVID-19 patients, the maximum duration of nasopharyngeal and fecal SARS-CoV-2 RNA shedding was longer than the recommended quarantine duration by the CDC. However, immediately after the patients were discharged from the hospital, there was no evidence of viral genetic materials; therefore, appropriate treatments were applied in this hospital. However, further, multi-center and prospective randomized controlled trials are recommended on a larger sample size to evaluate the effects of different drugs on viral shedding through body secretions.
Availability of data and materials
The datasets obtained during this study will be available upon request to the corresponding author.
Abbreviations
- CDC:
-
the centers for disease control and prevention
- RT-PCR:
-
real-time fluorescent quantitative polymerase chain reaction
References
Organization WH. World Health Organization coronavirus disease 2019 (COVID-19) situation report. 2020.
Organization WH. Coronavirus disease 2019 (COVID-19): situation report, 82. 2020.
Organization WH. WHO coronavirus disease (COVID-19) dashboard. 2020.
Gupta MK, Lipner SR. Personal protective equipment recommendations based on COVID-19 route of transmission. J Am Acad Dermatol. 2020;83(1):45–6. https://doi.org/10.1016/j.jaad.2020.04.068.
Harrison AG, Lin T, Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41:1100–15.
Heller L, Mota CR, Greco DB. COVID-19 faecal-oral transmission: Are we asking the right questions? Sci Total Environ. 2020;729:138919. https://doi.org/10.1016/j.scitotenv.2020.138919.
Setti L, Passarini F, De Gennaro G, Barbieri P, Perrone MG, Borelli M, et al. Airborne transmission route of COVID-19: why 2 meters/6 feet of inter-personal distance could not be enough. Int J Environ Res Public Health. 2020;17(8):2932. https://doi.org/10.3390/ijerph17082932.
Fenizia C, Biasin M, Cetin I, Vergani P, Mileto D, Spinillo A, et al. Analysis of SARS-CoV-2 vertical transmission during pregnancy. Nat Commun. 2020;11(1):1–10.
Fisher KA, Tenforde MW, Feldstein LR, Lindsell CJ, Shapiro NI, Files DC, et al. Community and close contact exposures associated with COVID-19 among symptomatic adults≥ 18 years in 11 outpatient health care facilities—United States, July 2020. MMWR Morb Mortal Wkly Rep. 2020;69(36):1258–64. https://doi.org/10.15585/mmwr.mm6936a5.
Peng L, Liu J, Xu W, Luo Q, Deng K, Lin B, et al. novel coronavirus can be detected in urine, blood, anal swabs and oropharyngeal swabs samples. MedRxiv. 2019. https://doi.org/10.1101/2020.02.21.20026179.
Zhiwei Y, Ganwen L, Xiaoling D, Guirong L, Gang L, Yusheng J. Three cases of novel coronavirus pneumonia with viral nucleic acids still positive in stool after throat swab detection turned negative. Chin J Dig. 2020;40:E002–E002. https://doi.org/10.3760/cma.j.issn.0254-1432.2020.0002.
Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y, et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26(4):502–5.
Centers for Disease Control and Prevention. Separate yourself from others if you have COVID-19;2020 [updated Nov. 3, 2020]. Available from: https://www.cdc.gov/coronavirus/2019-ncov/if-you-aresick/isolation.html.
Cento V, Colagrossi L, Nava A, Lamberti A, Senatore S, Travi G, et al. Persistent positivity and fluctuations of SARS-CoV-2 RNA in clinically-recovered COVID-19 patients. J Clean Prod. 2020;81(3):90–2. https://doi.org/10.1016/j.jinf.2020.06.024 (Epub 2020 Jun 20).
Danzetta ML, Amato L, Cito F, Di Giuseppe A, Morelli D, Savini G, et al. SARS-CoV-2 RNA persistence in Naso-Pharyngeal swabs. Microorganisms. 2020;8(8):1124. https://doi.org/10.3390/microorganisms8081124.
Gombar S, Chang M, Hogan CA, Zehnder J, Boyd S, Pinsky BA, et al. Persistent detection of SARS-CoV-2 RNA in patients and healthcare workers with COVID-19. J Clin Virol. 2020;129:104477. https://doi.org/10.1016/j.jcv.2020.104477.
Hanson KE, Caliendo AM, Arias CA, Englund JA, Lee MJ, Loeb M, et al. Infectious Diseases Society of America guidelines on the diagnosis of COVID-19. Clin Infect Dis. 2020. https://doi.org/10.1093/cid/ciaa1343.
Yagci AK, Sarinoglu RC, Bilgin H, Yanılmaz Ö, Sayın E, Deniz G, et al. Relationship of the cycle threshold values of SARS-CoV-2 polymerase chain reaction and total severity score of computerized tomography in patients with COVID 19. Int J Infect Dis. 2020;101:160–6. https://doi.org/10.1016/j.ijid.2020.09.1449.
Jameel T, Baig M, Gazzaz ZJ. Persistence of reverse transcription-polymerase chain reaction (RT-PCR) Positivity in COVID-19 recovered patients: a call for revised hospital discharge criteria. Cureus. 2020;12(7):9048. https://doi.org/10.7759/cureus.9048.
Hu Z, Song C, Xu C, Jin G, Chen Y, Xu X, et al. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci. 2020;63(5):706–11. https://doi.org/10.1007/s11427-020-1661-4.
Chang D, Zhao P, Zhang D, Dong J-H, Xu Z, Yang G, et al. Persistent viral presence determines the clinical course of the disease in COVID-19. J Allergy Clin Immunol Pract. 2020;8(8):2585–91. https://doi.org/10.1016/j.jaip.2020.06.015.
Carmo A, Pereira-Vaz J, Mota V, Mendes A, Morais C, da Silva AC, et al. Clearance and persistence of SARS-CoV-2 RNA in patients with COVID-19. J Med Virol. 2020;92(10):2227–31. https://doi.org/10.1002/jmv.26103 (Epub 2020 Jun 19).
Chen W, Lan Y, Yuan X, Deng X, Li Y, Cai X, et al. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg Microbes Infect. 2020;9(1):469–73. https://doi.org/10.1080/22221751.2020.1732837 (eCollection 2020).
Dheir H, Sipahi S, Yaylaci S, Genc AC, Genc FT, Genc AB, et al. Is the COVID-19 disease associated with de novo nephritic syndrome? Rev Assoc Méd Bras. 2020;66(9):1258–63. https://doi.org/10.1590/1806-9282.66.9.1258.
Ling Y, Xu S-B, Lin Y-X, Tian D, Zhu Z-Q, Dai F-H, et al. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J. 2020;133(9):1039–43. https://doi.org/10.1097/CM9.0000000000000774.
Dolz R, Vergara-Alert J, Pérez M, Pujols J, Majó N. New insights on infectious bronchitis virus pathogenesis: characterization of Italy 02 serotype in chicks and adult hens. Vet Microbiol. 2012;156(3–4):256–64. https://doi.org/10.1016/j.vetmic.2011.11.001 (Epub 2011 Nov 6).
Chacón RD, Astolfi-Ferreira CS, Chacón JL, Nuñez LF, David I, Ferreira AJP. A seminested RT-PCR for molecular genotyping of the Brazilian BR-I infectious bronchitis virus strain (GI-11). Mol Cell Probes. 2019;47:101426. https://doi.org/10.1016/j.mcp.2019.101426 (Epub 2019 Jul 28).
AlAli SY, AbdulRahman A, Yaghi O, Janahi E, AlQahtani M. SARS-Cov-2 viral load as an indicator for COVID-19 patients hospital stay. medRxiv. 2020. https://doi.org/10.1101/2020.11.04.20226365.
Rahman A, Al Ali S, Yaghi O, Otoom S, Atkin SL, Al-Qahtani M. The cycle of threshold measurement for COVID-19 infection and its implications in patient’s length of hospital stay. Saudi J Pathol Microbiol. 2020.
Fleming RMFM. Fleming inflammation and cardiovascular disease SARS-CoV-2 proposed treatment protocol. Initial COVID hydroxychloroquine failure responds to interferon α-2β and tocilizumab. J Clin Med Imag. 2020;5(3):1–3.
Fleming RM, Fleming MR. Preliminary results of tocilizumab and interferon a-2b treatment of SARS-CoV-2. Res Square. 2020. https://doi.org/10.21203/rs.3.rs-98566/v1.
Fleming RM, Fleming MR. FMTVDM quantitative nuclear imaging finds three treatments for SARS-CoV-2. Biomed J Sci Tech Res. 2021;33(4):26041–83. https://doi.org/10.26717/BJSTR.2021.33.005443.
Acknowledgements
We thank Prof. Dr. Reza Nassiri from Departments of Pharmacology and Toxicology, Michigan State University, USA, for comments that greatly improved the manuscript. The authors gratefully acknowledge the all personnel’s in the Gastrointestinal and Liver Disease Research Center and vice-chancellor for research of Guilan University of medical science and the staff at Razi Hospital.
Funding
None.
Author information
Authors and Affiliations
Contributions
FA: conception, literature search, study design, data collection, data analysis, data interpretation, writing and final approval; TYK, ST, LM, HAB, AJ, AP, EH: study design, data collection, writing and final approval; MK and MS and IJ: data collection, laboratory activities (PCR); FM-G: conception, literature search, data collection, data analysis, data interpretation, writing and final approval; MN: literature search, data collection, data interpretation, writing and final approval; SM: data analysis, data interpretation, writing and final approval; MA: data collection, literature search, writing and final approval. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was registered in the Research Department of Guilan University of Medical Sciences with the ethics code of IR.GUMS.REC.1399.013. This manuscript has not been published in whole or in part. All authors have read the manuscript and have agreed that the work is ready for submission and accept responsibility for its contents.
Consent for publication
Not applicable.
Competing interests
Authors have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Joukar, F., Yaghubi Kalurazi, T., Khoshsorour, M. et al. Persistence of SARS-CoV-2 RNA in the nasopharyngeal, blood, urine, and stool samples of patients with COVID-19: a hospital-based longitudinal study. Virol J 18, 134 (2021). https://doi.org/10.1186/s12985-021-01599-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-021-01599-9