Abstract
Aim
Inbreeding is thought to affect the morphometric parameters leading to lower health status among the progeny. The present study was aimed to investigate the repercussions of inbreeding on anthropometric traits, namely height, weight and body mass index (BMI).
Subjects and methods
The survey was conducted in two North Indian cities and total 813 individuals were randomly recruited from inbred and outbred families. The morphometric parameters of the subjects were measured using standard methods, BMI was calculated and categorized into underweight, normal weight and overweight. Family pedigrees were drawn and degree of inbreeding in terms of the inbreeding coefficient (F) was calculated.
Results
A significant decline in morphometric measures was observed among inbred individuals as compared to outbred ones. The mean differences (95% CI) were found significant for various inbred and first cousin categories as compared to outbred subjects (p < 0.05). We found the increased frequency of underweight individuals corresponding to the degree of inbreeding for different types of inbred categories (p = 2.086 × 10−9) and also for different subtypes of first cousin unions corresponding to their sex-linked inbreeding coefficients (p = 5.2 × 10−5). The regression slope and correlation coefficient revealed a fitness decline and depression in anthropometric measures (p < 0.05) with the increase in ‘F’ for all inbred groups and first cousin categories.
Conclusions
The present research confirms the adverse effects of inbreeding on morphometric parameters among inbred subjects. It has novelty in shedding light on the hitherto unreported differences in the consequences of inbreeding among different types of first cousin unions.
Highlights
-
Consanguinity acts as a risk factor among humans resulting in poor health status.
-
This empirical study deals with the effect of inbreeding on anthropometric measures.
-
Significant decrease in morphometric traits was observed among inbred individuals.
-
The risk for poor health status was found proportional to the degree of inbreeding.
-
We also reported the differences in inbreeding effects for various first cousin unions for the very first time.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
In epidemiologic investigations, height and weight are often used metrics to compute body mass index (BMI, kg/m2) as a surrogate measure for excess body fatness, which is a significant risk factor for cancer, cardiovascular disease, and overall mortality [1,2,3,4,5]. Due to its simplicity of collection and cheap cost, height and weight data is commonly self-reported on questionnaires in extensive prospective cohort studies. The accessibility and affordability of BMI has made it a straightforward method for tracking population health in the context of various hereditary or environmental factors [6]. At the same time, the waist, hip, and neck circumferences are additional anthropometric measures that are used to help in determining total weight and the distribution of body fat [7,8,9]. The height or stature of an individual itself is a quantitative and highly heritable complex trait, with 80 to 90% of the variation possibly resulting from genetic causes [6, 10]. Genome wide association study (GWAS) conducted on 5.4 million individuals from different ancestries showed 12,111 independent single nucleotide polymorphisms (SNPs) associated significantly with height, accounting for nearly all of the common SNP-based heritability [11].
The majority of research on height, weight, and BMI put a strong emphasis on social and economic variables. Childhood and teenage obesity are independently correlated with race/ethnicity, socio-economic status (SES), and psychological factors [6, 12]. A handful of studies have suggested that occupational or physical activity, socio-demographic characteristics, age, gender, environmental, and genetic factors may all influence BMI [6, 13,14,15,16]. BMI is a helpful tool for evaluating children’s growth and development and a recognized risk factor for numerous illnesses. A plethora of epidemiological researches shown that greater BMI is a significant risk factor for type II diabetes, hypertension [17,18,19], hypercholesterolemia [18], dyslipidaemia [20], mortality [17, 21,22,23]. It has also been reported that increased BMI has association with cardiovascular and cardio-metabolic disorders [20, 24], increased waist-to-hip ratio, increased muscle mass [18], osteoporosis, asthma, osteoarthritis, gout, bladder diseases, pancreatitis, dementia and non-alcoholic fatty liver disease [25]. In addition, many types of cancers such as, endometrial, breast, colon, prostate, kidney and bladder cancer have found to be linked with high BMI [18, 25, 26].
The nuptials between first cousins, second cousins, double first cousins, double second cousins, uncle-niece/aunt-nephew relationships or persons with shared biological ancestors is called inbreeding or consanguinity [27,28,29]. Consanguinity is quite common (approximately 20%) in various cultures and groups throughout the world, having the highest rates in the Middle East (20–50%), often exceeds 80% in certain regions [6, 30, 31]. In many cultures, these partnerships appear to bring about social and economic benefits, such as strengthening family bonds, increasing marital stability, and providing better support for the female partner [32,33,34,35]. The practice of inbreeding, endogamy, or consanguinity upsurges the homozygosity for autosomal recessive genetic disorders. Consequently, leading to decreased biological fitness of the population, called inbreeding depression (ID), which has become the primary focus of scientists in clinical studies [6, 36, 37].
The spouses belonging to first-degree cousins share 1/8th (12.5%) of their genes inherited from a common ancestor, so their progeny will be homozygous at 1/16th of all loci, signifying that they will inherit identical gene copies from both parents at these genomic sites. These extensive runs of homozygosity (ROH), which are identical by descent (IBD) or autozygosity, are dispersed across the genomes of descendants [38,39,40]. Therefore, consanguineous couples are at least ten times more likely to experience genetic diseases than non-consanguineous ones, involving many novel genetic syndromes and variations like, autosomal recessive disorders as well as X-linked traits [29, 34]. Consanguinity is linked to various chromosomal and microdeletion diseases, such as Prader-Willi syndrome (15q11-q13 deletion). It seems to reduce the ovarian reserve of female offspring and increase the frequency of severe male factor infertility, both of which appear to affect future generations’ ability to conceive [41,42,43,44]. According to the most recent research, consanguineous unions seem to raise the chance of producing a kid with genetic problems and the likelihood of certain chromosomal abnormalities, such as, higher rates of segmental aneuploidies [34]. These unions are also responsible for increasing the burden of disease carriers of speech and language-related disorders. A recent study has reported that short length ROHs are rich in unrelated individuals of consanguineous Pakistani population having no history of speech and language-related impairments [45]. A linkage scan from a Pakistani consanguineous family revealed significant linkage of stuttering with chromosome 3q13.2–3q13.33 under an autosomal recessive model of inheritance [46]. In an isolated Chilean population, genome-wide approach identified loci, possibly containing genetic variants underlying susceptibility to specific language impairment [47]. There are numerous reports suggesting the co-occurrence of more than one rare genetic disorders more commonly among consanguineous families, causing a variety of disease manifestations. For instance, homozygous pathogenic polymorphisms in ATP7B and CNGA1 were shown to be responsible for the two phenotypes in a Chinese patient who had both Wilson disease and retinitis pigmentosa [31, 48, 49].
Despite the involvement in diseases, inbreeding can also lead to depression in certain human traits such as cognitive behavior, growth parameters, lung function, and mortality [6, 10, 36, 37, 50,51,52,53]. It is well known that the detrimental consequences of inbreeding on the progeny increase as the degree of consanguinity (inbreeding coefficient, F) increase [6, 32, 36, 37, 54]. According to a recent study, there are disparities in the homozygosity patterns of four first-cousin union subtypes based on the differences in their sex-linked inbreeding coefficients (Fs) [39]. However, no research has been undertaken to compare the negative impacts of inbreeding among the four subtypes of first-cousin relationships. In light of this, the present study aimed to investigate the effects of inbreeding on physical characteristics or anthropometric measurements among individuals belonging to various types of consanguineous unions as well as multiple subtypes of first cousin marriages in comparison to outbred individuals from the northern part of the Indian subcontinent.
2 Methodology
2.1 Study subjects and data collection
A door-to-door survey was conducted from July 2018 through December 2021, from two North Indian cities, i.e., Aligarh (North–West district) and Ambedkar Nagar (North-east district), and the necessary information (Age, Gender, Caste, Locality, Religion etc.) were recorded using a standard questionnaire (Supplementary Fig. 1). From the Muslim populations of the mentioned area, we randomly recruited 813 healthy (not suffering any type of disease or health condition) individuals ranging from 6 to 64 years of age. Subjects who were diseased, suffering from congenital disabilities, did not attend school, were below or above the mentioned age, who withdrew from the test, who were not permitted by the guardians (in the case of children), or mentioned time constraints, were excluded.
2.2 Parental background assessment and pedigree analysis
The background of the individuals was assessed in terms of the residence (rural or urban or sub-urban) and the socio-economic status (SES). SES of the participants was measured by variables such as income, education, and occupation by employing the revised version of native Kuppuswamy’s socio-economic scale [55] (Supplementary Table 1). Based upon the scores in each variable, the categorization of SES was done into five classes: 26–29 as Upper, 16–25 as Upper Middle, 11–15 as Lower Middle, 5–10 as Upper Lower, and less than 5 as Lower.
Family pedigrees up to five generations back were drawn and the coefficient of inbreeding (F) was calculated by using Wright’s path coefficient method [56] following the order: Double first cousin or DFC (F = 0.125) > First cousin or FC (F = 0.0625) > First cousin once removed or FCR (F = 0.03125) > Second cousin or SC (F = 0.0156) > Second cousin once removed or SCR (F = 0.0078). In non-consanguineous families or distant marriages (taken as control), the value of ‘F’ is effectively zero (F = 0.000) (Supplementary Table 2 and 3).
2.3 Measurement of morphometric parameters
The standing height of the subjects without shoes was measured two times to the nearest 0.1 cm using a portable measuring tape (Perfect Measuring Tape Company, Toledo, Ohio). The individuals with light clothing and without shoes were weighed two times to the nearest 0.1 kg using the weighing machine (Omron HN-289 Automatic Digital Weighing Scale, Omron Corporation, Kyoto, Japan). The mean value of the two measurements was used for further analysis. The individuals were categorized based on their BMI values employing < 18.5 kg/m2 as underweight, 18.5–22.9 kg/m2 as normal weight, and > 22.9 kg/m2 as overweight [57,58,59,60].
2.4 Statistical analysis
All the statistical calculations were performed using GraphPad Prism 6, SigmaPlot 11.0, and Microsoft Excel version 2019, and Statistical significance was defined at the standard 5% level (p < 0.05). Different statistical analyses were conducted to reach the outcome. Qualitative data were presented as frequency and percentage, and the chi-square test difference was tested between the groups. Quantitative data were presented as mean ± standard deviation (SD), and differences were tested using the student’s t-test and one-way ANOVA. Additionally, the Mann–Whitney Rank Sum test, all pairwise Dunn’s tests, Bonferroni t-test, and Kruskal–Wallis one-way analysis of variance on ranks were applied wherever suitable. The differences in mean values ± standard error (SE) and 95% confidence interval (CI) among inbred categories from the outbred (control) individuals for height, weight and BMI were analyzed. The value of Cohen’s d was calculated to find out the practical significance of the research outcome. The correlation and regression analyses were performed for the estimation of inbreeding depression (ID) on the morphometric parameters, and the coefficients of correlation (r) and regression (R2) were calculated. The ID on physical parameters was calculated using the average coefficient of inbreeding (\(a\)) and percentage of depression in the trait was computed similar to the previous studies [61, 62], given as follows;
In Eq. (1), n indicates the number of individuals multiplied with inbreeding coefficients (F) for each inbred category. F = 0.00 tells non-inbred or outbred individuals, and N is the sum total of the individuals. In Eq. (2) \(\overline{x}\) shows the ‘mean’ value of the trait.
3 Results
3.1 Characteristics of the population surveyed
The study recruited 813 healthy individuals from two North Indian regions (Aligarh and Ambedkar Nagar). The individuals were broadly categorized into inbred and non-inbred or consanguineous and non-consanguineous, and further classified concerning different factors such as gender, age (in years), SES, and caste, and summarized into a master table (Table 1). The percentage of inbreeding was found to be approximately 24% of the total individuals. The association of breeding pattern with gender, age groups, SES, and castes was tested using chi-square statistics and revealed non-significant results for gender (p = 0.1946), age groups (p = 0.1537), and castes (p = 0.6394), but significant results (p = 0.00037) was observed in case of SES classes and breeding pattern (Table 1).
The population residing in the surveyed area displayed different types of inbreeding, having the maximum proportion of FC type marriages (53.89%) and the minimum percentage of DFC and SC (9.85%) of the total inbreeding (Table 2). Among various subtypes of first-cousin marriages, the maximum proportion belongs to the matrilateral cross (MC) type (35.58%), followed by matrilateral parallel (MP) and patrilateral parallel (PP), and minimum (13.46%) in patrilateral cross subtype (Table 3).
3.2 Morphometric differences among inbred and non-inbred groups relating to different factors
The participants were grouped concerning different factors, and the level of significance was checked for inbred and outbred categories for physical parameters (height, weight, and BMI). Figure 1 explains the differences in morphometric measurements among consanguineous and non-consanguineous subjects concerning the genders. Both males and females from two categories (inbred and non-inbred) showed significant differences (p < 0.05) for height (Fig. 1a). In contrast, significant results were observed only in the case of males for weight and BMI (Fig. 1b and c). Among different age groups, adolescents and adult individuals have significant differences (p < 0.05) in heights between inbred and non-inbred categories (Fig. 2a); similarly, individuals from adolescence, adulthood, and late adulthood have shown significant differences in their weight between the inbred and outbred categories (Fig. 2b). Although, the breeding pattern did not reveal any significant results among individuals of different ages concerning their BMI (Fig. 2c). The subject from three SES classes (Upper, Lower-middle, and Upper lower) and inbreeding has statistically significant results (p < 0.05) concerning their heights (Fig. 3a). Still, in the case of weight and BMI, significant results were seen only among the individuals who belonged to the lower middle class (Fig. 3b and c). Further analysis revealed differences in morphometric parameters concerning different castes and breeding patterns, specifying the significance in results for combined (both Ashrafs and Ajlafs) and separately for Ajlafs (p < 0.05) (Fig. 4).
Gender-wise differences in morphometric parameters among inbred and non-inbred individuals. The significant decline in the mean values observed for the trait in inbred individuals as compared to non-inbred ones. p-value was considered significant at < 0.05. a for height, * = 0.0401, ** = 0.0015; b for weight, *** = 0.0009; c for BMI, ** = 0.0046, NI : on-inbred/outbred, I: inbred, M : male, F: female
Differences concerning different age groups in anthropometric parameters among inbred and non-inbred individuals. The significant decline in the mean values observed for the trait in inbred individuals as compared to non-inbred ones. p-value was considered significant at < 0.05. a for height, * = 0.0183, ****≤0.0001; b for weight, ** = 0.0015, ** = 0.0044, * = 0.0237, c for BMI, no significant results were seen between the groups. NI: non-inbred/outbred, I: inbred, CH: childhood, AD: adolescence, AH: adulthood, LA: late adulthood
SES-based differences in morphometric parameters among inbred and non-inbred individuals. The significant decline in the mean values observed for the trait in inbred individuals as compared to non-inbred ones. p-value was considered significant at < 0.05. a for height, * = 0.0302, * = 0.0269, * = 0.0412; b for weight, * = 0.0101; c for BMI, * = 0.0294, NI: non-inbred/outbred, I: inbred, U: upper, UM: upper middle, LM: lower middle, UL: upper lower
Caste-based differences in physical parameters among inbred and non-inbred individuals. The significant decline in the mean values observed for the trait in inbred individuals as compared to non-inbred ones. p-value was considered significant at < 0.05. a for height, *** = 0.0002, **** = < 0.0001; b for weight, ** = 0.0013, *** = 0.0003; c for BMI, * = 0.0458, * = 0.0114, NI: non-inbred/outbred, I: inbred, Com: combined, Ash: ashraf, Ajl: ajlaf
3.3 Effect of inbreeding on body mass index (BMI)
3.3.1 Among all inbred categories
Table 4 presents the mean differences in physical parameters among various inbred groups compared to the outbred (control) ones. With the increase in the inbreeding coefficients, i.e., F = 0.0078 to F = 0.125, there was a persuasive decline in mean values for height and weight compared to the non-inbred (NI) group, i.e., F = 0.00. In contrast, some discrepancies can be seen in the case of BMI (Table 4). The mean difference in morphometric measures showed significant inflation with the increase in the degree of inbreeding (p < 0.05) and followed the order: DFC > FC > FCR > SC > SCR for height, DFC > FC > SCR > FCR > SC for weight and DFC > SCR > FC > FCR > SC for BMI (Table 4). This suggests an increased risk of depression in physical parameters among individuals with more significant inbreeding coefficients. The values of Cohen’s d (ranging from 0.00 to 0.79) are indicative of weak to strong effect of the phenomena in the population surveyed. The strongest effects were observed for DFC categories for all three traits studied here (Table 4).
The frequency distribution (in percentage) of individuals concerning different classes of BMI values (ranging from underweight to overweight) shows highly significant differences (p = 2.086 × 10−9) among the consanguineous group correlated with the non-consanguineous group (Table 5, Fig. 5). The higher frequency of low BMI scores (< 18.5 or underweight) was observed in the case of the DFC group, followed by FC, FCR, SC, and SCR, with the lowest frequency in the NI type. In contrast, the maximum frequency of high BMI score (> 22.9 or overweight) was seen in non-inbred individuals and the minimum in DFC type (Table 5). The line graph for inbred subjects gets almost flattened towards high BMI scores (18.5–22.9 or normal and > 22.9 or overweight), whereas it showed higher frequency peaks towards comparatively lower BMI values (< 18.5 or underweight) (Fig. 5).
BMI comparison with respect to the degree of inbreeding for different inbred categories. The graph depicting the percentage frequency distribution (p = 2.086 × 10−9) of BMI in relation to the inbreeding coefficient. The outbred individuals display high frequency peak on right side (presenting higher BMI values i.e., > 22.9) and lower frequency peaks on left side corresponding to comparatively low BMI (< 18.5 and 18.5–22.9). On contrary, elevated peaks are observed for low BMI values (left side) with increase in in breeding coefficient, and flattened towards high BMI values (right side of the graph) suggesting inbreeding depression on BMI
3.3.2 Among first cousin subtypes
Correspondingly, mean differences in morphometric measures among four subtypes of first cousins regarding the outbred (control) ones were calculated and presented in Table 6. With the increase of the sex-linked inbreeding coefficient, i.e., Fs = 0.00 to Fs = 0.1875, there was a convincing diminution in the mean values for height in comparison with the non-inbred group, i.e., F = 0.00, although, MC type show deviation from the trend for weight and BMI (Table 6). The mean difference in anthropometric parameters revealed a significant upsurge with the increment in the degree of inbreeding (p < 0.05) and followed the order: MP > MC > PP > PC for height. In contrast, the order MP > PP > MC > PC was followed by weight and BMI (Table 6). This suggests the elevated menace for depression in physical parameters among the progeny of first cousin individuals having the greater value of sex-linked inbreeding coefficients. The values of Cohen’s d (ranging from 0.026 to 0.539), suggesting weak to moderate effect of the phenomena in the population studied, having maximum values in DFC category for all the traits mentioned (Table 6).
The cataloguing of individuals from different subtypes of first cousins based on their BMI values (ranging from underweight to overweight) about the percentage frequency distribution revealed highly significant differences (p = 5.2 × 10−5) among the groups (Table 7, Fig. 6). The higher frequency of low BMI score (< 18.5 or underweight) was observed in the case of MP type followed by MC and PC, with the lowest frequency in PP type, contrary to the maximum frequency of high BMI score (> 22.9 or overweight) in PC type followed by PP type (Table 7). The line graph for FC subjects gets flattened towards higher BMI scores, i.e., > 22.9 or overweight, but shows comparatively higher frequency peaks towards lower BMI scores (< 18.5 or underweight). However, the subjects belonging to MC and MP subtypes show consistently low frequency of high values of BMI (18.5–22.9 or normal weight, > 22.9 or overweight), suggesting depression on morphometric parameters due to their higher values of sex-linked inbreeding coefficients (Fig. 6).
BMI comparison with respect to the degree of inbreeding for different subtypes of first cousin categories. The graph depicting the percentage frequency distribution (p = 5.2 × 10−5) of BMI classes in relation to the sex-linked inbreeding coefficient (Fs). The individuals of MP and MC categories display high frequency peaks on left side corresponding to lower BMI (< 18.5) as compared to PC and PP, but significantly low frequency peaks are present for higher BMI values (18.5–22.9, > 22.9) in case of MC and MP type individuals, while, PP and PC type showing comparatively higher peaks for high BMI
3.4 Inbreeding depression on morphometric parameters
3.4.1 Among all inbred categories
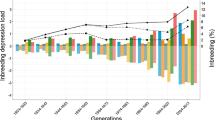
Besides the differences in percentage and mean values for height, weight, and BMI for different factors, we found analogous results in ANOVA and regression model with increasing inbreeding coefficients (Figs. 7, 8). For physical measurements (height, weight, and BMI), ANOVA revealed significant (p < 0.001) differences among the selected inbred and outbred individuals, with further analysis of the t-test between the groups also confirming the statistical significance of different levels for the morphometry of the subjects (Fig. 7). The regression and Pearson’s correlation coefficient (R2 and r) was found to be R2 = 0.9267, r = − 0.995 for height, R2 = 0.6125, r = − 0.943 for weight and R2 = 0.296, r = − 0.778 for BMI and all of which were statistically significant at p < 0.05 except BMI (p = 0.068). The regression slope and correlation coefficient (negative sign) show a fitness decline and depression in morphometric measures with the increase of inbreeding coefficients, respectively (Fig. 8).
Results of ANOVA revealed significant differences (p < 0.001) among the groups for physical parameters viz: Height, Weight and BMI. a for Height, NI with all inbred categories except SCR showed significant results, i.e., NI vs SC, p = 0.023; NI vs FCR, p = 0.013; NI vs FC, p < 0.001; NI vs DFC, p = 0.006, b for weight, t-test and ANOVA followed by Dunn’s method revealed significant differences between NI and FC with p < 0.001 and p < 0.05, NI and DFC with p = 0.001 and p < 0.05, respectively, c for BMI, only NI with DFC showed significant results for t-test (p < 0.01), others are not significant
Inbreeding depression on morphometric parameters for all inbred category. The regression analysis presents almost similar trend for Height, Weight and BMI. The slope depicts fitness decline (regression slope) with increase in the inbreeding coefficient (F = 0.00 to F = 0.125, or NI to DFC). The observed linear decline in fitness for physical parameters are found to be significant; a for Height, R2 = 0.9267, r = − 0.995, p = 0.003, b for Weight, R2 = 0.6125, r = − 0.943, p = 0.0046, c for BMI, R2 = 0.296, r = − 0.778, p = 0.068
The average coefficient of inbreeding (\(a\) total) for all the inbred categories surveyed was calculated to be 5.28. Table 8 represents the percent mean depression and percent depression in the physical parameters, having maximum values in the case of weight followed by BMI and height. The percentage depression for height, weight, and BMI values among individuals in case of high inbreeding coefficients provides a greater risk for inbreeding depression on physical parameters or being underweight.
3.4.2 Among first cousin subtypes
Although, the results of ANOVA on morphometric parameters, i.e., height, weight, and BMI of first cousin categories, revealed non-significant (p = 0.544, p = 0.133, and p = 0.096, respectively) differences among the groups (Fig. 9). The regression and Pearson’s correlation coefficient (R2 and r) were found to be R2 = 0.997, r = − 0.945 for height, R2 = 0.5682, r = − 0.678 for weight, and R2 = 0.1874, r = − 0.242 for BMI. The regression slope and correlation coefficient (negative sign) show a fitness decline and depression in physical parameters with increased sex-linked inbreeding coefficients, respectively (Fig. 10).
Inbreeding depression on anthropometric parameters for first cousin categories. The regression analysis presents almost similar trend for Height, Weight and BMI. The slope depicts fitness decline (regression slope) with increase in the sex-linked inbreeding coefficient (Fs = 0.00 to Fs = 0.1875, or PP/PC to MP). Significant regression can be seen in the traits but p-values are non-significant at p < 0.05. a for Height, R2 = 0.997, r = − 0.945, p = 0.055, b for Weight, R2 = 0.5682, r = − 0.678, p = 0.321, c for BMI, R2 = 0.1874, r = − 0.242, p = 0.758
For the first cousin category, the average coefficient of inbreeding (\(a\) FC) was calculated to be 10.22. The percent means depression and percent depression in the anthropometric measures were observed maximum in the case of weight, followed by BMI and height (Table 9). The percentage of depression for morphometrical values among individuals in case of high inbreeding coefficients (sex-linked) poses a greater risk for inbreeding depression or being underweight.
4 Discussion
The current study demonstrates the impact of inbreeding on height, weight, and BMI and the repercussions of these factors in the context of individual’s health. For this purpose, we incorporated 813 individuals, comprising 76.26% outbred and 23.74% inbred individuals, and collected their relevant data after their consent (Table 1). The inbreeding coefficient (F or the amount of homozygosity or autozygosity) was calculated using standard pedigree information [56] provided by the respondents (Tables 2, 3). Inbreeding promotes the homozygosity throughout the genome, and ID can be attributed to the increased homozygosity of (partially) recessive deleterious alleles, revealing their adverse effects on fitness and fitness-related traits [63]. The effect of inbreeding on physical parameters has been quantified using a regression-based fitness-related model (Figs. 8, 10). The closer the biological relationship between parents, the greater will be the proportion of the shared alleles and, therefore, the greater will be the probability that their offspring will receive identical copies of one or more deleterious recessive genes [34, 39, 64]. Previous analyses have also demonstrated the relationship between inbreeding load and higher inbreeding coefficients [6, 37, 65,66,67].
The rate of consanguinity among the Muslim population of the chosen area was determined to be 23.74% in the current study (Table 1), being consistent with other earlier findings [68,69,70,71,72]. In our study, the highest percentage of consanguineous marriage is seen of FC type followed by FCR and others (Table 2), analogous to the tradition of the Indian subcontinent, where FC marriages are the most prevalent form of consanguineous union [71, 72].
This study is an attempt to use body mass composition measures to investigate the interactions between genetic (i.e., inbreeding) and socio-demographic factors of individuals in the North Indian population. In our study, we observed some differences in the height and BMI among males and females (Fig. 1). Similar results from previous studies have shown that height in males tends to be higher as compared to females, while BMI was found to be higher in females than males [58, 59], although there is mixed research suggesting gender-based differences might be due to the influence of environment and other genes or may be due to sex hormones [16, 73, 74]. This study emphasizes height, weight, and BMI traits for the analysis of an individual's health because these traits have received little attention in studies of inbreeding but have been extensively studied in studies of socio-economic indicators [14, 75, 76]. In our study, the variance in body mass composition reveals minor relevance of racial or socio-demographic characteristics (Figs. 1–4) that the same geographical and cultural settings may alter; similar results have been reported by previous study conducted in Northern most part of India [6].
We discovered that inbreeding leads the health status of individuals to significantly decline and that offspring of inbred families are more likely to experience poor physical fitness based upon the varying degrees of their inbreeding coefficients (for autosomal locus). In contrast, individuals from non-consanguineous families exhibit higher values of morphometric measures (Table 5, Fig. 8) and, as a result, experience good health or vigor. Our findings agree that consanguineous marriages and other kinds of inbreeding have been linked to a noticeable decline in vigor or development of poor health outcomes among individuals [6]. Since, the FC marriages are the most prevalent form of consanguineous union [70, 71], it gained the interest of researchers to study the effects of inbreeding, especially on FC type inbreeding and its subtypes. It has been reported that the mental impairment among FC offspring is four times higher than that of the control group, and childhood mortality has been found to increase by 4–5% among the offspring of FC unions [37, 76]. Another recent study reported the differences in homozygosity patterns (ROHs) for sex-linked loci among four different types of FC unions [39]. Likewise, we also performed a separate analysis of FC inbred individuals to find the potential of inbreeding depression among different subtypes. The results showed significant differences in the frequency of lower BMI, with high values in matrilateral unions (MP and MC) and low values in patrilateral (PC and PP) unions (Table 7, Fig. 10), specifying the high chances of fitness decline among matrilateral offspring as compared to patrilateral ones.
In the present study, the ANOVA-based results on morphometric domains indicated significant differences in the context of inbreeding (Fig. 7), but it did not produce significant results among FC categories (Fig. 9). This may be due to the smaller sample size of FC type unions, although it revealed significant results for regression and correlation with respect to the sex-linked inbreeding coefficient (Fig. 10). Inbreeding was more important than other factors, including gender, age, and caste (Table 1, Figs. 1–4). Though the factors were insufficient to detect a substantial inbreeding effect, SES did demonstrate significant variations in physical parameters across inbred and non-inbred groups. Our research showed that the amount of inbreeding and the impact of consanguinity on morphometric traits are proportional to each other. According to the results, the offspring of DFC (on average) showed a significantly higher inbreeding load (mean difference) than those of first cousins (Table 4). This inbreeding load then continued to decrease with the lowering of inbreeding coefficients, with the least amount of depression being observed among offspring of non-consanguineous parents. Inbreeding-related depression of this magnitude was also documented in earlier research [37, 77, 78]. Similarly, in the case of different subtypes of FC unions, MP type (having a maximum value of sex-linked inbreeding coefficient) shows maximum depression and decreases with the decline in inbreeding coefficient of sex-linked locus, for physical parameters viz; height, weight, and BMI (Table 6), which is consistent with the previous study [39].
The likelihood of congenital disabilities and other severe medical disorders has been attributed to consanguinity. Research from inbred communities showed comparisons between the prevalence of birth abnormalities in the offspring of FC marriages and those of unrelated couples [6, 79, 80]. We discovered a substantial decline (below normal) in the height, weight, and BMI of kids in inbred households, demonstrating the impact of inbreeding reducing heterozygosity. The reason for depression in inbred populations is an upsurge in homozygosity (caused by inbreeding), resulting in limited genetic diversity, which leads to the expression of recessive alleles or loss of hybrid vigor of essential genes for height and weight. The effect of consanguinity in a Muslim converted population is reported here, there being no prolonged history of inbreeding as among the South Indian Hindus. This differentiates the population from South Indians, where genetic diseases have been otherwise reduced due to the elimination of harmful genes by prolonged inbreeding. The obvious effect of inbreeding on quantitative traits results into lowering of the means pertaining to general health and fitness of the population.
The present study conducted on 813 individuals from two regions of North Indian population have revealed the effects of inbreeding on their morphometric traits. More studies taking more population with larger sample size are needed for elaborating the health profile of the inbred and control group. Here, the reported differences in the effects of inbreeding among FC categories was based on small sample size (n = 104). Studies should be conducted on FC inbreds having large sample sizes to endorse our findings, as these types of unions are quite prevalent in different parts of the world. More studies for estimating the inbreeding depression among the offspring of FCR, SC, SCR and beyond are needed. Moreover, other morphometric parameters in relation to consanguinity can also be studied to assess the fitness and general health of the population in different parts of the world.
5 Conclusions
The present study not only contributes the evidence of inbreeding depression quantification on morphometric measures (height, weight, and BMI) among inbred categories but also showcases the differences in inbreeding depression among subtypes of first cousin unions. The dearth of the effect of factors other than inbreeding on these metrics suggests that the genetic component is more prominent over these parameters. The BMI offers a paradigm for comprehending and estimating the risk elements for health issues. More intriguingly, the depression appears to be more pronounced due to an increase in the extent of inbreeding (F). For a more thorough knowledge of their genetic underpinnings, we observed that inbred offspring (second cousin once removed) had somewhat higher scores for overweight. The study has novelty and inimitability as it avows hitherto unreported differences in the consequences of inbreeding (fitness decline) among several subtypes of first-cousin marriage. Since these parameters are fitness-related and complex, the effect of inbreeding on these traits would be beneficial to study in the context of individual’s health and fitness. Our research on inbreeding depression offers guidance to healthcare professionals and health policymakers in educating the public about its adverse effects and adhering to standardized, scientifically supported recommendations to help families with premarital counselling. Nevertheless, further research on larger sample size is needed to endorse our findings and determine the genetic factors working upon health status modifications.
Data availability
The datasets created during and/or analysed during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
Abbreviations
- BMI:
-
Body Mass Index
- GWAS:
-
Genome Wide Association Studies
- SNP:
-
Single nucleotide polymorphism
- SES:
-
Socio-economic status
- ROH:
-
Runs of homozygosity
- IBD:
-
Identity by descent
- F:
-
Inbreeding coefficient
- Fa:
-
Inbreeding coefficient for autosomal locus
- Fs:
-
Inbreeding coefficient for sex-linked locus
- ID:
-
Inbreeding depression
- SD:
-
Standard deviation
- ANOVA:
-
Analysis of variance
- SE:
-
Standard error
- CI:
-
Confidence interval
- DFC:
-
Double first cousin
- FC:
-
First cousin
- FCR:
-
First cousin once removed
- SC:
-
Second cousin
- SCR:
-
Second cousin once removed
- MP:
-
Matrilateral parallel
- MC:
-
Matrilateral cross
- PP:
-
Patrilateral parallel
- PC:
-
Patrilateral cross
References
Calle EE, Thun MJ, Petrelli JM, Rodriguez C, Heath CW. Body-mass index and mortality in a prospective cohort of U.S. adults. N Engl J Med. 1999;341(15):1097–105. https://doi.org/10.1056/NEJM199910073411501.
Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med. 2003;348(17):1625–38. https://doi.org/10.1056/NEJMoa021423.
Hodge JM, Shah R, McCullough ML, Gapstur SM, Patel AV. Validation of self-reported height and weight in a large, nationwide cohort of U.S adults. PLoS ONE. 2020;15(4):e0231229. https://doi.org/10.1371/journal.pone.0231229.
Patel AV, Hildebrand JS, Gapstur SM. Body mass index and all-cause mortality in a large prospective cohort of white and black U.S Adults. PLoS ONE. 2014;9(10):e109153. https://doi.org/10.1371/journal.pone.0109153.
Prospective Studies Collaboration. Body-mass index and cause-specific mortality in 900 000 adults: collaborative analyses of 57 prospective studies. Lancet Lond Engl. 2009;373(9669):1083–96. https://doi.org/10.1016/S0140-6736(09)60318-4.
Fareed M, Afzal M. Evidence of inbreeding depression on height, weight, and body mass index: a population-based child cohort study. Am J Hum Biol. 2014;26(6):784–95. https://doi.org/10.1002/ajhb.22599.
Barrios P, Martin-Biggers J, Quick V, Byrd-Bredbenner C. Reliability and criterion validity of self-measured waist, hip, and neck circumferences. BMC Med Res Methodol. 2016;16:49. https://doi.org/10.1186/s12874-016-0150-2.
Nyholm M, Gullberg B, Merlo J, Lundqvist-Persson C, Råstam L, Lindblad U. The validity of obesity based on self-reported weight and height: implications for population studies. Obes Silver Spring Md. 2007;15(1):197–208. https://doi.org/10.1038/oby.2007.536.
Olfert MD, et al. Self-reported vs measured height, weight, and bmi in young adults. Int J Environ Res Public Health. 2018;15(10):10. https://doi.org/10.3390/ijerph15102216.
McQuillan R, et al. Evidence of inbreeding depression on human height. PLOS Genet. 2012;8(7): e1002655. https://doi.org/10.1371/journal.pgen.1002655.
Yengo L, et al. A saturated map of common genetic variants associated with human height. Nature. 2022;610(7933):704–12. https://doi.org/10.1038/s41586-022-05275-y.
Singh GK, Kogan MD, Van Dyck PC, Siahpush M. Racial/ethnic, socioeconomic, and behavioral determinants of childhood and adolescent obesity in the United States: analyzing independent and joint associations. Ann Epidemiol. 2008;18(9):682–95. https://doi.org/10.1016/j.annepidem.2008.05.001.
Bener A, Saleh NM, Salameh KMK, Basha B, Joseph S, Al Buz R. Socio-demographic and consanguinity risk factors associated with low birthweight. JPMA J Pak Med Assoc. 2013;63(5):598–603.
Martin KR, Kuh D, Harris TB, Guralnik JM, Coggon D, Wills AK. Body mass index, occupational activity, and leisure-time physical activity: an exploration of risk factors and modifiers for knee osteoarthritis in the 1946 British birth cohort. BMC Musculoskelet Disord. 2013;14:219. https://doi.org/10.1186/1471-2474-14-219.
Ogden CL, Fryar CD, Carroll MD, Flegal KM. Mean body weight, height, and body mass index, United States 1960–2002. Adv Data. 2004;347:1–17.
Zillikens MC, et al. Sex-specific genetic effects influence variation in body composition. Diabetologia. 2008;51(12):2233–41. https://doi.org/10.1007/s00125-008-1163-0.
Jerant A, Franks P. Body mass index, diabetes, hypertension, and short-term mortality: a population-based observational study, 2000–2006. J Am Board Fam Med JABFM. 2012;25(4):422–31. https://doi.org/10.3122/jabfm.2012.04.110289.
Khanna D, et al. Body mass index (BMI): a screening tool analysis. Cureus. 2022;14:2. https://doi.org/10.7759/cureus.22119.
Zimmermann E, Bjerregaard LG, Gamborg M, Vaag AA, Sørensen TIA, Baker JL. Childhood body mass index and development of type 2 diabetes throughout adult life—a large-scale danish cohort study. Obesity. 2017;25(5):965–71. https://doi.org/10.1002/oby.21820.
Mazzoccoli G. Body composition: where and when. Eur J Radiol. 2016;85(8):1456–60. https://doi.org/10.1016/j.ejrad.2015.10.020.
Aune D, et al. BMI and all cause mortality: systematic review and non-linear dose-response meta-analysis of 230 cohort studies with 3.74 million deaths among 30.3 million participants. The BMJ. 2016;353: i2156. https://doi.org/10.1136/bmj.i2156.
Bhaskaran K, Santos Silva I, Leon DA, Douglas IJ, Smeeth L. Association of BMI with overall and cause-specific mortality: a population-based cohort study of 3·6 million adults in the UK. Lancet Diabetes Endocrinol. 2018;6(12):944–53. https://doi.org/10.1016/S2213-8587(18)30288-2.
Yoo HJ. Body mass index and mortality. J Obes Metab Syndr. 2017;26(1):3–9. https://doi.org/10.7570/jomes.2017.26.1.3.
Twig G, et al. Body-mass index in 2.3 million adolescents and cardiovascular death in adulthood. N Engl J Med. 2016;374(25):2430–40. https://doi.org/10.1056/NEJMoa1503840.
Önal AE. Introductory chapter: life, health and body mass index, in body-mass index and health. IntechOpen. 2018. https://doi.org/10.5772/intechopen.82142.
Aarestrup J, Gamborg M, Ulrich LG, Sørensen TIA, Baker JL. Childhood body mass index and height and risk of histologic subtypes of endometrial cancer. Int J Obes. 2016;40(7):7. https://doi.org/10.1038/ijo.2016.56.
Bittles A. Consanguinity and its relevance to clinical genetics. Clin Genet. 2001;60(2):89–98. https://doi.org/10.1034/j.1399-0004.2001.600201.x.
Bittles AH. The role and significance of consanguinity as a demographic variable. Popul Dev Rev. 1994;20(3):561. https://doi.org/10.2307/2137601.
Oniya O, Neves K, Ahmed B, Konje JC. A review of the reproductive consequences of consanguinity. Eur J Obstet Gynecol Reprod Biol. 2019;232:87–96. https://doi.org/10.1016/j.ejogrb.2018.10.042.
Bittles A, Black M. Consanguineous marriage and human evolution. Annu Rev Anthropol. 2010;39:193–207. https://doi.org/10.1146/annurev.anthro.012809.105051.
Temaj G, Nuhii N, Sayer JA. The impact of consanguinity on human health and disease with an emphasis on rare diseases. J Rare Dis. 2022;1(1):2. https://doi.org/10.1007/s44162-022-00004-5.
Fatma R, Chauhan W, Afzal M. The effect of consanguinity on reproductive fitness and selection intensity in the muslim population of Western Uttar Pradesh. India J Hum Ecol. 2021;74:1–3. https://doi.org/10.31901/24566608.2021/74.1-3.3321.
Hamamy H. Consanguineous marriages. J Community Genet. 2012;3(3):185–92. https://doi.org/10.1007/s12687-011-0072-y.
Melado L, et al. Features of chromosomal abnormalities in relation to consanguinity: analysis of 10,556 blastocysts from IVF/ICSI cycles with PGT-A from consanguineous and non-consanguineous couples. Sci Rep. 2023;13(1):1. https://doi.org/10.1038/s41598-023-36014-6.
Sahoo SA, Zaidi A, Anagol S, Mathieson I. Long runs of homozygosity are correlated with marriage preferences across global population samples. Hum Biol. 2022;93:3. https://doi.org/10.1353/hub.2021.0011.
Fareed M, Kaisar Ahmad M, Azeem Anwar M, Afzal M. Impact of consanguineous marriages and degrees of inbreeding on fertility, child mortality, secondary sex ratio, selection intensity, and genetic load: a cross-sectional study from Northern India. Pediatr Res. 2017;81(1):1. https://doi.org/10.1038/pr.2016.177.
Fareed M, Afzal M. Estimating the inbreeding depression on cognitive behavior: a population based study of child cohort. PLoS ONE. 2014;9(10): e109585. https://doi.org/10.1371/journal.pone.0109585.
Bennett RL, et al. Genetic counseling and screening of consanguineous couples and their offspring: recommendations of the national society of genetic counselors. J Genet Couns. 2002;11(2):97–119. https://doi.org/10.1023/A:1014593404915.
Fatma R, Chauhan W, Afzal M. The coefficients of inbreeding revealed by ROH study among inbred individuals belonging to each type of the first cousin marriage: a preliminary report from North India. Genes Genomics. 2023. https://doi.org/10.1007/s13258-023-01367-9.
Woods CG, et al. Quantification of homozygosity in consanguineous individuals with autosomal recessive disease. Am J Hum Genet. 2006;78(5):889–96. https://doi.org/10.1086/503875.
Inhorn MC, Kobeissi L, Nassar Z, Lakkis D, Fakih MH. Consanguinity and family clustering of male factor infertility in Lebanon. Fertil Steril. 2009;91(4):1104–9. https://doi.org/10.1016/j.fertnstert.2008.01.008.
Postma E, Martini L, Martini P. Inbred women in a small and isolated Swiss village have fewer children. J Evol Biol. 2010;23(7):1468–74. https://doi.org/10.1111/j.1420-9101.2010.02013.x.
Seher T, et al. Is parental consanguinity associated with reduced ovarian reserve? Reprod Biomed Online. 2015;31(3):427–33. https://doi.org/10.1016/j.rbmo.2015.06.003.
Shawky RM, Elsayed SM, Zaki ME, Nour El-Din SM, Kamal FM. Consanguinity and its relevance to clinical genetics. Egypt J Med Hum Genet. 2013;14(2):157–64. https://doi.org/10.1016/j.ejmhg.2013.01.002.
Yasmin T, Andres EM, Ashraf K, Basra MAR, Raza MH. Genome-wide analysis of runs of homozygosity in pakistani controls with no history of speech or language-related developmental phenotypes. Ann Hum Biol. 2023;50(1):101–8. https://doi.org/10.1080/03014460.2023.2180087.
Raza MH, Riazuddin S, Drayna D. Identification of an autosomal recessive stuttering locus on chromosome 3q132–3q1333. Hum Genet. 2010;128(4):461. https://doi.org/10.1007/s00439-010-0871-y.
Villanueva P, et al. Genome-wide analysis of genetic susceptibility to language impairment in an isolated Chilean population. Eur J Hum Genet EJHG. 2011;19(6):687–95. https://doi.org/10.1038/ejhg.2010.251.
Lal D, et al. Increased probability of co-occurrence of two rare diseases in consanguineous families and resolution of a complex phenotype by next generation sequencing. PLoS ONE. 2016;11(1): e0146040. https://doi.org/10.1371/journal.pone.0146040.
Ye Z, Jia X, Liu X, Zhang Q, Wang K, Chen M. Case report: the first reported concurrence of Wilson disease and bilateral retinitis pigmentosa. Front Med. 2022. https://doi.org/10.3389/fmed.2022.877752.
Afzal M. Consequences of consanguinity on cognitive behavior. Behav Genet. 1988;18(5):583–94. https://doi.org/10.1007/BF01082310.
Clark DW, et al. Associations of autozygosity with a broad range of human phenotypes. Nat Commun. 2019;10(1):4957. https://doi.org/10.1038/s41467-019-12283-6.
Joshi PK, et al. Directional dominance on stature and cognition in diverse human populations. Nature. 2015;523(7561):7561. https://doi.org/10.1038/nature14618.
Yengo L, et al. Detection and quantification of inbreeding depression for complex traits from SNP data. Proc Natl Acad Sci USA. 2017;114(32):8602–7. https://doi.org/10.1073/pnas.1621096114.
Fareed M, Afzal M. Genetics of consanguinity and inbreeding in health and disease. Ann Hum Biol. 2017;44(2):99–107. https://doi.org/10.1080/03014460.2016.1265148.
Saleem S. Modified kuppuswamy socioeconomic scale updated for the year 2020. Ind J Forens Commun Med. 2020. https://doi.org/10.18231/J.IJFCM.2021.001.
Wright S. Coefficients of inbreeding and relationship. Am Nat. 1922;56(645):330–8. https://doi.org/10.1086/279872.
Aziz N, Kallur SD, Nirmalan PK. Implications of the revised consensus body mass indices for asian indians on clinical obstetric practice. J Clin Diagn Res. 2014. https://doi.org/10.7860/JCDR/2014/8062.4212.
Fatma R, Chauhan W, Riyaz S, Rasheed K, Afzal M. Genetic association analysis of rs662799 (–1131A > G) polymorphism of APOA5 gene with morphometric and physio-metric traits using multiplex PCR. Egypt J Med Hum Genet. 2023;24(1):19. https://doi.org/10.1186/s43042-023-00398-x.
Fatma R, Chauhan W, Shahi M, Afzal M. “Association of BDNF gene missense polymorphism rs6265 (Val66Met) with three quantitative traits, namely, intelligence quotient, body mass index, and blood pressure: a genetic association analysis from North India. Front Neurol. 2023. https://doi.org/10.3389/fneur.2022.1035885.
Pan W-H, Yeh W-T. How to define obesity? evidence-based multiple action points for public awareness, screening, and treatment: an extension of Asian-Pacific recommendations. Asia Pac J Clin Nutr. 2008;17(3):370–4.
Badaruddoza B, Afzal M. Inbreeding depression and intelligence quotient among north Indian children. Behav Genet. 1993;23(4):343–7. https://doi.org/10.1007/BF01067435.
Vandenberg SG. Assortative mating, or who marries whom? Behav Genet. 1972;2(2–3):127–57. https://doi.org/10.1007/BF01065686.
Yengo L, Yang J, Keller MC, Goddard ME, Wray NR, Visscher PM. Genomic partitioning of inbreeding depression in humans. Am J Hum Genet. 2021;108(8):1488–501. https://doi.org/10.1016/j.ajhg.2021.06.005.
Charlesworth D, Willis JH. The genetics of inbreeding depression. Nat Rev Genet. 2009;10:11. https://doi.org/10.1038/nrg2664.
Michael L. The genetic interpretation of inbreeding depression and outbreeding depression. Evol Int J Org Evol. 1991;45:3. https://doi.org/10.1111/j.1558-5646.1991.tb04333.x.
Richards CM. Inbreeding depression and genetic rescue in a plant metapopulation. Am Nat. 2000;155(3):383–94. https://doi.org/10.1086/303324.
Acharya S, Sahoo H. Consanguineous marriages in India: prevalence and determinants. J Health Manag. 2021;23(4):631–48. https://doi.org/10.1177/09720634211050458.
Angural A, et al. Review: understanding rare genetic diseases in low resource regions like Jammu and Kashmir–India. Front Genet. 2020. https://doi.org/10.3389/fgene.2020.00415.
Bittles AH, Black ML. Consanguinity, human evolution, and complex diseases. Proc Natl Acad Sci USA. 2010;107(suppl_1):1779–86. https://doi.org/10.1073/pnas.0906079106.
Bittles AH, Hussain R. An analysis of consanguineous marriage in the Muslim population of India at regional and state levels. Ann Hum Biol. 2000;27(2):163–71. https://doi.org/10.1080/030144600282271.
Khan SY. Consanguinity and inbreeding coefficient f in Aligarh city, India: a cross sectional study. Pesqui Bras Em Odontopediatria E Clín Integr. 2019;19: e4401. https://doi.org/10.4034/PBOCI.2019.191.40.
Bonthuis M, Jager KJ, Abu-Hanna A, Verrina E, Schaefer F, van Stralen KJ. Application of body mass index according to height-age in short and tall children. PLoS ONE. 2013;8(8): e72068. https://doi.org/10.1371/journal.pone.0072068.
Singh SK, Dubey PP. A Comparative analysis of BMI among males and females aged between 15-75 years. Int J Contemp Med Res. 2018;5(1):3.
Brown DE, Hampson SE, Dubanoski JP, Murai AS, Hillier TA. Effects of ethnicity and socioeconomic status on body composition in an admixed, multiethnic population in Hawaii. Am J Hum Biol. 2009;21(3):383–8. https://doi.org/10.1002/ajhb.20889.
Georges E, Mueller WH, Wear ML. Body fat distribution: associations with socioeconomic status in the hispanic health and nutrition examination survey. Am J Hum Biol. 1991;3(5):489–501. https://doi.org/10.1002/ajhb.1310030509.
Rudan I, et al. Inbreeding and risk of late onset complex disease. J Med Genet. 2003;40(12):925–32. https://doi.org/10.1136/jmg.40.12.925.
Badaruddoza B. Effect of inbreeding on wechsler intelligence test scores among North Indian children. Asia Pac J Public Health. 2004;16(2):99–103. https://doi.org/10.1177/101053950401600204.
Verweij KJH, et al. The association of genotype-based inbreeding coefficient with a range of physical and psychological human traits. PLoS ONE. 2014;9(7): e103102. https://doi.org/10.1371/journal.pone.0103102.
Shieh JTC, Bittles AH, Hudgins L. Consanguinity and the risk of congenital heart disease. Am J Med Genet A. 2012;158A(5):1236. https://doi.org/10.1002/ajmg.a.35272.
Zlotogora J. Genetic disorders among palestinian arabs: 1. effects of consanguinity. Am J Med Genet. 1997;68(4):472–5. https://doi.org/10.1002/(sici)1096-8628(19970211)68:4%3c472::aid-ajmg20%3e3.0.co;2-o.
Acknowledgements
Authors are thankful to the chairperson, Department of Zoology, Aligarh Muslim University, for providing essential facilities. Maulana Azad National Fellowship (MANF-2018-19-UTT-100618) under University Grants Commission (UGC) to the first author is highly acknowledged. The authors are also grateful to all of the research participants for their cooperation.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
RF, Conceptualization; Study design; Data curation; Data analysis; Investigation; Methodology; Resources; original draft; Writing—review and editing, WC, Investigation; Conceptualization; Writing—review and editing, MA, Supervision; Study design; Validation; Visualization: Conceptualization; Formal analysis.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study has been performed in accordance with the Declaration of Helsinki and have been reviewed and approved by the Institutional Ethics Committee of Jawaharlal Nehru Medical College (IEC-JNMC), Aligarh Muslim University, India (Approval code: 314/FM/IEC). Informed consent forms have been obtained from all participants in this study, either directly from the participant or from his/her legal guardian in case of below the age of consent (< 16 years of age).
Consent for publication
Not applicable.
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Fatma, R., Chauhan, W. & Afzal, M. Assessment of inbreeding depression on morphometric traits among North Indian population cohorts. Discov Public Health 21, 14 (2024). https://doi.org/10.1186/s12982-024-00132-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12982-024-00132-x