Abstract
Background
The 70-gene signature (70-GS, MammaPrint) test has been recommended by the main guidelines to evaluate prognosis and chemotherapy benefit of hormonal receptor positive human epidermal receptor 2 negative (HR + /Her2−) early breast cancer (BC). However, this expensive assay is not always accessible and affordable worldwide. Based on our previous study, we established nomogram models to predict the binary and quartile categorized risk of 70-GS.
Methods
We retrospectively analyzed a consecutive cohort of 150 female patients with HR + /Her2− BC and eligible 70-GS test. Comparison of 40 parameters including the patients’ medical history risk factors, imaging features and clinicopathological characteristics was performed between patients with high risk (N = 62) and low risk (N = 88) of 70-GS test, whereas risk calculations from established models including Clinical Treatment Score Post-5 years (CTS5), Immunohistochemistry 3 (IHC3) and Nottingham Prognostic Index (NPI) were also compared between high vs low binary risk of 70-GS and among ultra-high (N = 12), high (N = 50), low (N = 65) and ultra-low (N = 23) quartile categorized risk of 70-GS. The data of 150 patients were randomly split by 4:1 ratio with training set of 120 patients and testing set 30 patients. Univariate analyses and multivariate logistic regression were performed to establish the two nomogram models to predict the the binary and quartile categorized risk of 70-GS.
Results
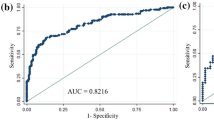
Compared to 70-GS low-risk patients, the high-risk patients had significantly less cardiovascular co-morbidity (p = 0.034), more grade 3 BC (p = 0.006), lower progesterone receptor (PR) positive percentage (p = 0.007), more Ki67 high BC (≥ 20%, p < 0.001) and no significant differences in all the imaging parameters of ultrasound and mammogram. The IHC3 risk and the NPI calculated score significantly correlated with both the binary and quartile categorized 70-GS risk classifications (both p < 0.001). The area under curve (AUC) of receiver-operating curve (ROC) of nomogram for binary risk prediction were 0.826 (C-index 0.903, 0.799–1.000) for training and 0.737 (C-index 0.785, 0.700–0.870) for validation dataset respectively. The AUC of ROC of nomogram for quartile risk prediction was 0.870 (C-index 0.854, 0.746–0.962) for training and 0.592 (C-index 0.769, 0.703–0.835) for testing set. The prediction accuracy of the nomogram for quartile categorized risk groups were 55.0% (likelihood ratio tests, p < 0.001) and 53.3% (p = 0.04) for training and validation, which more than double the baseline probability of 25%.
Conclusions
To our knowledge, we are the first to establish easy-to-use nomograms to predict the individualized binary (high vs low) and the quartile categorized (ultra-high, high, low and ultra-low) risk classification of 70-GS test with fair performance, which might provide information for treatment choice for those who have no access to the 70-GS testing.
Similar content being viewed by others
Introduction
Breast cancer (BC) is the commonest malignancy worldwide and the leading cause of cancer death in Chinese women younger than 45 years [1,2,3]. In 2020, nearly 2,261,419 women were newly diagnosed breast cancer and 684,996 died from breast cancer, with a cumulative lifetime (age range: 0–74 years) risk of 5.20% [1, 4]. Hormonal receptor positive human epidermal receptor 2 negative (HR + /Her2−) comprises of approximately 60% of all BC [5], and multiple gene expression profiles are frequently used to evaluated the recurrence risk and benefit from adjuvant chemotherapy for HR + /Her2− early breast cancer (EBC) patients [6,7,8]. Currently, the 70-gene signature (70-GS, MammaPrint) test and 21-gene recurrence score (RS) have been validated in large prospective cohort, and recommended by the National Comprehensive Cancer Network (NCCN) guidelines as the mainstream of gene expression assays for HR + /Her2− EBCs [9].
Van’t Veer et al. analyzed treatment-naive EBC samples with DNA microarray analysis and found this 70-GS covering 7 pathways related to tumor proliferation, angiogenesis, invasion and migration which could predict risk of distance metastases [10]. In MINDACT trial, 46% of patients at clinical high risk (C-high) were assessed as genetic low risk (G-low) by MP 70-GS, and this group of patients could spare chemotherapy safely. A prospective multicenter study including 660 HR + /Her2− EBC patients showed that MP 70-GS changed half of the physician-intended recommendation of adjuvant chemotherapy [11]. Another study also demonstrated that use of the 70-GS changed patients’ inclination to receive adjuvant chemotherapy and facilitated decision-making [12]. Patients with 70-GS ultralow risk manifested good prognosis which was even distinctive from the 70-GS low risk, with 8-year breast cancer-specific survival (BCSS) rate of 99.6%, and distant metastasis-free interval (DMFI) rate of 97% [13] and might potentially be candidates for further de-escalation of treatment including the duration endocrine therapy [13, 14]. In the neoadjuvant setting, the adaptive randomized I-SPY2 trial and the observational prospective NBRST trial showed that the MP 70-GS high and ultrahigh risk could be associated with pathological complete response (pCR) rate and determine the chemo-sensitivity and long-term outcomes as predictive and prognostic biomarker [15, 16].
Previously we established immunohistochemistry 3 (IHC3) model based on the 21-gene RS and survival data to evaluate the personalized prognosis of HR + /Her2− EBCs and guide treatment choice [17]. We also combined the Clinical Treatment Score post-5 years (CTS5) model and 21-gene RS to develop a novel nomogram for prognosis prediction [18]. Study on BC patients among African-American females (AAF) who has unfavorable outcome compared to Caucasians showed that the 21-gene RS and 70-GS offered different prognostic information [19]. Another study revealed that 70-GS could provide useful information in addition to 21-gene assay resulting in changes of treatment decision in 33.6% of HR + /Her2− BC patients [20]. Given the expenses for the 70-GS assay, it is not always available and affordable worldwide, particularly in developing countries. There is little information about distribution of 70-GS risk among Chinese women and prediction models for 70-GS risk.
In this study, we planned to establish a nomogram model based on individualized medical history, imaging features and clinicopathological characteristics to predict the binary (high/low) and quartile categorized (ultrahigh, high, low, ultralow) risk of 70-GS test in HR + /Her2− EBCs among Chinese from a consecutive clinical cohort.
Patients and methods
Ethics statement
This retrospective study was approved by the Ethics Committee of the Peking Union Medical College Hospital (PUMCH), Chinese Academy of Medical Sciences.
Patient population
There were 150 consecutive female patients diagnosed with HR + /Her2− breast cancer and received treatment in Dept. Breast Surgery, PUMCH from November 2019 to March 2022. The 70-GS (MammaPrint) test was performed by ZhenHe Genecast Biotechnology Ltd, sole and exclusive appointed partner of 70-GS assay in China by Agendia. Patients’ medical history, reports of ultrasound (US) and mammogram (MG) and clinicopathological characteristics were reviewed collected (Fig. 1).
Comparison of medical history risk factors, imaging features and clinicopathological characteristics between high vs low risk of 70-GS test
Comparison of 40 parameters including the patients’ medical history risk factors, imaging features and clinicopathological characteristics was performed between patients with high risk (N = 62) and low risk (N = 88) of 70-GS test. Imaging features including MG density, micro-calcification cluster, nodule/mass and breast imaging reporting and data system (BI-RADS) category, US aspect ratio, boundary, morphology, hyperechoity, multicentricity/multifocality, blood flow, lymph node condition and BI-RADS category were extracted from imaging reports and coded for comparison (Fig. 1).
Comparisons of risk calculations from established models among patients with different categories of 70-GS risk
Comparisons of risk calculations from established models including Adjuvant! Online (AOL) version 8.0 (Additional file 1: Fig. S1) [21], CTS5 [18, 22], IHC3 [17] and Nottingham prognostic index (NPI) [23] were performed between patients with high risk (N = 62) and low risk (N = 88) of 70-GS binary risk classification, as well as among patients with ultra-high (N = 12, defined as 70-GS score < − 0.569) [24], high (N = 50, 70-GS score − 0.569–0), low (N = 65, 70-GS score 0–0.355) and ultra-low risk (N = 23, 70-GS score > 0.355) of 70-gene quartile categorized risk classification [13].
Establishment and validation of nomogram models to predict binary and quartile categorized risk of 70-GS
The data of 150 patients were randomly split by 4:1 ratio and the training set included data from 120 patients and testing set 30 patients. Univariate analyses and multivariate logistic regression were performed both based on binary 70-GS risk classification (high vs low risk), and based on quartile categorized risk classification (ultra-high, high, low and ultra-low risk). Two nomograms were established to predict the binary and quartile risk categories of 70-GS (Fig. 1).
Statistical analysis
The quantitative variables were compared with t-test, the categorical variables with chi-square tests. Univariate analysis was performed to identify variables associated with 70-GS risk. Multivariate logistic regression was used to develop the nomogram models. Risk predictors were selected using both stepwise regression analyses which based on Akaike information criterion (AIC), clinical importance and our previous study. Area under curve (AUC) of receiver operating characteristic (ROC) curves and C-index with 95% confidence interval (CI) was calculated to evaluate accuracy and discrimination of nomogram models. Calibration curve were used for visual inspection of calibration. The decision curve analysis (DCA) was used to reveal the potential clinical utility only for nomogram model of binary risk prediction. Statistical analyses were performed using R (4.0.3) software. All the statistical tests were two-sided, and statistical significance was defined as p value < 0.05.
Results
Comparison of medical history risk factors, imaging features and clinicopathological characteristics between 70-GS high vs low risk patients
Compared to 70-GS low-risk patients, the 70-GS high-risk patients had less cardiovascular co-morbidity (12.9% vs 27.3%, p = 0.034), more grade 3 BC (19.4% vs 4.5%, p = 0.006), lower progesterone receptor (PR) positive percentage (53.92 ± 36.49% vs 68.83 ± 30.43%, p = 0.007), more Ki67 high BC (≥ 20%, 87.1% vs 45.5%, p < 0.001) (Table 1). There were no significant differences in age, body mass index (BMI), childbirth, menarche, menopause, screen-detected BC, bilateral BC, all the included imaging parameters, TNM stage, multi-focality, lymphovascular invasion (LVI), estrogen receptor (ER) positivity and Her2 expression (Table 1).
Comparisons of risk calculations from established models among patients with different categories of 70-GS risk
There was no significant difference in CTS5 score and in percentage of high-risk patients evaluated by AOL, CTS5 and NPI models between 70-GS binary risk groups of patients (Table 2) or in quartile categorized risk classification of patients (Table 3). There were more high-risk patients evaluated by IHC3 model in the 70-GS high-risk group (43.5% vs 11.4%, p < 0.001) (Table 2), and the percentage of IHC3 risk decreased accordingly among 70-GS ultra-high, high, low and ultra-low risk subgroups of patients (83.3%, 34.0%, 15.4% and 0.0%, p < 0.001) (Table 3). The NPI score also decreased accordingly with the 70-GS risk (Table 3).
Establishment and validation of nomogram models to predict binary and quartile categorized risk of 70-GS
The risk factors identified by the univariate analyses and multivariate logistic regression included cardiovascular co-morbidity, histological grade, PR positive percentage and Ki67 index for both nomograms (Figs. 2, 4). The points for each factor were marked on the scale and the total points for each individual could indicate the possibility of high risk (Fig. 2) based on binary risk classification (high vs low) as well as the possibility of ultra-high, high or low risk (Fig. 4) based on quartile categorized risk classification (ultra-high, high, low and ultra-low risk) of 70-GS test.
The calibration plots indicated the predicted 70-GS risk generated by the nomograms had a good consistency with the original 70-GS risk tested (Figs. 3, 5). The DCA indicated that when the threshold for predicted probability of high risk (binary risk classification) was within the range of 0.2–0.8, the nomogram model would add more net benefit than “all or none” strategy. The AUC of ROC curves of the nomogram for binary high risk prediction were 0.826 (C-index 0.903, 95%CI 0.799–1.000) for training and 0.737 (C-index 0.785, 95%CI 0.700–0.870) for validation dataset respectively (Table 4, Figs. 2, 3). The AUC of ROC of nomogram for quartile risk prediction was 0.870 (C-index 0.854, 95%CI 0.746–0.962) for training and 0.592 (C-index 0.769, 95%CI 0.703–0.835) for testing set (Table 4, Figs. 4, 5). The prediction accuracy of the nomogram for quartile categorized risk groups were 55.0% (likelihood ratio tests, p < 0.001) and 53.3% (p = 0.04) for training and validation, which more than double the baseline probability of 25%. The AIC was 128.54 (training) and 46.16 (testing) for binary risk nomogram and 247.07 (training) and 73.85 (testing) for quartile risk nomogram.
Receiver operating characteristic (ROC) curve (A), calibration curve (C) and decision curve analysis (DCA) (E) of the training set (N = 120) as well as the ROC curve (B), calibration curve (D) and DCA analysis (F) of the testing set (N = 30) from the established nomogram model (Fig. 2C) based on binary risk classification (high vs low risk) of 70-gene signature test
Receiver operating characteristic (ROC) curve (A) and calibration curve (C) of the training set (N = 120) and counterparts (B, D) of the testing set (N = 30) from the established nomogram model (Fig. 4C) based on quartile categorized risk classification (ultra-high, high, low and ultra-low risk) of 70-gene signature test
Discussion
Multi-gene assays are used worldwide for prognostic evaluation and predictive information beyond histological parameters for escalation and de-escalation of individualized treatment of HR + /Her2− BC patients. The 21-gene recurrence score (RS) has been validated as quantifying the likelihood of distant recurrence in tamoxifen treated patients with HR + /Her2− BC [25] as well as the potential benefit from adjuvant chemotherapy according to the TAILORx and RxPONDER trial [7, 26]. Study showed that the prognostic and predictive value of the 21-gene RS was consistent in different countries [27], however, there might be difference in the 21-gene assay performance across racial groups with better performance among white compared with African American and Hispanic individuals. [28]
The 21-gene RS could be predicted by nomograms and machine-learning models based on clinicopathological parameters [29,30,31,32]. The 70-GS further stratifies HR + /Her2− BC patients with clinically high-risk according to AOL model into binary (high vs low) or quartile categorized (ultra-high, high, low and ultra-low) risk classifications [6, 33]. Study revealed that the 70-GS could provide additional information regarding patients classified as intermediate risk by the 21-gene assay resulting in changes of treatment decision in 33.6% of HR + /Her2− BC patients [20]. Chemotherapy was added or withheld by the treating physician with more confidence based on the results of the 70-GS test [20]. Given the considerable expenses of the 70-GS assay (approximately 19,800 RMB Yuan, USD 2828$), there was also endeavor to establish deep learning model to predict the binary 70-GS risk from microscopic pathological whole slide images (WSIs) [34]. However, easy-to-use prediction models for 70-GS risk are still lacking.
To our knowledge our study was the first to establish user-friendly nomograms based on imaging and clincopathological parameters to predict the 70-GS risk both in binary (high vs low) and quartile categorized (ultra-high, high, low and ultra-low) risk groups among Chinese population. It might reduce the cost of tumor genetic testing and address optimal management of early breast cancer patients. Moreover, it was in a format that can be translated to patients that would simplify decision-making on their part. Risk factor indicating hormonal conditions such as childbirth, menarche and menopause were not associated with 70-GS risk because all the patients’ BCs were already hormonal receptor positive. However, the co-morbidity of cardio-vascular disease provided additional information of the patient’s general condition and was associated with a lower 70-GS risk (Table 1, Figs. 2, 4). Interestingly, there were no significant differences in all the included imaging features of ultrasound and mammogram between 70-GS high vs low risk patients (Table 1). Not surprisingly, the most ‘classic’ clincopathological parameters associated with 70-GS risk were still histological grade, PR and Ki-67 index (Table 1, Figs. 2, 4), which was in accordance with studies building nomogram predicting 21-gene RS [17, 29, 30].
Majority (71.3%) of patients were clinical high risk according to AOL model. However, study showed that AOL model over-estimated survival of Asian BC patients [35], which might explain why there were 43 (28.7%) patients with AOL low risk and were still judged by their physician as ‘clinical’ high risk and received 70-GS test (Table 2). The CTS5 risk model was developed based on the data from the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial and the BIG (Breast International Group) 1–98 trial to estimate risk of late distant recurrence with parameters including age, tumor size, grade and lymph node [22]. Our previous study developed a nomogram combining both CTS5 and 21-gene RS could improve the evaluation of HR + /Her2− BC patients [18]. The NPI is used to determine prognosis for invasive BC of all subtypes which combines nodal status, tumor size and histological grade, stratifies patients with breast cancer into good, moderate, and poor prognostic groups with validation in large cohort [23, 36, 37]. The IHC3 model was developed in our previous study with parameters including Ki-67 index, PR positive percentage, tumor size and grade and it improved the evaluation of prognosis of HR + /Her2 BC patients compared to 21-gene RS [17]. Notably, the IHC3 risk and the NPI calculated score instead of the NPI risk group significantly correlated with both the binary (high vs low) and quartile categorized (ultra-high, high, low and ultra-low) risk classifications (both p < 0.001, Tables 2, 3). Thus, although the IHC3 model was developed to evaluate the lymph-node negative BC patients, it might also be used to evaluate patient with 1–3 positive nodes combined with the NPI score. For example, if a HR + /Her2 BC patient was evaluated as AOL clinical high-risk and IHC3 low-risk with a NPI score < 4.0, she might potentially spare or de-escalate chemotherapy (Table 3). Furthermore, if a HR + /Her2 BC patient was judged by both of our 70-GS nomogram as low-risk then she might also potentially spare or de-escalate chemotherapy (Figs. 2, 4).
The main reason for constructing a quartile risk model after the establishment and evaluation of the binary risk model is to improve the discrimination and recognition of those patients who were ultra-low risk (70-GS score 0.355–1) or ultra-high risk (70-GS score − 1 ~ − 0.569), who showed clinical prognostic significance and were justified for personalized treatment [13,14,15,16]. Although ROC curve is usually for accuracy evaluation of models predicting binary/dichotomic results [38], we managed to calculate ROC curves both for training and testing set for nomogram model predicting quartile risk (Fig. 3, Table 4). However, the DCA analysis is for evaluating alternative diagnostic and prognostic strategies only for models predicting binary/dichotomic results [39]. The threshold probability used in DCA analysis is used to determine both whether a patient is defined as test-positive or negative and to model the clinical consequences of true and false positives using a clinical net benefit function [40]. Therefore we did not perform DCA analysis for models predicting categorized/multiple classification.
Our study also has several limitations. First, it was a retrospective study with limited sample size, and the testing set is small with only 30 patients, which limited interpretation of the results and also made it difficult for establishment of prediction model with artificial intelligence. Second, the survival data was still unavailable in the currently study, so the prediction model could only be validated with 70-GS risk categories without the actual follow-up outcome. Therefore, our next-step research would be modification on the nomogram models with increased sample size and survival outcome. Third, the imaging features were extracted from the text from the reports of ultrasound and mammogram and there were no actual images analyzed by deep learning and integrated into prediction model. Fourth, the treatment information was not included in the currently study, yet different treatment may affect the prognosis.
Conclusion
We established easy-to-use nomogram models to predict the individualized binary (high vs low) and the quartile categorized (ultra-high, high, low and ultra-low) risk classification of 70-GS test with acceptable performance, which could guide treatment decision making for those who have no access to the 70-GS testing.
Availability of data and materials
The datasets analysed during the current study are available from the corresponding author on reasonable request.
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33.
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32.
Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM, Goss PE. Breast cancer in China. Lancet Oncol. 2014;15(7):e279-289.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.
Kamal AH, Loprinzi CL, Reynolds C, Dueck AC, Geiger XJ, Ingle JN, Carlson RW, Hobday TJ, Winer EP, Goetz MP. Breast medical oncologists’ use of standard prognostic factors to predict a 21-gene recurrence score. Oncologist. 2011;16(10):1359–66.
Cardoso F, van’t Veer LJ, Bogaerts J, Slaets L, Viale G, Delaloge S, Pierga JY, Brain E, Causeret S, DeLorenzi M, et al. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. N Engl J Med. 2016;375(8):717–29.
Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, Geyer CE Jr, Dees EC, Goetz MP, Olson JA Jr, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018;379(2):111–21.
Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360(8):790–800.
Gradishar WJ, Moran MS, Abraham J, Aft R, Agnese D, Allison KH, Anderson B, Burstein HJ, Chew H, Dang C, et al. Breast cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2022;20(6):691–722.
van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6.
Kuijer A, Straver M, den Dekker B, van Bommel ACM, Elias SG, Smorenburg CH, Wesseling J, Linn SC, Rutgers EJT, Siesling S, et al. Impact of 70-gene signature use on adjuvant chemotherapy decisions in patients with estrogen receptor-positive early breast cancer: results of a prospective cohort study. J Clin Oncol. 2017;35(24):2814–9.
van Steenhoven JEC, den Dekker BM, Kuijer A, van Diest PJ, Nieboer P, Zuetenhorst JM, Imholz ALT, Siesling S, van Dalen T. Patients’ perceptions of 70-gene signature testing: commonly changing the initial inclination to undergo or forego chemotherapy and reducing decisional conflict. Breast Cancer Res Treat. 2020;182(1):107–15.
Lopes Cardozo JMN, Drukker CA, Rutgers EJT, Schmidt MK, Glas AM, Witteveen A, Cardoso F, Piccart M, Esserman LJ, Poncet C, et al. Outcome of patients with an ultralow-risk 70-gene signature in the MINDACT trial. J Clin Oncol. 2022;40(12):1335–45.
Opdam M, van der Noort V, Kleijn M, Glas A, Mandjes I, Kleiterp S, Hilbers FS, Kruger DT, Bins AD, de Jong PC, et al. Limiting systemic endocrine overtreatment in postmenopausal breast cancer patients with an ultralow classification of the 70-gene signature. Breast Cancer Res Treat. 2022;194(2):265–78.
Pusztai L, Yau C, Wolf DM, Han HS, Du L, Wallace AM, String-Reasor E, Boughey JC, Chien AJ, Elias AD, et al. Durvalumab with olaparib and paclitaxel for high-risk HER2−negative stage II/III breast cancer: results from the adaptively randomized I-SPY2 trial. Cancer Cell. 2021;39(7):989-998e985.
Whitworth P, Beitsch PD, Pellicane JV, Baron PL, Lee LA, Dul CL, Nash CH 3rd, Murray MK, Richards PD, Gittleman M, et al. Age-independent preoperative chemosensitivity and 5-year outcome determined by combined 70- and 80-gene signature in a prospective trial in early-stage breast cancer. Ann Surg Oncol. 2022;29(7):4141–52.
Zhang Y, Zhou Y, Mao F, Yao R, Sun Q. Ki-67 index, progesterone receptor expression, histologic grade and tumor size in predicting breast cancer recurrence risk: a consecutive cohort study. Cancer Commun. 2020;40(4):181–93.
Wang C, Xu Y, Lin Y, Zhou Y, Mao F, Zhang X, Shen S, Zhang Y, Sun Q. Comparison of CTS5 risk model and 21-gene recurrence score assay in large-scale breast cancer population and combination of CTS5 and recurrence score to develop a novel nomogram for prognosis prediction. Breast. 2022;63:61–70.
Nunes RA, Wray L, Mete M, Herbolsheimer P, Smith KL, Bijelic L, Boisvert ME, Swain SM. Genomic profiling of breast cancer in African-American women using MammaPrint. Breast Cancer Res Treat. 2016;159(3):481–8.
Tsai M, Lo S, Audeh W, Qamar R, Budway R, Levine E, Whitworth P, Mavromatis B, Zon R, Oldham D, et al. Association of 70-gene signature assay findings with physicians’ treatment guidance for patients with early breast cancer classified as intermediate risk by the 21-gene assay. JAMA Oncol. 2018;4(1):e173470.
Ravdin PM, Siminoff LA, Davis GJ, Mercer MB, Hewlett J, Gerson N, Parker HL. Computer program to assist in making decisions about adjuvant therapy for women with early breast cancer. J Clin Oncol. 2001;19(4):980–91.
Dowsett M, Sestak I, Regan MM, Dodson A, Viale G, Thurlimann B, Colleoni M, Cuzick J. Integration of clinical variables for the prediction of late distant recurrence in patients with estrogen receptor-positive breast cancer treated with 5 years of endocrine therapy: CTS5. J Clin Oncol. 2018;36(19):1941–8.
Haybittle JL, Blamey RW, Elston CW, Johnson J, Doyle PJ, Campbell FC, Nicholson RI, Griffiths K. A prognostic index in primary breast cancer. Br J Cancer. 1982;45(3):361–6.
Park JW, Liu MC, Yee D, Yau C, van’t Veer LJ, Symmans WF, Paoloni M, Perlmutter J, Hylton NM, Hogarth M, et al. Adaptive randomization of neratinib in early breast cancer. N Engl J Med. 2016;375(1):11–22.
Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–26.
Kalinsky K, Barlow WE, Gralow JR, Meric-Bernstam F, Albain KS, Hayes DF, Lin NU, Perez EA, Goldstein LJ, Chia SKL, et al. 21-gene assay to inform chemotherapy benefit in node-positive breast cancer. N Engl J Med. 2021;385(25):2336–47.
Carlson JJ, Roth JA. The impact of the Oncotype Dx breast cancer assay in clinical practice: a systematic review and meta-analysis. Breast Cancer Res Treat. 2013;141(1):13–22.
Ibraheem A, Olopade OI, Huo D. Propensity score analysis of the prognostic value of genomic assays for breast cancer in diverse populations using the National cancer data base. Cancer. 2020;126(17):4013–22.
Yoo SH, Kim TY, Kim M, Lee KH, Lee E, Lee HB, Moon HG, Han W, Noh DY, Han SW, et al. Development of a nomogram to predict the recurrence score of 21-gene prediction assay in hormone receptor-positive early breast cancer. Clin Breast Cancer. 2020;20(2):98-107e101.
Yu J, Wu J, Huang O, He J, Zhu L, Chen W, Li Y, Chen X, Shen K. A nomogram to predict the high-risk RS in HR+/HER2−breast cancer patients older than 50 years of age. J Transl Med. 2021;19(1):75.
Harowicz MR, Robinson TJ, Dinan MA, Saha A, Marks JR, Marcom PK, Mazurowski MA. Algorithms for prediction of the Oncotype DX recurrence score using clinicopathologic data: a review and comparison using an independent dataset. Breast Cancer Res Treat. 2017;162(1):1–10.
Baltres A, Al Masry Z, Zemouri R, Valmary-Degano S, Arnould L, Zerhouni N, Devalland C. Prediction of Oncotype DX recurrence score using deep multi-layer perceptrons in estrogen receptor-positive, HER2−negative breast cancer. Breast Cancer. 2020;27(5):1007–16.
Piccart M, van’t Veer LJ, Poncet C, Lopes Cardozo JMN, Delaloge S, Pierga JY, Vuylsteke P, Brain E, Vrijaldenhoven S, Neijenhuis PA, et al. 70-gene signature as an aid for treatment decisions in early breast cancer: updated results of the phase 3 randomised MINDACT trial with an exploratory analysis by age. Lancet Oncol. 2021;22(4):476–88.
Phan NN, Hsu CY, Huang CC, Tseng LM, Chuang EY. Prediction of breast cancer recurrence using a deep convolutional neural network without region-of-interest labeling. Front Oncol. 2021;11:734015.
Bhoo-Pathy N, Yip CH, Hartman M, Saxena N, Taib NA, Ho GF, Looi LM, Bulgiba AM, van der Graaf Y, Verkooijen HM. Adjuvant! online is overoptimistic in predicting survival of Asian breast cancer patients. Eur J Cancer. 2012;48(7):982–9.
Galea MH, Blamey RW, Elston CE, Ellis IO. The Nottingham prognostic index in primary breast cancer. Breast Cancer Res Treat. 1992;22(3):207–19.
Balslev I, Axelsson CK, Zedeler K, Rasmussen BB, Carstensen B, Mouridsen HT. The Nottingham prognostic index applied to 9149 patients from the studies of the Danish breast cancer cooperative group (DBCG). Breast Cancer Res Treat. 1994;32(3):281–90.
Nahm FS. Receiver operating characteristic curve: overview and practical use for clinicians. Korean J Anesthesiol. 2022;75(1):25–36.
Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26(6):565–74.
Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53.
Funding
This work was supported by the Natural Science Foundation of China (No. 81001183), the Science & Technology Research Project of Returned Visiting Scholar, Ministry of Human Resources and Social Security (2015), the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (CIFMS) 2021-I2M-1-014, National High Level Hospital Clinical Research Funding (Grant No. 2022-PUMCH-A-165 and 2022-PUMCH-B-039).
Author information
Authors and Affiliations
Contributions
BP, YX, RY and XC: research idea generation, study design, data collection and analysis and manuscript writing. XZ, ZH, FG, CZ and JL: data collection and patients' follow-up. YZ, CW and SS: methodology and model development, patients’ treatment and data collection. YL and QZ: ultrasound relevant data collection writing-review and editing. XR: pathology relevant data collection and manuscript editing. LK: mammography relevant data collection and writing-review. YZ and QS: Project administration, supervision, and writing-review.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All procedures performed in our studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors. For this type of study, formal consent is not required. This retrospective study was approved by the Ethics Committee of the Peking Union Medical College (PUMC) Hospital, Chinese Academy of Medical Sciences.
Competing interests
The authors have declared that no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Fig. S1.
The Adjuvant! Online (AOL) version 8.0 risk model used in the MINDACT trial as well as in this study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Pan, B., Xu, Y., Yao, R. et al. Nomogram prediction of the 70-gene signature (MammaPrint) binary and quartile categorized risk using medical history, imaging features and clinicopathological data among Chinese breast cancer patients. J Transl Med 21, 798 (2023). https://doi.org/10.1186/s12967-023-04523-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-023-04523-7