Abstract
Here we described the virological and serological assessment of 23 COVID-19 patients hospitalized and followed up in Milan, Italy, during the first wave of COVID-19 pandemic. Nasopharyngeal (NPS), anal swabs, and blood samples were collected from 23 COVID-19 patients, at hospital admission, and periodically up to discharge, for a median time of 20 days (3–83 days). RNA was isolated and tested for SARS-CoV-2 by qRT-PCR; anti-SARS-CoV-2 IgM and IgG antibody titers were evaluated in serum samples by ELISA. SARS-CoV-2 genome was detected in the NPS swabs of the 23 patients, at the admission, and 8/19 (42.1%) were still positive at the discharge. Anal swabs were positive to SARS-CoV-2 RNA detection in 20/23 (86.9%) patients; 6/19 (31.6%) were still positive at discharge. The mean time of RNA negative conversion was 17 days (4–36 days) and 33 days (4–77 days), for NPS and anal swabs, respectively. SARS-CoV-2-RNA was detected in the blood of 6/23 (26.1%) patients. Thirteen/23 (56.5%) and 17/23 (73.9%) patients were seropositive for IgM and IgG, respectively, at the admission, and the median IgM and IgG levels significantly (p < 0.05) increased after 13 days. Although the limited cohort size, our report provides evidence that SARS-CoV-2 is shed through multiple routes, with important implications in healthcare settings.
Similar content being viewed by others
Background
Coronavirus disease 2019 (COVID-19) is a rapidly escalating world pandemic due to Severe Acute Respiratory Sindrome-CoronaVirus-2 (SARS-CoV-2) infection. SARS-CoV-2 was isolated for the first time at the beginning of 2020, at Wuhan Hospital, in China (Zhu et al. 2020). Italy, in particular Lombardy region, was one of the pandemic epicenters during the first wave (Villani et al. 2020).
The SARS-CoV-2 genome shows high sequence similarity with other coronaviruses, the Betacoronaviruses SARS-CoV, and Middle East Respiratory Syndrome-CoronaVirus (MERS-CoV) (Koyama et al. 2020). The positive single-strand RNA (ssRNA) is about 30 kbp, and the genome contains the open reading frames (ORFs), coding for the spike (S gene), envelope (E gene), membrane (M gene), and nucleocapsid (N gene) proteins, the ORF1ab polyproteins, and several accessory proteins, known as ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF9c, and ORF10 (Alanagreh et al. 2020).
As for the other human coronaviruses, SARS-CoV-2 is transmitted through respiratory droplets, as well as direct contact (Meselson 2020). However, emerging evidences showed that SARS-CoV-2 causes a systemic infection, due to the presence of the ACE-2 receptors in different body districts (Hikmet et al. 2020).
The gut involvement by SARS-CoV-2 (Jin et al, 2021), and its presence in the feces and in wastewater (Ahmad et al. 2021; Zhang et al. 2021), raises hypothesis about the role of fecal shedding in viral transmission. Additionally, several studies have sought SARS-CoV-2 RNA in blood, with variable results (Ahmed Moustafa 2020; Andersson et al. 2020; Lin et al. 2021; Novazzi et al. 2020; Peng et al. 2020; Sun et al. 2020; Wang et al. 2020; Zhang et al. 2020a, b; Loubaki et al. 2021).
Immune response to SARS-CoV-2 is characterized by humoral immunity, with the presence of both IgM and IgG antibodies, which are produced 6 to 15 days after the COVID-19 onset (Liu et al. 2020b): particularly, antibodies were detected in < 40% of COVID-19 patients after 1 week from the manifestation of symptomatology, and in the totality of the patients after 15 days (Zhao et al. 2020; Masiá et al. 2021).
The main purpose of this study was to describe the virological and serological assessment of 23 COVID-19 patients hospitalized in Milan, Italy, during the first epidemic wave and followed up to 83 days after the diagnosis.
Material and methods
Study design
This single-center prospective observational study was conducted on 23 patients hospitalized at the Istituto Clinico Città Studi (ICCS) hospital in Milan (Lombardy, Italy), in April and May 2020. Sex, age, diagnosis at the Emergency Room (ER) admission, report of gastrointestinal symptoms during the follow-up, and discharge type are summarized in Table 1. Nasopharyngeal swabs (NPS) were collected from each patient at the admission time (T0), whenever possible every 72 h, and at the discharge, for subsequent molecular SARS-CoV-2 tests. Diagnostic assessment of SARS-CoV-2 in NPS was conducted at the Department of Biomedical Science for Health, University of Milan, and the positive NPS confirmed the COVID-19 diagnosis in the 23 enrolled patients. Additionally, anal swabs and peripheral blood samples were collected at the admission (T0) and every 72 h. Serum samples were collected at the admission and, where possible, after 13 days. Clinical specimens were collected upon approval of the Local Ethical Committee and signature of the informed consent (Fondazione Ca’ Granda, Ospedale Maggiore, Milano, Italy approved the protocol 456_2020, on May 2020).
RNA isolation from clinical specimens
RNA was extracted from NPS, with the commercial method (QIAamp Viral RNA Mini kit, QIAGEN) following the manufacturer’s instructions, while it was isolated starting from 150 µL anal swab medium, using the NucleoSpin RNA virus kit (Macherey–Nagel, Germany), according to the manufacturer’s instructions.
Blood samples, previously stored in 700 µL QIAzol reagent (Qiagen, Germany), were thawed, mixed, and incubated for 5 min at room temperature. Then, 140 µL of chloroform were added, the tube was shaken vigorously for 15 s, and incubated for 3 min. The mixture was centrifuged at 12,000 g for 15 min at 4 °C. The aqueous upper phase, containing RNA, was transferred into a new tube, and the RNA isolation was performed according to the QIAamp RNA Blood Mini kit manufacturer’s protocol (Qiagen, Germany). RNA quality was evaluated using NanoPhotometer NP80 (Implen, Germany).
Real-time PCR for SARS-CoV-2 N1 gene amplification
SARS-CoV-2 N1 gene presence was assessed by means of AgPath-ID One-Step RT-PCR assay (ThermoFisher, USA), using the 7500 Real Time PCR system instrument (ThermoFisher, USA). The primer and probe sequences have been published by CDC (Table 2). Each sample was tested for SARS-CoV-2 positivity using the CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel for the qualitative detection of nucleic acid from the 2019-nCoV (N1 gene).
The standard curve was constructed by using a serially diluted plasmid pEX-A128-nCoV_all (Eurofins, Luxemburg), containing part of the SARS-CoV-2 genome (3 × 107–3 × 101 copies/μL). Samples were analyzed in duplicate, and a negative control, consisting of mix supplemented of water, was added. The limit of viral genome detection was evaluated as 3 copies/reaction.
PCR for SARS-CoV-2 RBD and ORF8 genomic regions
The Receptor Binding Domain (RBD, part of the S gene) and ORF8 sequences were amplified from the positive samples, using the one-Step RT-PCR kit (ThermoFisher, USA). The nucleotide sequence and the position of each set of primers are reported in Table 3. The reaction was performed in a final volume of 50 µL, composed as follows: 25 µL of 2 × Reaction mix, 2 µL of SuperScript III RT/Platinum Taq Mix (ThermoFisher, USA), 1 µL of Forward primer (10 µM), 1 µL of Reverse primer (10 µM), and a variable volume from 1 to 10 µL of RNA samples, depending on SARS-CoV-2 load. The PCR steps were as follows: 1 cycle at 50 °C for 15 min and at 95 °C for 2 min, followed by 40 cycle at 95 °C for 15 s; the annealing temperatures were 54 °C for RBD and 57 °C for ORF8 for 30 s, while the polymerization temperature was 68 °C for 40 s. Finally, the last step was conducted at 68 °C for 5 min. Right sizes of the RBD and ORF8 amplification products (lengths: 1000 bp, and 594 bp, respectively) were checked on 1.5% agarose gels in a 0.5 × TBE buffer, by electrophoresis. PCR products were purified using the QIAquick Gel Extraction Kit (QIAgen), following the manufacturer’s instructions. Automatic sequencing of the fragments was performed by an external facility (Eurofins Genomics, Germany), with the Sanger method. The homology of sequence was searched through the NCBI site (USA) using BLAST, comparing the amplified fragments to the reference Wuhan-Hu-1 strain (NC_045512.2). Multiple sequence alignment (MSA) of all nucleotide sequences were carried out in the EMBL-EBI Clustal Omega server to investigate sequence conservation (Sievers et al. 2011). The phylogenetic tree was constructed in the Interactive Tree of Life (iTOL) online tool (Ciccarelli et al. 2006).
Anti SARS-CoV-2 IgM and IgG detection
Anti SARS-CoV-2 IgM and IgG antibody titers were evaluated in serum samples using the MAGLUMI™ 2019-nCoV IgG and IgM kits on a MAGLUMI™ 800 instrument (Snibe, China), following manufacturers’ protocol. IgM and IgG serum titers were expressed as AU/mL, with manufacturer’s cutoff > 1 AU/mL.
Statistical analysis
Categorical and numerical variables were described using medians, confidence interval 95% (CI95), and interquartile range (IQR). Data were compared using Fisher’s exact test for SARS-CoV-2 prevalence’s and distributions or Mann–Whitney U test for SARS-CoV-2 viral loads as appropriate. Significances were defined as p-value < 0.05. Viral load at follow-up was quantified considering, for patients with multiple measurements, the last available. For anal swabs, multiple measurements were available for 15 patients and the log geometric means were reported at each follow-up time.
Results
Study population
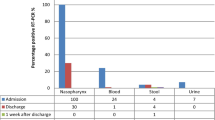
Twenty-three patients were enrolled in the study. Median age of patients was 67 years (range: 43–89); 52.2% (12/23) were female. Twelve/23 (52.2%) patients have pneumonia, 9/23 (39.1%) patients had respiratory failure, and 1/23 (4.3%) had undefined pneumonia. Patients were followed up from the admission to the discharge, for a median time of 20 days (range: 3–83 days). A careful evaluation of the possible presence of gastrointestinal disorders and symptoms was conducted by evaluating the daily medical and nursing diary of each patient. The gastrointestinal disorders taken into consideration were alvus disorders (diarrhea and constipation), nausea, loss of appetite, and epigastralgia. Sixteen/23 (69.6%) patients did not present any gastrointestinal symptoms during the hospitalization time; 4/23 (17.4%) had constipation and nausea for a few days, while 3/23 (13.0%) suffered of diarrhea. The discharge was due to transfer to other quarantine structures (12/23, 52.2%), to remission (7/23, 30.4%), or to death (4/23, 17.4%). Among the transferred patients, 6/12 (50%) were still SARS-CoV-2 positive at the time of the discharge, and were lost to the follow up (Table 1; and S1).
SARS-CoV-2 prevalence in NPS
The analytical description of the results obtained analyzing the clinical specimens longitudinally collected from the patients is summarized in Tables S1, S2, and S3 in the supplementary material section.
COVID-19 diagnosis was confirmed in all the enrolled patients by the SARS-CoV-2 positivity of the NPS collected at the hospital admission. Four patients deceased, so the prevalence at the discharge was evaluated on 19 patients. Eleven/19 (57.9%, CI95 22%) had a negative NPS at the discharge; the mean time of nucleic acid negative conversion was 33 days (range: 4–77 days). Interestingly, three patients showed alternance of results in NPS during the follow-up.
SARS-CoV-2 prevalence in anal swabs
SARS-CoV-2 RNA was detected in the anal swabs of 20/23 (86.9%, CI95 14%) patients: 7/23 (30.4%, CI95 18%) patients showed SARS-CoV-2 positivity at T0, 10/23 (43.5%, CI95 20%) became SARS-CoV-2 positive during the follow up, and 6/19 (31.6%, CI95 21%) were still positive at the discharge. The analytical description of the results in each anal swab is summarized in Tables S1, S2, and S3 in the supplementary material section. The mean time of nucleic acid negative conversion was 17 days (range: 4–36 days). Of note, 7/23 (30.4%) patients experienced gastrointestinal symptoms; all but one showed SARS-CoV-2 anal swab positivity, and 2 of them deceased.
SARS-CoV-2 prevalence in blood
SARS-CoV-2-RNA was present in the blood of 4/23 (17.4%, CI95 15%) patients at the admission, while 2/23 (8.7%, CI95 12%) were SARS-CoV-2-RNA during the follow-up, and only one (1/19, 5.3%, CI95 9.8%) was still positive at the discharge. Of note, SARS-CoV-2-RNA was found in both anal swab and blood of 6/23 (26.0%, CI95 17.8%) patients; 2 of them deceased.
SARS-CoV-2 viral load in biological samples
At the time of admission, geometric mean of viral loads were 1.58 × 105 copies/mL (95%CI: 4.84 × 104–5.18 × 105 copies/mL), 2.99 × 102 copies/mL (95%CI: 1.37 × 102–6.53 × 102 copies/mL), and 1.88 × 102 copies/mL (95%CI: 4.12 × 10–85.66 × 102 copies/mL) in NPSs, anal swabs and blood samples, respectively.
Viral load analysis during the follow up showed the following geometric mean values: 1.17 × 104 copies/mL (95%CI: 1.7 × 103–8.10 × 104 copies/mL), 3.53 × 102 copies/mL (95%CI: 1.46 × 102–8.55 × 102 copies/mL), and 1.50 × 102 copies/mL (95%CI: 2.88–7.81 × 103 copies/mL) in NPSs, anal swabs, and blood samples, respectively. The SARS-CoV-2 load in anal swabs at each follow-up time is reported in Fig. 1. Analytical description of viral load of the NPSs and anal swabs collected from each patient during the follow-up is shown in Fig. 1S, and in Table 4S.
Description of SARS-CoV-2 load in anal swabs collected during the time of hospitalization from each patient. The solid black line shows the log geometric mean calculated on patients with positive tests. The calculation was performed if at least two patients showed positivity. The two dashed black lines are the 95% confidence interval of the log geometric mean. The red squares represent the percentage of patients testing positive out of the number tested (reported immediately above the x axis)
SARS-CoV-2 ORF8 and RBD sequence analysis
In order to confirm the SARS-CoV-2 detection, sequence analysis of the SARS-CoV-2 RBD and ORF8 genomic regions was conducted on 7 and 3 RNA, respectively, isolated from anal swabs. The RBD sequences showed high homology with the Wuhan-Hu-1 strain (accession numbers NC_045512.2, YP_009724390.1), and the phylogenetic tree is shown in Fig. 2, while the three analyzed ORF8 sequences showed 100% homology with Wuhan-Hu-1 strain.
Anti-SARS-CoV-2 IgM and IgG titers
Anti-SARS-CoV-2 IgM and IgG titers were evaluated at the admission, and where possible (18/23 patients, 78.3%) after 13 days. A total of 13/23 (56.5%) and 17/23 (73.9%) patients were seropositive for IgM and IgG, respectively, at the admission; in particular, 11 patients presented both IgM and IgG against SARS-CoV-2 in the serum. The concomitant positivity was maintained up to the day 13 in all the 11 patients; the two patients showing only IgM at T0 showed a simultaneously IgM and IgG positivity at the subsequent follow-up; finally, among the 6 patients who tested IgG positive at the T0, four showed a simultaneous IgM and IgG positivity after 13 days. The median and geometric mean IgM and IgG level were 1.83 AU/mL (range: 0.11–14.10 AU/mL), 1.59 AU/ml (95%CI: 0.79–3.22), and 6.70 AU/mL (range: 0.06–42.44 AU/mL), 4.70 AU/ml (95%CI: 2.03–10.90) at the admission, and significantly increased up to median values of 10.97 AU/mL (range: 0.75–30.00 AU/mL, IQR 25.14), 9.88 AU/ml (95%CI: 5.80–16.83) and 34.01 AU/mL (2.00–45.00 AU/mL), and 24.19 AU/ml (95%CI: 16.41–35.67) after 13 days, respectively (p < 0.01 for IgM and p = 0.01 for IgG). Analytical description of IgG and IgM levels in the serum collected from each patient, at T0 and after 13 days, is shown in Fig. 3.
Discussion
Here, we described the virological and serological assessments of 23 COVID-19 patients admitted at the Istituto Clinico Città Studi, a general hospital, located in Milan, center city, Lombardy, Italy, during the first epidemic wave, providing additional evidence on SARS-CoV-2 shedding.
The focus was made on respiratory, fecal, and blood samples, collected during the patients’ hospitalization, until discharge. SARS-CoV-2 genome was found in all the clinical specimens, with similar frequencies in NPS and anal swabs, but much lower frequency in blood samples. As expected, the viral load was significantly higher in the NPS than in the other specimens (Li et al. 2020a).
Transmission of SARS-CoV-2 can occur through direct, indirect, or close contact with infected people, through infected secretions such as saliva and respiratory secretions or their respiratory droplets, which are expelled when an infected person coughs, sneezes, talks, or sings (Liu et al. 2020a). However, different possible modes of transmission for SARS-CoV-2, including contact, droplet, airborne, fomite, fecal–oral, bloodborne, mother-to-child, and animal-to-human transmission, have been proposed. Growing evidence has revealed positive detection of viral RNA in anal swabs and fecal samples of patients with COVID-19, at different rates (Novazzi et al. 2020; Wu et al. 2020a; Zhang et al. 2020b). The presence of SARS-CoV-2 RNA in feces samples seemed to peak later (2–5 days), than in NPS swabs, and to persist longer than in respiratory specimens (Jiang et al. 2020; Lin et al. 2021; Lyu et al. 2021; Prakash et al. 2021; Tang et al. 2020; Xing et al. 2020). However, although the frequent presence of SARS-CoV-2 RNA in fecal or rectal swabs, evidence of infectious viruses shedding in feces are limited, and viable virus particles have been rarely reported in feces-related specimens from COVID-19 patients (Xiao et al. 2020), so that some authors are questioning whether the SARS-CoV-2 fecal–oral transmission may be plausible (Pedersen et al. 2021).
In our study, the frequencies of SARS-CoV-2 RNA detection in anal swabs (86.9%) and of the gastrointestinal symptoms (30.4%) reported in the enrolled patients were greater compared to what observed in other published studies (Zhang et al. 2021). These disparate values may be attributed to the limited number of patients enrolled in our study, or to the sampling time, or the sample type across studies, or the severity of the COVID-19 affecting our patients, and requiring hospitalization of the patients. The persistent positivity was similar in respiratory and feces-related samples, but the median time of negativization was higher in anal than in NPS swabs, and within the average durations, that have been previously reported (Cheung et al. 2020; Wu et al. 2020b). Long-term post-discharge follow-up would be warranted to evaluate the risk of transmission, due to routes different from the respiratory one, and also in the absence of respiratory symptoms, as proposed by Lyu and colleagues (Lyu et al. 2021).
Interestingly, independent studies conducted on young mild symptomatic subjects reported SARS-CoV-2 detection in anal swabs, but not in other clinical specimens from the same patients (Peng et al. 2020; Sun et al. 2020; Zhang et al. 2020a). To this regard, during the same frame of time, four patients admitted at our hospital, but not included in the present study, because with SARS-CoV-2 negative NPSs, showed persistence viral positivity in the anal swabs. About one-third of patients with SARS-CoV-2 positive anal swabs showed gastrointestinal symptoms, such as nausea and diarrhea, and two of them deceased.
Although based on a limited number of patients, our results corroborated the numerous reports of SARS-CoV-2 genome finding in wastewater all over the world (Patel et al. 2021), and the detection of viral genomes in wastewater increasing with the number of COVID-19-related hospitalizations, (Medema et al. 2020), but did not give any information about the infectivity of the shedding viruses, that were not cultured.
However, based on the evident and frequent replication of the virus in the GI tract, the fecal–oral route should be considered as a potential transmission route for SARS-CoV-2, especially in specific environments. Particular attention should be paid in healthcare settings, where COVID-19 patients are forced to share the facilities or to stay in the bed constantly, without the possibility of using the bathroom, but with the continuous use of diapers. The repeated use of shared toilets, together with the infrequent cleaning, might result in progressive accumulation of virus, on the toilet seats, sink, and door handle (Amoah et al. 2021; Chia et al. 2020; Ding et al. 2021; Li et al. 2020b; Ong et al. 2020), where the virus may persist for up to 4 days (Chin et al. 2020). Additionally, the risk of SARS-CoV-2 fecal contamination may be much higher in poor-setting countries, characterized by poor sanitation (Jones et al. 2020).
Besides the positivity in the NPS and anal swabs, about 26% of the patients enrolled in our study, tested positive in the blood samples, with viral loads comparable to that reported in the anal swabs, and persistence of viremia in one patient. A systematic review of the literature identified that the blood from up to 76% of COVID-19 patients, in critical care settings, may present detectable SARS-CoV-2 RNA. However, in the majority of the cases, SARS-CoV-2 viremia was not detected, and the load was so low that it was outside the threshold, usually reported as positive in diagnostic tests (Andersson et al. 2020, L). Nonetheless, as underlined by Andersson and colleagues, the recognition that SARS-CoV-2 RNA may be detected and quantified in blood raised concerns about the safety for personnel handling samples in clinical and research environments (Andersson et al. 2020; Loubaki et al. 2021).
Finally, IgM and IgG serum titres were evaluated in the serum samples of each patient at the hospital admission, and after 13 days of hospitalization. Our results showed that the most patients were seropositive for IgM and/or IgG, already at the admission, due to the fact that hospitalization probably occurred some days after the onset of the infections. As expected, the antibody titers significantly increased over the time, reaching the almost complete seroconversion to IgG, about 17–19 days after symptoms onset. This is in line with reports from the literature, concluding that the rate and timing of IgM and IgG seroconversion were inconsistent across the independent analyzed studies, but most seroconverted for IgG within 2 weeks and 100% within 4 weeks (O Murchu et al. 2020; Yadav et al. 2021).
Conclusions
The main limit of the described study was given by the small cohort size, and by the severe health conditions of the patients, requiring the hospitalization. This implies that the study gave a picture of the SARS-CoV-2 shedding in the most severe cases of COVID-19, and did not reflect the heterogeneity of the mild and asymptomatic infected population.
However, it should be taken into account that the collection of biological samples was carried out from consecutively enrolled patients, at the very beginning of the pandemic. Unfortunately, the hospital in which the patients were recruited was immediately overwhelmed by the unexpected pandemic wave, with unimaginable numbers of hospitalized patients in serious or critical conditions. This event put a strain on the work of doctors and health personnel who could not continue with the systematic collection of biological samples, and who had to limit the collection to nasopharyngeal swabs, obviously essential from a diagnostic point of view.
Although based on limited numbers of subjects and samples, these findings could be helpful for corroborating the model proposed by Sun and colleagues, stating that SARS-CoV-2 infection course could start in the upper respiratory infection (mainly by respiratory droplets), continue in the lower respiratory infection (mainly presented as pulmonary infection), followed by viremia and diffusion to other target organs (including the gastro intestinal tract) (Sun et al. 2020). Furthermore, our results might also be of support for the idea that the spread of SARS-CoV-2 in hospitals and elderly residences, where patients are often constantly bedridden and are not autonomous in the management of their stools, could occur through fecal contamination, involving healthcare personnel in the infection, who usually is able to protect themselves from droplets, wearing masks, but less easily during the direct care and hygiene of patients. Same attentions should be paid when gastrointestinal tract diagnostic or surgical procedures are performed (Deidda et al. 2021), and when stools or vomitus samples are handled. This implies (a) the mandatory use of Personal Protective Equipments (PPE), including disposable coat, mask, gloves, glasses or face shield, and hair cap by healthcare personnel; (b) the promotion and enforcing of hand-hygiene measures, among health personnel and patients; (c) the definition of protocols for the disinfection and decontamination of the surfaces, the room, and the equipments, based on the use of sodium-hypochlorite or similar disinfectants; (d) the definition of strict protocols for the medical procedures, that are considered to be at risk of infection; and (e) when possible, the definition of a washroom design, providing limitation of the contamination risks (i.e., adequate ventilation, non-touch flush buttons, automatic doors) (Vardoulakis et al. 2021).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article (and its additional file).
References
Ahmad J, Ahmad M, Usman ARA, Al-Wabel MI (2021) Prevalence of human pathogenic viruses in wastewater: a potential transmission risk as well as an effective tool for early outbreak detection for COVID-19. J Environ Manage 298:113486
Ahmed Moustafa RKA (2020) Traces of SARS-CoV-2 RNA in the blood of COVID-19 patients. medRxiv
Alanagreh L, Alzoughool F, Atoum M (2020) The human coronavirus disease COVID-19: its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens 29;9(5):331
Amoah ID, Pillay L, Deepnarian N, Awolusi O, Pillay K, Ramlal P, Kumari S, Bux F (2021) Detection of SARS-CoV-2 RNA on contact surfaces within shared sanitation facilities. Int J Hyg Environ Health 236:113807
Andersson MI, Arancibia-Carcamo CV, Auckland K, Baillie JK, Barnes E, Beneke T, Bibi S, Brooks T, Carroll M, Crook D, Dingle K, Dold C, Downs LO, Dunn L, Eyre DW, Gilbert Jaramillo J, Harvala H, Hoosdally S, Ijaz S, James T, James W, Jeffery K, Justice A, Klenerman P, Knight JC, Knight M, Liu X, Lumley SF, Matthews PC, McNaughton AL, Mentzer AJ, Mongkolsapaya J, Oakley S, Oliveira MS, Peto T, Ploeg RJ, Ratcliff J, Robbins MJ, Roberts DJ, Rudkin J, Russell RA, Screaton G, Semple MG, Skelly D, Simmonds P, Stoesser N, Turtle L, Wareing S, Zambon M (2020) SARS-CoV-2 RNA detected in blood products from patients with COVID-19 is not associated with infectious virus. Wellcome Open Res 5:181
BLAST: Basic local alignment search tool. https://blast.ncbi.nlm.nih.gov/Blast.cgi. Accessed in September 2020
Centers for Disease Control and Prevention. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html. Accessed in March 2020
Cheung KS, Hung IFN, Chan PPY, Lung KC, Tso E, Liu R, Ng YY, Chu MY, Chung TWH, Tam AR, Yip CCY, Leung KH, Fung AY, Zhang RR, Lin Y, Cheng HM, Zhang AJX, To KKW, Chan KH, Yuen KY, Leung WK (2020) Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and Meta-analysis. Gastroenterology 159:81–95
Chia PY, Coleman KK, Tan YK, Ong SWX, Gum M, Lau SK, Lim XF, Lim AS, Sutjipto S, Lee PH, Son TT, Young BE, Milton DK, Gray GC, Schuster S, Barkham T, De PP, Vasoo S, Chan M, Ang BSP, Tan BH, Leo YS, Ng OT, Wong MSY, Marimuthu K, Team SNCOR (2020) Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nat Commun 11:2800
Chin AWH, Chu JTS, Perera MRA, Hui KPY, Yen HL, Chan MCW, Peiris M, Poon LLM (2020) Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe 1:e10
Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P (2006) Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287
Deidda S, Tora L, Firinu D, Del Giacco S, Campagna M, Meloni F, Orrù G, Chessa L, Carta MG, Melis A, Spolverato G, Littera R, Perra A, Onali S, Zorcolo L, Restivo A (2021) Gastrointestinal coronavirus disease 2019: epidemiology, clinical features, pathogenesis, prevention, and management. Expert Rev Gastroenterol Hepatol 15:41–50
Ding Z, Qian H, Xu B, Huang Y, Miao T, Yen HL, Xiao S, Cui L, Wu X, Shao W, Song Y, Sha L, Zhou L, Xu Y, Zhu B, Li Y (2021) Toilets dominate environmental detection of severe acute respiratory syndrome coronavirus 2 in a hospital. Sci Total Environ 753:141710
Hikmet F, Méar L, Edvinsson Å, Micke P, Uhlén M, Lindskog C (2020) The protein expression profile of ACE2 in human tissues. Mol Syst Biol 16:e9610
Jiang X, Luo M, Zou Z, Wang X, Chen C, Qiu J (2020) Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J Med Virol 92:1807–1809
Jin B, Singh R, Ha SE, Zogg H, Park PJ, Ro S (2021) Pathophysiological mechanisms underlying gastrointestinal symptoms in patients with COVID-19. World J Gastroenterol 27:2341–2352
Jones DL, Baluja MQ, Graham DW, Corbishley A, McDonald JE, Malham SK, Hillary LS, Connor TR, Gaze WH, Moura IB, Wilcox MH, Farkas K (2020) Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci Total Environ 749:141364
Koyama T, Platt D, Parida L (2020) Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ 98:495–504
Li H, Ren L, Zhang L, Wang Y, Guo L, Wang C, Xiao Y, Rao J, Wang X, Liu Y, Huang C, Gu X, Fan G, Lu B, Cao B, Wang J (2020a) High anal swab viral load predisposes adverse clinical outcomes in severe COVID-19 patients. Emerg Microbes Infect 9:2707–2714
Li YH, Fan YZ, Jiang L, Wang HB (2020b) Aerosol and environmental surface monitoring for SARS-CoV-2 RNA in a designated hospital for severe COVID-19 patients. Epidemiol Infect 148:e154
Lin W, Xie Z, Li Y, Li L, Wen C, Cao Y, Chen X, Ou X, Hu F, Li F, Tang X, Cai W (2021) Association between detectable SARS-COV-2 RNA in anal swabs and disease severity in patients with coronavirus disease 2019. J Med Virol 93:794–802
Liu J, Liao X, Qian S, Yuan J, Wang F, Liu Y, Wang Z, Wang FS, Liu L, Zhang Z (2020a) Community transmission of severe acute respiratory syndrome coronavirus 2, Shenzhen, China, 2020. Emerg Infect Dis 26:1320–1323
Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, Wang Q, Tan L, Wu W, Tang S, Xiong Z, Zheng S (2020b) Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol 58(6):e00461-e520
Loubaki L, Gantner P, Pagliuzza A, Fausther-Bovendo H, Kobinger G, Chomont N, Germain M (2021) Testing for the presence of SARS-CoV-2 RNA in presymptomatic blood donors. Transfusion 61(2):649–651
Lyu Y, Wang D, Li X, Gong T, Xu P, Liu L, Sun J (2021) Continued nucleic acid tests for SARS-CoV-2 following discharge of patients with COVID-19 in Lu’an. China. Peerj 9:e11617
Masiá M, Telenti G, Fernández M, García JA, Agulló V, Padilla S, García-Abellán J, Guillén L, Mascarell P, Asenjo JC, Gutiérrez F (2021) SARS-CoV-2 seroconversion and viral clearance in patients hospitalized with COVID-19: viral load predicts antibody response. Open Forum Infect Dis 8(2):ofab005
Medema GHL, Elsinga G, Italiaander R, Brouwer A (2020) Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ Sci Technol Lett 7(7):511–516
Meselson M (2020) Droplets and aerosols in the transmission of SARS-CoV-2. N Engl J Med 382:2063
Novazzi F, Cassaniti I, Piralla A, Di Sabatino A, Bruno R, Baldanti F (2020) SARS-CoV-2 positivity in rectal swabs: implication for possible transmission. J Glob Antimicrob Resist 22:754–755
O Murchu E, Byrne P, Walsh KA, Carty PG, Connolly M, De Gascun C, Jordan K, Keoghan M, O’Brien KK, O’Neill M, Smith SM, Teljeur C, Ryan M, Harrington P (2020) Immune response following infection with SARS-CoV-2 and other coronaviruses: A rapid review. Rev Med Virol 31(2):e2162
Ong SWX, Tan YK, Chia PY, Lee TH, Ng OT, Wong MSY, Marimuthu K (2020) Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA 323:1610–1612
Patel M, Chaubey AK, Pittman CU, Mlsna T, Mohan D (2021) Coronavirus (SARS-CoV-2) in the environment: Occurrence, persistence, analysis in aquatic systems and possible management. Sci Total Environ 765:142698
Pedersen RM, Tornby DS, Bang LL, Madsen LW, Skov MN, Jensen TG, Johansen IS, Andersen TE (2021) Rectally shed SARS-CoV-2 lacks infectivity: time to rethink faecal-oral transmission? Nat Rev Gastroenterol Hepatol 18:669
Peng L, Liu J, Xu W, Luo Q, Chen D, Lei Z, Huang Z, Li X, Deng K, Lin B, Gao Z (2020) SARS-CoV-2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J Med Virol 92:1676–1680
Prakash S, Shukla S, Mishra H, Prakash O, Khan DN, Pandey A, Reddy DH, Jain A (2021) SARS-CoV-2 -RNA persists longer in faecal sample as compared to nasal and throat swab samples of COVID-19 patients’; an observational study. Indian J Med Microbiol 39:122–124
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539
Sun M, Guo D, Zhang J, Teng HF, Xia J, Liu P, Ge QX, Wang MY (2020) Anal swab as a potentially optimal specimen for SARS-CoV-2 detection to evaluate hospital discharge of COVID-19 patients. Future Microbiol 15:1101–1107
Tang A, Tong ZD, Wang HL, Dai YX, Li KF, Liu JN, Wu WJ, Yuan C, Yu ML, Li P, Yan JB (2020) Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg Infect Dis 26:1337–1339
Vardoulakis S, Espinoza Oyarce DA, Donner E (2021) Transmission of COVID-19 and other infectious diseases in public washrooms: a systematic review. Sci Total Environ 803:149932
Villani L, McKee M, Cascini F, Ricciardi W, Boccia S (2020) Comparison of deaths rates for COVID-19 across Europe during the first wave of the COVID-19 pandemic. Front Public Health 8:620416
Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W (2020) Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323:1843–1844
Wu J, Liu J, Li S, Peng Z, Xiao Z, Wang X, Yan R, Luo J (2020a) Detection and analysis of nucleic acid in various biological samples of COVID-19 patients. Travel Med Infect Dis 37:101673
Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X, Yin H, Xiao Q, Tang Y, Qu X, Kuang L, Fang X, Mishra N, Lu J, Shan H, Jiang G, Huang X (2020b) Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol 5:434–435
Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H (2020) Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 158:1831-1833.e3
Xing YH, Ni W, Wu Q, Li WJ, Li GJ, Wang WD, Tong JN, Song XF, Wing-Kin Wong G, Xing QS (2020) Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. J Microbiol Immunol Infect 53:473–480
Yadav AK, Ghosh S, Kotwal A, Kaushik SK, Bobdey S, Sahu R, Kapoor S, Faujdar DS, Teli PT, Anand V (2021) Seroconversion among COVID-19 patients admitted in a dedicated COVID hospital: a longitudinal prospective study of 1000 patients. Med J Armed Forces India 77(Suppl 2):S379–S384
Zhang B, Liu S, Dong Y, Zhang L, Zhong Q, Zou Y, Zhang S (2020a) Positive rectal swabs in young patients recovered from coronavirus disease 2019 (COVID-19). J Infect 81:e49–e52
Zhang W, Du RH, Li B, Zheng XS, Yang XL, Hu B, Wang YY, Xiao GF, Yan B, Shi ZL, Zhou P (2020b) Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect 9:386–389
Zhang Y, Cen M, Hu M, Du L, Hu W, Kim JJ, Dai N (2021) Prevalence and persistent shedding of fecal SARS-CoV-2 RNA in patients with COVID-19 infection: a systematic review and meta-analysis. Clin Transl Gastroenterol 12:e00343
Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, Qian S, Hong C, Wang F, Liu Y, Wang Z, He Q, Li Z, He B, Zhang T, Fu Y, Ge S, Liu L, Zhang J, Xia N, Zhang Z (2020) Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin Infect Dis 71:2027–2034
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W (2020) A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733
Acknowledgements
All the authors would like to remember their beloved and esteemed colleague, Mrs. Loredana Cavicchini, who devoted all her work time to the research, and lately, to the COVID-19 research. She was one of the young victims of SARS-CoV-2, and this paper is dedicated to her. The authors also would like to thank all the medical and healthcare staff of the Istituto Clinico Città Studi, who have been working with dedication, ability, enthusiasm, and spirit of sacrifice since the beginning of the pandemic.
Funding
This project was partially supported by MIUR, grant PRIN 2017 to PF, and by the Italian Ministry of Health, grant COVID-2020–12371849 to SD.
Author information
Authors and Affiliations
Contributions
Conceptualization: P.F., E.P., and S.D.; methodology: M.D., L.S, L.C., F.A., and C.G.; investigation: L.S., M.D., R.T., S.P., B.I., and N.B.; resources: P.B., N.C., P.F. and S.D.; data curation: S.B., E.P., A.M., and P.F.; writing—original draft preparation: L.S., and M.D.; writing—review and editing: P.F., S.D., E.P. N.B., F.A. and S.P.; visualization: L.S., and M.D..; supervision: S.D., P.F., N.B.; funding acquisition: S.D., and P.F. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Clinical specimens were collected upon approval of the Local Ethical Committee and signature of the informed consent (Fondazione Ca’ Granda, Ospedale Maggiore, Milano, Italy, approved the protocol 456_2020, on May 2020).
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Signorini, L., Dolci, M., Castelnuovo, N. et al. Longitudinal, virological, and serological assessment of hospitalized COVID-19 patients. J. Neurovirol. 28, 113–122 (2022). https://doi.org/10.1007/s13365-021-01029-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13365-021-01029-0