Abstract
Background
The identification of novel functional biomarkers is essential for recognizing high-risk patients, predicting recurrence, and searching for appropriate treatment. However, no prognostic biomarker has been applied for colorectal cancer (CRC) in the clinic.
Methods
Integrated with transcriptomic data from public databases, multi-omics examinations were conducted to search prognostic biomarkers for CRC. Moreover, the potential biological functions and regulatory mechanism of these predictive genes were also explored.
Results
In this study, we revealed that three mitochondrial genes were associated with the poor prognosis of CRC. Integrated analyses of transcriptome and proteome of CRC patients disclosed numerous down-regulated mitochondrial genes at both mRNA and protein levels, suggesting a vital role of mitochondria in carcinogenesis. Combined with the bioinformatics studies of transcriptomic datasets of 538 CRC patients, three mitochondrial prognostic genes were eventually selected out, including HIGD1A, SUCLG2, and SLC25A24. The expression of HIGD1A exhibited a significant reduction in two subtypes of adenoma and six subtypes of CRC, while the down-regulation of SUCLG2 and SLC25A24 showed more advantages in rectal mucinous adenocarcinoma. Moreover, we unveiled that these three genes had common expressions and might collaboratively participate in the synthesis of ribosomes. Our original multi-omics datasets, including DNA methylation, structural variants, chromatin accessibility, and phosphoproteome, further depicted the altered modifications on their potential transcriptional factors.
Conclusions
In summary, HIGD1A, SUCLG2, and SLC25A24 might serve as predictive biomarkers for CRC. The biological activities they involved in and their upstream regulators we uncovered would provide a functional context for the further-in-depth mechanism study.
Graphic abstract

Similar content being viewed by others
Background
Colorectal cancer (CRC) is the fourth leading cause of all cancer mortality. It kills nearly 900,000 people a year and accounts for 10% of cancer-inducing deaths [1]. CRC has an annual mortality rate of over 40%, despite the recent emergence of promising new therapies, such as targeted- and immune-therapy [2, 3]. One of the most important reasons for the high death rate is that only the early-stage patients could benefit from treatment, but no significant improvement for patients with distant metastases [4]. Therefore, it is necessary to search for effective predictive biomarkers to identify high-risk patients and develop appropriate treatment strategies to improve their prognosis.
Abnormal mitochondrial function is closely related to CRC [5,6,7,8,9,10,11]. At the opening stage of CRC, mitochondria show excessive consumption and the loss of mitochondria subsequently drives transformation of tumor [6, 7]. In recent years, some studies have identified several mitochondrial genes as potential prognostic biomarkers. However, most studies lack a large-sample cohort, or rarely compare the genes expression between various subtypes of CRC, or the biomarkers have higher correlations with other tumors [12,13,14,15,16], which means more work still need to be done before clinical application.
In this study, intestinal mucosa from eight CRC patients [four early-stage patients (I–II), two late-stage patients (III–IV), and two late-stage patients with systematic metastasis] were collected and analyzed on multiplex dimensionalities. Integrated with the transcriptome data of 538 samples from the Cancer Genome Mapping Project (TCGA) and our original proteomic data, we searched for independent prognostic factors (IPFs) of CRC, and disclosed three mitochondrial genes for predicting the outcomes of CRC patients. Through co-expression enrichment analyses, the biological processes they involved in were also explored. Furthermore, system examinations, including DNA methylation, structural variants, chromatin accessibility, and phosphoproteome, uncovered their potential upstream regulators’ molecular alterations.
Methods
Patients
Eight samples of colorectal adenocarcinoma were collected for investigation. The inclusion criteria were: age ≤ 80 years old, histologically confirmed CRC, no serious major organ dysfunction, and no chemotherapy or radiotherapy. Exclusion criteria were: age ≥ 81 years or older, severe major organ dysfunction, prior chemo- or radiation treatment, or a family history of CRC. In each of the colon cancer tissues studied in this study, two independent pathologists confirmed the morphology of CRC and paracancerous normal colorectal tissue using frozen sections stained with hematoxylin and eosin. All clinical information was obtained from the electronic medical records. Patients participating in this research were voluntary and have been informed and signed informed consent. This project was approved by the ethics committee of Shenzhen People’s Hospital.
Intestinal mucosa from eight CRC patients were collected in a pre-determined method [17]. In brief, tissues within 1 h after surgery were included and collected. The tumorous tissues were taken from the colon segment, and the normal adjacent colonic mucosa within 5 cm of the tumor was gained. The samples were laid in liquid nitrogen for more than 3 h and then stored at − 80 °C.
Studying cohort from TCGA
RNA-Seq datasets of 51 healthy people and 647 CRC patients were downloaded from TCGA (colonic or rectal intestinal mucosa were examined in the RNA-Seq analyses). Samples with incomplete survival time were excluded. Thus 51 healthy peoples and 538 CRC patients in total were collected for the following study.
Protein isolation and trypsin digestion
With liquid nitrogen, all the tumorous tissues and the normal adjacent mucosa tissues from the eight CRC patients were ground into cell powder. Mixed with the lysis buffer [8 M urea and 1% protease inhibitor cocktaiIII (Merck Millipore, 156535140)], the mixture was broken at a high-intensity by ultrasound equipment (Scientz), and kept on ice at all times. The debris was then centrifuged at 12,000×g at 4 °C for 10 min. Finally, the upper layers of the collection were collected, and the protein concentration was measured using the BCA kit (Beyotime P0011-1) according to the manufacturer’s instructions.
The equivalent amount of protein of each sample was taken for enzymatic hydrolysis, and an appropriate amount of standard protein was added. The volume was adjusted by the lysate, mixed with trichloroacetic acid (TCA), to a final concentration 20%. Subsequently, the protein stayed at 4 °C for 2 h. After centrifuged at 4500×g for 5 min, the precipitation was collected and washed with precooled acetone 2–3 times. TEAB was used to dissolve the dried precipitation (final concentration = 200 mM). Dispersed by ultrasound, the mixture was added with trypsin at a ratio of 1:50 (protease: protein, m/m) overnight. Finally, Dithiothreitol (DTT) was put into the solution (final concentration = 5 mM), incubated at 56 °C for 30 min, and then was mixed with Iodoacetamide (IAA) (final concentration = 11 mM), standing at room temperature in the dark for 15 min.
Liquid chromatography mass spectrometry (LC–MS)/MS analyses for proteome and phosphoproteome investigation
The LC–MS/MS analyses were following a standard protocol [18]. The digested proteins (peptides) were dissolved in liquid chromatography (HPLC) mobile phase A and then separated using NanoElute ULTRA-high performance liquid phase system. Mobile phase A is an aqueous solution containing 0.1% formic acid, then loaded into a reversed-phase analytical column (75-μm i.d., 15-cm length columns for proteomic analyses; 100-μm i.d. and 25-cm length columns for phosphoproteomic analyses). Subsequently, for proteomic analysis, 98% acetonitrile with 0.1% formic acid was added into the solvent gradient, the concentration of which was then increased from 6 to 24% for the first 70 min, and increased from 24 to 35% for 14 min, and to 80% for 3 min, and then maintained at 80% for the last 3 min. For phosphoproteomic analysis, acetonitrile with 0.1% formic acid was added into the gradient, the concentration of which was increased then from 2 to 22% for 50 min, and increased from 22 to 35% for 2 min, and increased to 90% for 3 min, and was then maintained at 90% for the last 5 min.
The peptide segments were separated by an ultra-high performance liquid system and then ionized by the capillary ion source. Then the peptide segments were analyzed by timsTOF Pro mass spectrometry. The ion source voltage was set at 1.6 kV, and TOF was used to detect and analyze the peptide parent ion and its secondary fragments. The scanning range of secondary mass spectrometry was set to 100–1700 m/z. The data acquisition mode used the parallel cumulative Serial fragmentation (PASEF) mode. After a first-stage mass spectrometry acquisition, a secondary spectrogram with the parent ion charge in the range of 0–5 was collected in PASEF mode ten times. The dynamic elimination time of tandem mass spectrometry scanning was set as 30 s to avoid repeated parent ion scanning.
Database searching, protein annotation, and functional enrichment
Maxquant (V1.6.6.0) was used to analyze the MS/MS data. Using the database Homo_sapiens_9606_SP_(2019)1115 (20,380 sequences), the reverse library was added to calculate the false positive rate (FDR) caused by random matching. In addition, common contamination repositories were also added to the database to eliminate contaminated proteins' influence on the results. Trypsin/P was used for the enzyme digestion, and the missing digit was set as 2. The minimum peptide length was set to 7 amino acid residues, and the maximum number of modified peptides was set as 5. In the first and main search, the mass tolerance of precursor ions was 20 PPM, and fragment ions were 0.02 Da. The fixed modification was the alkylation of cysteine, and the variable modification was oxidation of methionine. Finally, the FDR for protein identification and PSM identification was set at 1%.
GO annotations were performed using the Uniprot-Goa database, supplemented by InterProScan (an algorithm based on protein sequences). For KEGG annotations, the identified proteins were annotated primarily using KAAS V.2.0 (KEGG online service tool). The proteins were then matched to the KEGG pathway using KEGG Mapper V2.5. Finally, subcellular enrichment was carried out using Wolfpsort V.0.2 software.
The database
Level 3 RNA-Seq datasets (processed and standardized data) of normal and CRC tissues were downloaded from TCGA, and samples with incomplete survival time were excluded. Individual files were merged into matrix files, and gene names were converted from Ensembl IDs to gene symbols via the Ensembl database. The datasets of normal and CRC peoples were extracted from the symbol matrix. The survival analyses (Kaplan–Meier chart) and progression-free survival (PFS) were performed using an R package named “Survival V.” The mRNA expression data of HIGD1A, SUCLG2 and SLC25A24 in 410 patients with CRC were obtained from Oncomine. A list of nuclear-coding mitochondrial genes (NCMGs) was downloaded from MitoCarta [11, 19]. The experimental-tested transcription factors for NCMGs were searched from hTFtarget. The protein–protein interactions (PPI) were analyzed using an online tool GENEMANIA.
IPFs screening
Univariate and multivariate regression Cox analyses were performed on the mRNA expression datasets to explore IPFs for CRC patients. A P-value of less than 0.05 was regarded as significant.
Statistical analyses
Student’s t test was used to analyze the significance of differential expression of proteins detected out by proteomic investigations. Fisher's exact test was used to analyze the significance of enrichment analyses. A P-value of less than 0.05 was regarded as significant.
Results
The integrated analyses of proteome and transcriptome disclosed three mitochondrial genes as prognostic biomarkers of CRC
Since proteins are the basic working units for the human body, we first conducted proteomics examinations on eight CRC patients (four stage I–II patients, two stage III–IV patients, and two stage III–IV patients with systemic metastasis) (Table 1, Fig. 1a). Samples from two patients of each stage were pooled and then performed. As a result, 5451 proteins were quantifiable, in which 598 genes were down-regulated and 496 genes were up-regulated in CRC tissues versus healthy tissues (Fig. 1b). Through GO enrichment of the down-regulated proteins, a large number of mitochondrial proteins were found (Fig. 1c), indicating that mitochondria played an essential role in the development of CRC. And then, we investigated if these expression changes in CRC tissues versus normal adjacent tissues were also occurring at the transcriptional level. Transcriptomic datasets of 538 CRC patients were downloaded from the TCGA (Table 2), and the differentially expressed genes were analyzed. As a result, 10,632 genes were up-regulated, and 5482 genes were down-regulated in CRC tissues versus normal adjacent tissues (Fig. 1d). Through the enrichment analyses of down-regulated genes, the mitochondrial genes were also shown significant expression changes (Fig. 1e). The above results indicated less transcription of mitochondrial genes and a sharp reduction of the mitochondrial proteome. Consistent with previous researches, the inactivity of mitochondria in the opening stage of CRC participates in cellular transformation [6, 7]. Next, we analyzed the transcriptome dataset of 538 CRC patients from TCGA. As a result, 203 IPFs were discovered using univariate and multivariate regression Cox analyses. (Additional file 1: Table S1). After overlapping the 5482 down-regulated mRNA in CRC, and 598 down-regulated proteins in CRC, and the IPFs, we discovered that the expression of six IPFs were both changed at the mRNA and protein levels (Fig. 1f), in which three candidates were mitochondrial genes, including HIGD1A, SUCLG2, and SLC25A24 (Fig. 1g, Additional file 2).
The transcriptome-proteome investigations revealed three mitochondrial genes as potentially prognostic biomarkers for human CRC. a Schematic overview of the research. b The numbers of differentially expressed proteins in CRC. c GO enrichment of the down-regulated proteins. d The numbers of differentially-expressed mRNAs in CRC. Genes with a fold change > 1.5, p < 0.05 were regarded as the differentially-expressed genes. e GO enrichment of the down-regulated mRNAs. f Six IPFs were down-regulated at both mRNA and protein level. g SLC25A24, HIGD1A and SUCLG2 were screened out as IPFs for CRC
To further confirm the protein expression of the three genes in human CRC, we also mined their protein expression in The Human Protein Atlas. We found that the expression of HIGD1A was significantly down-regulated or absent in most CRC patients, and SUCLG2 and SLC25A24 were also down-regulated in partial patients (Fig. 2a–c).
The protein levels of HIGD1A, SUCLG2 and SLC25A24 were decreased in CRC. The immunohistochemical measurement of a HIGD1A (normal = 3, tumor = 21), b SUCLG2 (normal = 6, tumor = 45) and c SLC25A24 (normal = 3, tumor = 21) in the normal and colon adenocarcinoma tissues. The immunohistochemical results were obtained from The Human Protein Atlas. Normal normal adjacent tissues, and CRC CRC tissues
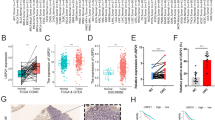
For a detailed study, the mRNA expression of HIGD1A, SUCLG2, and SLC25A24 was also analyzed in 538 CRC patients and healthy people (n = 51) from TCGA. Consequently, HIGD1A, SUCLG2, and SLC25A24 were all down-regulated in CRC (Fig. 3a) and exhibited a stably declined expression throughout the tumor progression (Fig. 3b). Furthermore, the relationship between their expression and the overall survival rate (OS) of patients was also investigated with a follow-up threshold of 12 years. After grouped by the median value of expression of the target gene, the OS of patients was significantly declined in low-expression groups (Fig. 3c). Meanwhile, we analyzed the OS in colon and rectal cancer, respectively. The results showed that all three genes were significantly associated with the OS of colon cancer, but only SUCLG2 was associated with the OS of rectal cancer (Additional file 2). Furthermore, the PFS analyses revealed that the decreased expression of HIGD1A and SLC25A24 accelerated the tumor malignancy (Fig. 3d).
Subsequently, to examine the predictive effect of HIGD1A, SUCLG2, and SLC25A24 on CRC survival when they act as signature, we grouped patients into a high-risk group and low-risk group by the median value of expression of the three genes. The results showed that patients with low expression of the three genes were at high risks (Fig. 4a), and there was a significant difference in the OS between the high-risk group and the low-risk group (Fig. 4b), and the AreaUnderRoc (AUC) curve also showed that the signature has a good value in classification (Fig. 4c).
HIGD1A, SUCLG2, and SLC25A24 acted as a signature could differentiate CRC patients at high risk from low risk. a CRC patients were divided into high- and low-risk groups according to the expression of three genes. b The overall survival rate of patients in high- and low-risk groups. c The AUC curve showed effect of the signature for classification
The expression of the three genes could complement and verify each other in various subtypes of colorectal tumors
Next, to further understand the potentials of HIGD1A, SUCLG2, and SLC25A24 as the IPFs for CRC, we investigated their mRNA expression in unique datasets of CRC and different subtypes of colorectal tumors through the Oncomine database. As a result, the three genes were all down-expressed in 20, five and two independent CRC studies (Fig. 5a). In one independent study, HIGD1A, SUCLG2, and SLC25A24 were all significantly reduced in various subtypes of colorectal tumors, including cecum adenocarcinoma (n = 22), colon adenocarcinoma (n = 101), colon mucinous adenocarcinoma (n = 22), rectal adenocarcinoma (n = 60), rectal mucinous adenocarcinoma (n = 6), and rectosigmoid adenocarcinoma (n = 3) (Fig. 5b–d). Furthermore, another investigation uncovered that the expression of HIGD1A was significantly reduced in two subtypes of adenoma and six subtypes of CRC, but the difference in rectal mucinous adenocarcinoma was significant only in one analysis. On the other hand, SUCLG2 and SLC25A24 were not significantly down-regulated as HIGD1A in adenoma and five subtypes of CRC, while SUCLG2 exhibited consistently decreased expression in both two independent studies of rectal mucinous adenocarcinoma, and SLC25A24 also showed down-regulation in one out of two independent studies (Fig. 5e). This suggested that SUCLG2 and SLC25A24 might be used as helper genes for HIGD1A in clinical diagnosis, which might improve the diagnostic accuracy of rectal mucinous adenocarcinoma.
HIGD1A, SUCLG2, and SLC25A24 could complement each other’s detection results in various subtypes of benign and malignant colorectal tumors. The mRNA expression of a SLC25A24, HIGD1A and SUCLG2 in unique CRC datasets of Oncomine, and b SLC25A24, c HIGD1A, and d SUCLG2 in different subtypes of colorectal tumors. e The expression rank of HIGD1A, SUCLG2, and SLC25A24 in various subtypes of colorectal tumors. The colors represent the gene-rank percentile for the investigations within the cell
Co-expression analyses demonstrated that the three genes might collaboratively participate in the biogenesis of ribosomes or protein translation
As we have known, the three selected-out genes all locate in mitochondria. HIGD1A is identified as a regulator of cytochrome c oxidase and modulates respiratory functions [20, 21]. SUCLG2 is a subunit of succinyl-CoA synthetase, involving the tricarboxylic acid cycle (TCA) [22, 23]. SLC25A24 plays as a carrier protein for the transport of ATP-Mg for phosphate exchanges [24, 25]. Whereas, how they functioned in CRC is not clear. Therefore, we analyzed the co-expressed genes of the three genes basing on the transcriptome datasets of CRC patients. The results showed that the three genes HIGD1A, SUCLG2, and SLC25A24 were co-expressed with each other (Fig. 6a), indicating that they might simultaneously participate in the same biological process or have synergistic effects. Next, we classified all the differentially-expressed proteins based on expression pattern of proteomics detected by the proteomic examinations. We found that HIGD1A and SLC25A24 belong to the same cluster (Fig. 6b), and there were 552 proteins co-expressed with the two genes (Additional file 1: Table S1). SUCLG2 belongs to another cluster, and 741 proteins were expressed together with SUCLG2 (Fig. 6c). Subsequently, we performed KEGG analyses of these co-expressed proteins and observed that a large number of ribosomal proteins were enriched (Fig. 6d, e). The enriched ribosomal proteins, ulteriorly, include cellular and mitochondrial ribosomal proteins. This suggested that the down-regulation of the expression of these three genes might affect the biogenesis of cellular and mitochondrial ribosome. Nevertheless, the ribosome-related proteins co-expressed with HIGD1A and SLC25A24 were utterly different from those co-expressed with SUCLG2. This implied that there might be complementary functions between HIGD1A, SLC25A24, and SUCLG2. These results indicated that the three genes were likely to regulate the ribosomes' biosynthesis collaboratively. Furthermore, we explored the protein–protein interaction of the enriched ribosomal proteins. As a result, most of these proteins have physical interactions, and the potentially central proteins were RPS23 and RPS24 (Fig. 6f, g).
HIGD1A, SUCLG2, and SLC25A24 potentially participated in the ribosome’s biogenesis in a collaborative manner. a The mRNAs of HIGD1A, SUCLG2, and SLC25A24 were likely to be co-expressed with each other in human CRC. b A cluster of proteins co-expressed with HIGD1A and SLC25A24, and c a cluster of proteins co-expressed with SUCLG2. KEGG analyses of d HIGD1A and SLC25A24 co-expressed proteins, and e SUCLG2 co-expressed proteins. Protein–protein interaction networks of f HIGD1A and SLC25A24 co-expressed ribosomal proteins, and g SUCLG2 co-expressed ribosomal proteins
Subsequently, we analyzed the tumor-related biological functions the three genes potentially involved in using single-cell-sequencing datasets from CancerSEA. As a result, expression of the three genes exhibited differences in unique CRC cells (Fig. 7a–c). Noticeably, HIGD1A was negatively correlated with invasion, and positively correlated with differentiation, angiogenesis and apoptosis. SUCLG2 was negatively correlated with proliferation and quiescence. SLC25A24 was positively correlated with cell differentiation and negatively correlated with DNA repair (Fig. 7d–f). Because the expression of these three genes is reduced in CRCs, it is likely that they were potential suppressors in CRC, which related to tumor initiation and progression.
HIGD1A, SUCLG2 and SLC25A24 were potentially associated with a series of tumor-related biological processes such as invasion, angiogenesis and apoptosis. a–c Difference expression of HIGD1A, SUCLG2 and SLC25A24 was shown in unique CRC cells; d–f HIGD1A, SUCLG2 and SLC25A24 were potentially related to multi-functions of cells. **p < 0.01, *p < 0.05
Changes in phosphorylation and chromatin accessibility of the upstream transcriptional factors revealed the underlying mechanisms for the down-regulation of these three genes
Next, to investigate the reasons for the reduction of the three genes, six to eight CRC samples were examined on multiplex dimensionalities, including DNA methylation, structural variation, chromatin accessibility and phosphorproteome. We knew that transcription factors played a crucial role in the expression of downstream gene, so we downloaded all transcriptional factors of the three genes that were experimentally verified in the colon from the TFtarget, and investigated their molecular changes using our multiplatform datasets. As a result, six transcriptional factors, showed phosphorylation changes, including CREB1, ARID1A, CDK12, YAP1, MYH11, and CBX3 (Fig. 8a, b). Moreover, we found that the common transcriptional factors of these three genes, SP1, SP2, KLF5, and GLIS1, lacked DNA binding sites in CRC through analyzing chromatin accessibility (Fig. 8c). The results revealed that the changes of phosphorylation on transcription factors or the absence of binding motifs of the transcription factors on DNA might be reasons for the decreased expression of the three genes.
Comprehensive investigations unveiled altered amount of phosphorylation modifications or binding motifs on the three mitochondrial genes' potential transcriptional factors. a The regulatory functions of transcription factors CREB1, ARID1A, CDK12, YAP1, MYH11, and CBX3 on HIGD1A, SUCLG2, and SLC25A24. Blue color represents a regulatory relationship. b Differentially expressed phosphorylation of the potential transcriptional factors. c The specific binding motifs in normal tissues and their corresponding transcriptional factors. ***p value < 0.001, **p value < 0.01, and *p value < 0.05
Discussion
In this study, we found that the mitochondrial proteome was significantly reduced in CRC. Subsequently, we screened out three mitochondrial IPFs for CRC, including HIGD1A, SUCLG2 and SLC25A24. We verified the expression of these three genes in two colorectal adenomas and six CRC subtypes. Furthermore, the enrichment analyses of these three genes’ co-expressed proteins revealed that their role in the regulation of ribosomal synthesis. Finally, multi-omics studies uncovered different phosphorylation and open chromatin of their potential transcriptional factors, which elucidated the possible mechanism of their decreased expression (Fig. 9).
Integrated analysesof human CRC uncovered three mitochondrial genes potentially associated with patients’ outcomes. Through the multiple platform investigations of the three genes, we demonstrated their expression in various subtypes of colorectal tumors, predicted biological processes they involved in, and disclosed the molecular changes of their potential transcriptional factors. TFs transcriptional factors
Reliable prognostic biomarkers may help providing more effective treatments for patients at high risk. However, because patients with different clinical characteristics have great heterogeneity, and the identification of prognostic markers requires long-term follow-up observation of patients, which costs a lot of manpower and financial resources, so far, no prognostic marker genes have been applied in the diagnosis of CRC in clinical practice. In CRC progression, 50% of patients face metastasis and recurrence [26], highlighting the importance of discovering of prognostic markers. In this study, we found three mitochondrial genes as potential CRC prognostic biomarkers, which provided new opportunities for clinical diagnosis and treatment.
Meanwhile, mitochondrial dysfunction plays an important role in the development of CRC. In recent years, mitochondrial targeting drugs have attracted much more attention. Some drugs have been approved by the Food and Drug Administration (FDA), including ABT-199 (A BCL2-specific agent), etc. [27]. However, there is still a lack of targeted agents available in the treatment of CRC. Through the analysis of CRC tissues versus normal tissues by omics combined with public database, a large number of CRC related genes have been obtained with high throughput, so that we may have a deeper understanding of the pathogenesis of CRC and obtain evidence for looking for new drug targets.
Protein is the basic unit of biological processes in cells. Through proteomic testing of eight CRC samples, we found a reduction of mitochondrial proteins. This is consistent with previous reports of reduced mitochondrial activity at the early CRC [6, 7]. In tumorigenesis, mitochondrial changes start from cell transformation and continue throughout all stages of CRC. The use of mitochondrial genes as prognostic factors is likely to apply to CRC patients at all phases, from the early stage to the late stage, to the stage of metastasis. We know that the amounts of mRNA and protein of a gene are associated with many factors, including mRNA modification [28, 29], translation efficiency [30, 31], and protein stability and degradation [32, 33]. A change in the expression of a gene on mRNA may not result in a difference in the protein level that would affect the body's physiological activities. In other words, changes in both mRNA and protein levels suggest that the gene may have a more substantial effect on the body than those expressed only at the mRNA level. Simultaneously, if the gene is used as a prognostic marker, the mRNA and protein levels can be detected together for mutual verification. In this study, the protein levels of HIGD1A, SUCLG2, and SLC25A24 screened by us were decreased in CRC patients. Meanwhile, transcriptomic analyses of 538 patients showed that HIGD1A, SUCLG2, and SLC25A24 were also significantly down-regulated in mRNA levels, indicating that these three genes may play a more critical role in the occurrence, development, and prognosis of CRC. Moreover, since all three genes were altered in mRNA and protein levels, future clinical applications can be detected at the mRNA or protein level or both, which makes them more promising as prognostic genes.
SUCLG2, HIGD1A, and SLC25A24, three outcome-associated genes of CRC are functional proteins in mitochondria. They take participate in the biological processes in mitochondria including TCA, cell apoptosis and anaerobic environment [20,21,22,23,24,25]. SUCLG2 encodes a succinyl-CoA synthetic enzyme, participating in the formation of succinic acid which is an intermediate product of TCA, and is also one of the fermentation products of anaerobic metabolism [22, 23]. The function of HIGD1A is a positive regulator of cytochrome C oxidase and can induce apoptosis under hypoxia. It is associated with hypoxia microenvironment of cells, and when hypoxia-induced factors 1 (HIF-1) lacks, expression of HIGD1A will increases, cooperated with electron transport proteins reducing oxygen consumption and ROS production [20, 21]. SLC25A24 is a carrier protein that exchanges ATP-Mg for phosphate [24, 25].
We found that the three genes might have synergistic expression, and they were positively correlated with the generation or function of the ribosome and mitochondrial ribosome; that is, the reduction of the proteins of the three genes may lead to the decrease in ribosome biogenesis, the weakening of activity, and the decline or slowing down of protein translation. Ribosomogenesis is the most complex and energy-consuming biological process in the body and plays a close role in tumorigenesis [34,35,36,37,38,39]. Therefore, it can be speculated that, at least at the CRC cell transformation stage, one of the consequences of reduced mitochondrial function may be to induce a reduction in ribosome production, thereby reducing the energy consumption of tumor cells. But whether there is any interaction or synergy between the two biological processes remains unknown. On the other hand, the transcriptome and proteome analyses discovered that more than 16,000 mRNAs but only 1097 proteins were differentially expressed in CRC. This also may be related to the slow translation in CRC cells.
Conclusions
In summary, the present study found that reduced expression of three mitochondrial genes was associated with poor outcomes (overall survival rates, etc.) of CRC patients, and revealed their potential synergistic effect in the modulation of ribosomes. The findings serve candidate genes for the prognosis of CRC and provide a deeper understanding of the pathogenesis.
Availability of data and materials
The mass spectrometry proteomics and phosphoproteomics data have been unloaded and deposited onto the ProteomeXchange Consortium through the PRIDE partner repository: PXD021314, PXD021318.
Abbreviations
- CRC:
-
Colorectal cancer
- IPFs:
-
Independent prognostic factors
- TCGA:
-
The Cancer Genome Atlas Program
- NCMGs:
-
The nuclear-coding mitochondrial genes
- OS:
-
Overall survival rate
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- PFS:
-
Progression-free survival analyses
- PPI:
-
Protein–protein interactions
- TFs:
-
Transcriptional factors
- PASEF:
-
Parallel accumulation serial fragmentation
- FDR:
-
False positive rate
References
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–80.
Rawla P, Barsouk A, Hadjinicolaou AV, Barsouk A. Immunotherapies and targeted therapies in the treatment of metastatic colorectal cancer. Med Sci. 2019;7:83.
Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16:361–75.
Oh HH, Joo YE. Novel biomarkers for the diagnosis and prognosis of colorectal cancer. Intest Res. 2020;18:168–83.
de Araujo LF, Fonseca AS, Muys BR, Plaça JR, Bueno RB, Lorenzi JC, et al. Mitochondrial genome instability in colorectal adenoma and adenocarcinoma. Tumour Biol. 2015;36:8869–79.
Haberman Y, Karns R, Dexheimer PJ, Schirmer M, Somekh J, Jurickova I, et al. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat Commun. 2019;10:38.
Jackson DN, Theiss AL. Gut bacteria signaling to mitochondria in intestinal inflammation and cancer. Gut Microbes. 2020;11:285–304.
Zong WX, Rabinowitz JD, White E. Mitochondria and cancer. Mol Cell. 2016;61:667–76.
Moreno-Sánchez R, Rodríguez-Enríquez S, Marín-Hernández A, Saavedra E. Energy metabolism in tumor cells. FEBS J. 2007;274:1393–418.
Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663–74.
Li H, Slone J, Huang T. The role of mitochondrial-related nuclear genes in age-related common disease. Mitochondrion. 2020;53:38–47.
Gao JY, Song BR, Peng JJ, Lu YM. Correlation between mitochondrial TRAP-1 expression and lymph node metastasis in colorectal cancer. World J Gastroenterol. 2012;18:5965–71.
Yang YC, Chien MH, Lai TC, Su CY, Jan YH, Hsiao M. Monoamine oxidase B expression correlates with a poor prognosis in colorectal cancer patients and is significantly associated with epithelial-to-mesenchymal transition-related gene signatures. Int J Mol Sci. 2020;21:2813.
Lascorz J, Bevier M, Schönfels WV, Kalthoff H, Aselmann H, Beckmann J, et al. Polymorphisms in the mitochondrial oxidative phosphorylation chain genes as prognostic markers for colorectal cancer. BMC Med Genet. 2012;13:31.
Lin PC, Lin JK, Yang SH, Wang HS, Li AF, Chang SC. Expression of beta-F1-ATPase and mitochondrial transcription factor A and the change in mitochondrial DNA content in colorectal cancer: clinical data analysis and evidence from an in vitro study. Int J Colorectal Dis. 2008;2312:1223–32.
Pan T, Liu J, Xu S, Yu Q, Wang H, Sun H, et al. ANKRD22, a novel tumor microenvironment-induced mitochondrial protein promotes metabolic reprogramming of colorectal cancer cells. Theranostics. 2020;10:516–36.
van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–45.
Meier F, Brunner AD, Koch S, Koch H, Lubeck M, Krause M, et al. Online parallel accumulation-serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol Cell Proteom. 2018;17:2534–45.
Rath S, Sharma R, Gupta R, Ast T, Chan C, Durham TJ, et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021;49:D1541–7.
Nagao T, Shintani Y, Hayashi T, Kioka H, Kato H, Nishida Y, et al. Higd1a is a positive regulator of cytochrome c oxidase. Proc Natl Acad Sci USA. 2015;112:1553–8.
Hayashi T, Asano Y, Shintani Y, Aoyama H, Kioka H, Tsukamoto O, et al. Higd1a improves respiratory function in the models of mitochondrial disorder. FASEB J. 2020;34:1859–71.
Chinopoulos C, Batzios S, van den Heuvel LP, Rodenburg R, Smeets R, Waterham HR, et al. Mutated SUCLG1 causes mislocalization of SUCLG2 protein, morphological alterations of mitochondria and an early-onset severe neurometabolic disorder. Mol Genet Metab. 2019;126:43–52.
Dobolyi A, Bagó AG, Gál A, Molnár MJ, Palkovits M, Adam-Vizi V, et al. Localization of SUCLA2 and SUCLG2 subunits of succinyl CoA ligase within the cerebral cortex suggests the absence of matrix substrate-level phosphorylation in glial cells of the human brain. J Bioenergy Biomembr. 2015;47:33–41.
Writzl K, Maver A, Kovačič L, Martinez-Valero P, Contreras L, Satrustegui J, et al. De novo mutations in SLC25A24 cause a disorder characterized by early aging, bone dysplasia, characteristic face, and early demise. Am J Hum Genet. 2017;101:844–55.
Harborne SP, King MS, Crichton PG, Kunji ER. Calcium regulation of the human mitochondrial ATP-Mg/Pi carrier SLC25A24 uses a locking pin mechanism. Sci Rep. 2017;7:45383.
Lichtenstern CR, Ngu RK, Shalapour S, Karin M. Immunotherapy, inflammation and colorectal cancer. Cells. 2020;9:61.
Chiu HY, Tay EXY, Ong DST, Taneja R. Mitochondrial dysfunction at the center of cancer therapy. Antioxid Redox Signal. 2020;32:309–30.
Lee Y, Choe J, Park OH, Kim YK. Molecular mechanisms driving mRNA degradation by m 6 a modification. Trends Genet. 2020;36:177–88.
Liang W, Lin Z, Du C, Qiu D, Zhang Q. mRNA modification orchestrates cancer stem cell fate decisions. Mol Cancer. 2020;19:38.
Xu M, Tao Z, Wang S, Jiang Y, Qu M. Suppression of oncogenic protein translation via targeting eukaryotic translation initiation factor 4E overcomes chemo-resistance in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2019;512:902–7.
Koppenhafer SL, Goss KL, Terry WW, Gordon DJ. mTORC1/2 and protein translation regulate levels of CHK1 and the sensitivity to CHK1 inhibitors in ewing sarcoma cells. Mol Cancer Ther. 2018;17:2676–88.
Costa L, Corre S, Michel V, Le Luel K, Fernandes J, Ziveri J, et al. USF1 defect drives p53 degradation during Helicobacter pylori infection and accelerates gastric carcinogenesis. Gut. 2020;69:1582–91.
Schapira M, Calabrese MF, Bullock AN, Crews CM. Targeted protein degradation: expanding the toolbox. Nat Rev Drug Discov. 2019;18:949–63.
Sharifi S, da Costa HFR, Bierhoff H. The circuitry between ribosome biogenesis and translation in stem cell function and ageing. Mech Ageing Dev. 2020;189:111282.
Correll CC, Bartek J, Dundr M. The nucleolus: a multiphase condensate balancing ribosome synthesis and translational capacity in health, aging and ribosomopathies. Cells. 2019;8:869.
Karbstein K. Mitochondria teach ribosome assembly. Science. 2019;365:1077–8.
Slimane SN, Marcel V, Fenouil T, Catez F, Saurin JC, Bouvet P, et al. Ribosome biogenesis alterations in colorectal cancer. Cells. 2020;9:E2361.
Pelletier J, Thomas G, Volarević S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer. 2018;18:51–63.
Gilles A, Frechin L, Natchiar K, Biondani G, Loeffelholz OV, Holvec S, et al. Targeting the human 80S ribosome in cancer: from structure to function and drug design for innovative adjuvant therapeutic strategies. Cells. 2020;9:629.
Acknowledgements
The authors would like to thank CRC patients who donated their specimen to the study.
Funding
This work was supported by the Science and Technology Planning Project of Guangdong Province, China (No. 2017B020209001), the Key Research and Development Program of Guangdong Province (No. 2019B020229001), the National Natural Science Foundation of China (No. 81671596, 82003172), the Guangdong Basic and Applied Basic Research Foundation (No. 2019A1515111138) and China Postdoctoral Science Foundation (No. 2020M673065).
Author information
Authors and Affiliations
Contributions
WZ, YD and YX: conceived the study; LHY: study design; CZ: collected specimens and tested the quality; WZ, DET, LWL, WXC and LGX: data extraction and elaboration. All authors: data interpretation and discussion. WZ, WED and YD: manuscript writing. All authors: manuscript editing. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All patients participating in this study were informed, signed informed consents and voluntarily participated. This project was approved by the ethical committee of Shenzhen People’s Hospital (LL-KY-2019213).
Consent for publication
Not applicable.
Competing interests
The authors declared no potential competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Down-regulated proteins in CRC patients; down-regulated mRNAs in CRC patients; independent prognostic factors for CRC; co-expressed proteins of HIGD1A, SUCLG2 and SLC25A24; ribosome proteins co-expressed with HIGD1A, SUCLG2 and SLC25A24; experimentally-tested transcriptional factors of HIGD1A, SUCLG2 and SLC25A24 in human colon.
Additional file 2.
The cox analyses of HIGD1A, SUCLG2 and SLC25A24; The overall survival rates of HIGD1A, SUCLG2 and SLC25A24 in colon and rectum cancer.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, W., Lin, L., Xia, L. et al. Multi-omics analyses of human colorectal cancer revealed three mitochondrial genes potentially associated with poor outcomes of patients. J Transl Med 19, 273 (2021). https://doi.org/10.1186/s12967-021-02939-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-021-02939-7