Abstract
Background
Experimental studies suggested that intercellular adhesion molecule 4 (ICAM-4) might be implicated in ischemic stroke, but the population-based evidence on the relationship between ICAM-4 and ischemic stroke were limited. Herein, we performed a two-sample Mendelian randomization (MR) analysis to investigate the associations of genetically determined plasma ICAM-4 with the risks of ischemic stroke and its subtypes.
Methods
A total of 11 single-nucleotide polymorphisms associated with ICAM-4 were selected as instrumental variables based on the genome-wide association studies (GWAS) with 3,301 European individuals. Summary-level data about ischemic stroke and its subtypes were obtained from the Multi-ancestry GWAS launched by the International Stroke Genetics Consortium. We used the inverse-variance weighted method followed by a series of sensitivity analyses to evaluate the associations of genetically determined ICAM-4 with the risks of ischemic stroke and its subtypes.
Results
Genetically determined higher ICAM-4 levels were significantly associated with increased risks of ischemic stroke (in the IVW method fitted to multiplicative random effects model: odds ratio [OR] per standard deviation [SD] increase, 1.04; 95% confidence interval [CI], 1.01–1.07; P = 0.006; in the IVW analysis with fixed effects model: OR per SD increase, 1.04; 95% CI, 1.01–1.07; P = 0.003) and cardioembolic stroke (in multiplicative random effects model: OR per SD increase, 1.08; 95% CI, 1.02–1.14; P = 0.004; in fixed effects model: OR per SD increase, 1.08; 95% CI, 1.03–1.13; P = 0.003). There was no association of ICAM-4 with the risks of large artery stroke and small vessel stroke. MR-Egger regression showed no directional pleiotropy for all associations, and the sensitivity analyses with different MR methods further confirmed these findings.
Conclusions
We found positive associations of genetically determined plasma ICAM-4 with the risks of ischemic stroke and cardioembolic stroke. Future studies are needed to explore the detailed mechanism and investigate the targeting effect of ICAM-4 on ischemic stroke.
Similar content being viewed by others
Background
Stroke is the leading cause of death and severe disability around the world [1]. In 2019, there were 101 million stroke cases globally, of which ischemic stroke was 77.19 million and accounted for more than 70%.1 Smoking, excessive alcohol consumption, obesity, hypertension, hyperlipidemia, and diabetes are well-established risk factors for ischemic stroke [1, 2]. Although interventions against these traditional risk factors have obtained substantial progress, ischemic stroke is still a major public health problem worldwide [1, 2]. Therefore, further studies are needed to identify novel biomarkers for the prevention and treatment of ischemic stroke.
Intercellular adhesion molecule 4 (ICAM-4), also known as the Landsteiner-Wiener blood group glycoprotein, was a member of the ICAM family expressed primarily on erythrocytes and erythroid precursor cells [3,4,5]. Compared to other members of ICAMs, ICAM-4 could bind various integrins (e.g., CD11a/CD18, CD11b/CD18, α4β1, αIIbβ3, and αv integrins) on cell surface [3, 5]. Previous studies suggested that the interaction of ICAM-4 with integrins played a critical role in cell adhesion, hemostasis and thrombosis [6,7,8]. In addition, ICAM-4 was reported to mediate the abnormal adhesion of sickle cell to endothelial cells, and then induced platelet-erythrocyte aggregation and blocked blood flow [9]. Platelet-erythrocyte aggregation and endothelial cell dysfunction were well known implicated in the thrombosis, [6,7,8, 10] so we hypothesized that ICAM-4 played a key role in ischemic stroke etiology. However, to date, there is limited population-based research on the effect of ICAM-4 in the risk of ischemic stroke.
Mendelian randomization (MR) is an emerging epidemiologic method in which genetic variants associated with the exposure of interest are served as instrumental variables to assess the causal effect of a lifelong exposure on diseases [11, 12]. MR design had previously been used to assess the effects of circulating cytokines, serum bilirubin, lipoprotein lipid, and apolipoproteins on the risk of ischemic stroke [13,14,15]. Besides, two-sample MR design could greatly increase the scope of MR analysis with advantages of two independent samples [16]. Hence, we explored the associations of genetically determined ICAM-4 levels and the risks of ischemic stroke and its subtypes (cardioembolic stroke [CES], large artery stroke [LAS], and small vessel stroke [SVS]) via a two-sample MR study.
Methods
Study design
The present study was reported using the Strengthening the Reporting of Observational Studies in Epidemiology using Mendelian Randomization (STROBE-MR) guideline [17]. As shown in Fig. 1, we designed a two-sample MR study to systematically investigate the associations of genetically determined ICAM-4 levels and the risks of ischemic stroke and its subtypes. We selected single-nucleotide polymorphisms (SNPs) that achieved genome-wide significance (P < 5.0 × 10− 8) for the ICAM-4 levels identified by Sun et al. as instrumental variables in the MR analysis [18]. The summary genetic data about ischemic stroke and its subtypes were obtained from the Multi-ancestry Genome-Wide Association Study launched by the International Stroke Genetics Consortium (MEGASTROKE) [19]. All participants in the present MR analysis were subjects of European ancestry. The protocol and data were approved by the ethics committee of the original genome-wide association studies (GWASs), and written informed consent was obtained from each participant prior to data collection.
An overview of the present mendelian randomization study design. The assumption is that (1) instrumental variables are associated with intercellular adhesion molecule 4 levels, (2) instrumental variables are not associated with confounders, (3) and instrumental variables affect ischemic stroke or its subtypes only through the effects on intercellular adhesion molecule 4 levels
Abbreviations: SNP, single-nucleotide polymorphism; IVW, inverse-variance weighted; MR-RAPS, mendelian randomization robust adjusted profile score
Instrumental variables for ICAM-4
The summary genetic data about ICAM-4 were derived from a genomic atlas analysis of the human plasma proteome conducted by Sun et al. [18] They randomly selected two non-overlapping sub-cohorts from INTERVAL study, which comprised about 50,000 participants nested within a randomized trial of varying blood donation intervals [18, 20]. After genetic quality control, Sun et al. analyzed 2,994 proteins in 3,301 individuals from European with approximately 10.5 million SNPs (available from the IEU GWAS database: https://gwas.mrcieu.ac.uk/). The SNPs that were identified to be associated with plasma ICAM-4 levels at the genome-wide significance level (P < 5.0 × 10− 8) and were not in linkage disequilibrium (LD) with other SNPs (r2 < 0.1 within a clumping window of 500 kb) were selected as genetic instruments for plasma ICAM-4 levels. In addition, if the ICAM-4-associated SNP was not available in the MEGASTROKE dataset, a proxy SNP (r2 > 0.8) was selected by default based on a 1000 Genomes European reference panel. Overall, a total of 11 SNPs were selected as the genetic instruments for plasma ICAM-4 in this MR study (Table 1).
Data source for ischemic stroke and its subtypes
In the present study, genetic association data of ischemic stroke and its subtypes were obtained from the previously published GWAS released by the MEGASTROKE project, which was a large-scale international collaboration launched by the International Stroke Genetics Consortium [19]. This dataset was based on a meta-analysis of 29 European-ancestry GWASs with 8 million SNPs, involving 34,217 ischemic stroke cases and 406,111 controls. Among these ischemic stroke cases, 7,193 cases were CES, 4,373 cases were LAS, and 5,386 cases were SVS according to the Trial of Org 10,172 in Acute Stroke Treatment criteria [21].
Statistical analysis
A two-sample MR analysis was performed to estimate the associations of genetically determined plasma ICAM-4 with the risks of ischemic stroke and its subtypes using summarized data of the SNP-ICAM-4 and SNP-ischemic stroke. We used F statistics with the formula F = (\(\frac{N-K-1}{K}\)) (\(\frac{{R}^{2}}{1-{R}^{2}}\)) to measure the strength of instrumental variables, where R2 was the proportion of variation in plasma ICAM-4 levels explained by the instrumental SNPs, N was the sample size, and K was the number of SNPs used as genetic instruments of plasma ICAM-4 [22]. In general, a mean F statistic greater than 10 ensured negligible bias from weak instruments [23]. In addition, an online web tool named mRnd (https://shiny.cnsgenomics.com/mRnd/) was used to calculate the statistical power of the present MR study [24].
Conventionally, under the causal null hypothesis that all instrumental variables were valid and pleiotropic balanced, the inverse-variance weighted (IVW) method with the greatest statistical power was the most efficient way to combine them into a single variant-specific causal estimate and it generally was used as the primary analysis method in MR study [25,26,27]. Therefore, we used the IVW method as the main analysis to evaluate the associations of genetically determined plasma ICAM-4 with the risks of ischemic stroke and its subtypes in the present MR study. The heterogeneity among genetic instruments was assessed via Cochran’s Q statistic, which was a weighted sum of the squared distances of the variant-specific estimates from the overall IVW estimate [27, 28]. A large value of the Q statistic meant that the variant-specific ratio estimates differ by more than expected due to chance alone and significant P-value with Q-statistic was usually set to 0.05 [28, 29]. To avoid possible causal estimates bias that might be caused by fitting different models, we conducted the random-effect IVW model and fixed-effect IVW model to further explore the association between ICAM-4 and ischemic stroke [27, 30].
In the sensitivity analyses, we employed various MR methods (the penalized IVW, [31] maximum likelihood, [32] MR-Robust Adjusted Profile Score [MR-RAPS], [33] and MR-Egger regression [34]) to assess the robustness of our findings. The penalized IVW penalized the weights of SNPs with pleiotropy [31]. The maximum likelihood method was applied to provide relatively reliable estimates in the presence of measurement error in the SNP-exposure effect [32]. The MR-RAPS analysis could solve the bias of horizontal pleiotropy and weak instruments [33]. The MR-Egger regression method could evaluate the average pleiotropic effects across all SNPs via the intercept term [34].
Results were presented as odds ratios (ORs) with 95% confidence intervals (CIs). A Bonferroni-corrected significance level of 2-sided P < 0.0125 (0.05/4 [ischemic stroke and 3 ischemic stroke subtypes]) was considered as the statistically significant evidence for a causal association. All analyses were performed using R software (version 4.1.1; R Development Core Team) with R packages named gtx, TwoSampleMR, and MendelianRandomization.
Results
A total of 11 SNPs were used as genetic instruments for plasma ICAM-4 levels in the present study, and the details of genetic instruments were provided in Table 1. 11 genetic instruments identified together explained 34.90% variances of plasma ICAM-4 levels, and the statistical power in this MR study ranged from 97 to 100% (Table S1). The F-statistics of instrumental variables was 160.60, indicating that there was no weak instrument bias (Table S1).
Causal effects of ICAM-4 levels on ischemic stroke and its subtypes
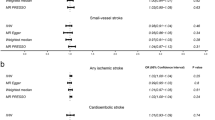
The results from the Cochran’s Q test (Table S2) contained the Q statistic, degrees of freedom, and Q statistic associated P-value, and we observed no evidence of heterogeneity between genetic variants (all P > 0.05). As illustrated in Fig. 2, the IVW analysis with multiplicative random effects model demonstrated that genetically determined ICAM-4 levels were positively associated with the risks of ischemic stroke (OR per SD [standard deviation] increase: 1.04; 95% CI: 1.01–1.07; P = 0.006) and CES (OR per SD increase: 1.08; 95% CI: 1.02–1.14; P = 0.004). In contrast, there was no significant association of genetically determined ICAM-4 with LAS (OR per SD increase: 1.06; 95% CI: 1.00-1.12; P = 0.043) and SVS (OR per SD increase: 1.03; 95% CI: 0.97–1.09; P = 0.302). In the IVW analysis fitting fixed effects model, genetically determined high ICAM-4 levels were associated with increased risks of ischemic stroke (OR per SD increase: 1.04; 95% CI: 1.01–1.07; P = 0.003) and CES (OR per SD increase: 1.08; 95% CI: 1.03–1.13; P = 0.003) but not with LAS (OR per SD increase: 1.06; 95% CI: 0.99–1.13; P = 0.090) and SVS (OR per SD increase: 1.03; 95% CI: 0.97–1.09; P = 0.338). Associations between each instrumental variant for ICAM-4 levels and the risks of ischemic stroke and CES were presented in Fig. 3.
Main analysis for associations of genetically determined ICAM-4 levels with ischemic stroke and its subtypes
Abbreviations: ICAM-4, intercellular adhesion molecule 4; IVW, inverse-variance weighted; SNP, single-nucleotide polymorphism; SE: standard error; OR, odds ratio; 95% CI, 95% confidence interval
Associations between genetic instruments of ICAM-4 and the risks of ischemic stroke and cardioembolic stroke. The line indicates the estimate for the associations of intercellular adhesion molecule 4 (ICAM-4) levels with ischemic stroke and cardioembolic stroke using inverse-variance weighted method. Circles indicate associations of each genetic variant related to ICAM-4 levels with the risks of ischemic stroke and cardioembolic stroke. Genetic error bars indicate 95% confidence intervals
A, ischemic stroke; B, cardioembolic stroke
Sensitivity analyses
We conducted sensitivity analyses with a series of MR methods to assess the robustness of our findings (Table 2). Genetically determined ICAM-4 was positively associated with the risk of ischemic stroke in the sensitivity analyses with the penalized IVW method (OR per SD increase:1.04; 95% CI:1.01–1.07; P = 0.006), maximum likelihood method (OR per SD increase: 1.04; 95% CI:1.01–1.07; P = 0.003), and MR-RAPS method (OR per SD increase: 1.04; 95% CI:1.01–1.07; P = 0.003). Similarly, the penalized IVW analysis (OR per SD increase: 1.08; 95% CI:1.02–1.14; P = 0.004), maximum likelihood analysis (OR per SD increase: 1.08; 95% CI:1.03–1.14; P = 0.003), and MR-RAPS analysis (OR per SD increase: 1.08; 95% CI:1.03–1.14; P = 0.003) showed that genetically determined high ICAM-4 levels were associated with an increased risk of CES. In addition, MR-Egger regression suggested that there was no directional pleiotropy for these associations (all P > 0.05; Table 2).
Discussion
To the best of our knowledge, this is the first study with sufficient statistical power to investigate the associations of genetically determined ICAM-4 levels with the risks of ischemic stroke and its subtypes. In this MR study with 446,696 European participants, we found that genetically determined high ICAM-4 levels were associated with increased risks of ischemic stroke and CES, but not LAS or SVS. Sensitivity analyses using a series of MR methods further confirmed these identified associations.
As the key molecules involved in progression of the ischemia, the upregulation of ICAMs was able to enhance leukocyte-endothelial cell interactions and induce neutrophils infiltrate into damaged brain tissue, thereby aggravating the damage of blood-brain barrier [10]. ICAM-4, an important member of the ICAMs, was well-established to be implicated in hemostasis and thrombosis [6, 7]. Data from animal studies had revealed that ICAM-4 contributed to the adhesion of endothelial cells, thrombosis, and vaso-occlusion through interacting with activated platelets and leukocytes [35, 36]. Vivian et al. found that ICAM-4 could induce massive erythrocytes incorporation into thrombus and activate platelets, while blockading ICAM-4 could cause a reduction of fibrin and thrombus [8]. Therefore, it has been suggested that ICAM-4 may be associated with the development of thrombosis and ischemic stroke, but the population-based evidence is limited so far.
We conducted a systematical MR study to investigate the associations of genetically determined ICAM-4 levels and the risks of ischemic stroke and its subtypes. In this MR study, genetically determined high ICAM-4 levels were observed to be associated with the increased risk of ischemic stroke, suggesting that it might be a promising predictive marker for ischemic stroke. Besides, in the analysis of ischemic stroke subtypes, we found that there were possible mechanism-specific detrimental effects of genetically determined ICAM-4 on CES but not LAS or SVS. Given that ischemic stroke subtypes might differ for the genetic pathophysiological mechanisms, [37] CES had a higher inflammatory environment, macrophage, and platelet content in thrombus than the other stroke subtypes [38, 39]. Therefore, we speculated that elevated ICAM-4 levels were significantly associated with increased risks of ischemic stroke and CES via mediating aggregation and abnormal adhesion of inflammatory cells, macrophages, and platelets. Further studies are warranted to explore the detailed mechanism underlying the association of ICAM-4 with ischemic stroke and CES.
Our findings had significant public health and clinical implications. The present MR study was the first to provide population-based evidence for the associations of ICAM-4 levels with the risks of ischemic stroke and its subtypes from a genetic point of view. Collectively, our study showed that elevated ICAM-4 levels increased the risks of ischemic stroke and CES, suggesting that ICAM-4 might be acted as a promising biomarker to identify high-risk individuals for active monitoring and early intervention of ischemic stroke. In addition, it was of clinical interest to explore whether targeting ICAM-4 or its downstream effectors could reduce the risks of ischemic stroke, especially CES.
The present study had several methodological strengths. First, MR design followed the genetic rule that parental alleles were randomly assigned to offspring and possessed reasonable causal order [11, 12]. Therefore, the implementation of MR approach in this study diminished the interference of confounding factors and reverse causation on the results, which might be more convincing than observational studies [11, 12]. In addition, we used the most comprehensive and the largest available GWASs about ICAM-4 levels, ischemic stroke, and its subtypes, [18, 19] which enabled us to provide a valid appraisal of the associations with the high statistical power. Finally, the significant associations observed in this MR study were subjected to multiple corrected and a series of sensitivity analyses further confirmed our findings.
Our study had several limitations that needed to be interpreted. Firstly, MR analysis might be influenced by instrument bias and potential pleiotropy. However, the F-statistic for the genetic instruments in the present study was greater than 10, suggesting that there was no weak instrument bias. Furthermore, the MR-Egger regression suggested no directional pleiotropy for identified associations in this MR study. Secondly, the present study estimated the lifetime effect of plasma ICAM-4 in the risks of ischemic stroke and its subtypes, so the results should not be directly extrapolated to assess the effect of any potential clinical intervention targeting ICAM-4. Finally, the summary GWAS data we used merely concerned European individuals, so we should cautiously utilize our conclusion in racially and ethnically diverse populations. However, this restriction decreased the possibility of spurious associations due to population stratification bias. Further studies are needed to confirm our findings among individuals of non-European ancestry.
Conclusions
We found positive associations of genetically determined high plasma ICAM-4 levels with the risks of ischemic stroke and CES. Further studies are needed to verify our findings and explore the detailed mechanism underlying the detrimental effects of ICAM-4 on the risk of ischemic stroke.
Data availability
The public summary statistic data for ICAM-4, ischemic stroke and ischemic stroke subtypes during the current study are available from the IEU GWAS database (https://gwas.mrcieu.ac.uk/). Statistical code analysed during the current study are available from the corresponding author on reasonable request. (email: Zhengjin22163@fudan.edu.cn or zbzhu@suda.edu.cn).
Abbreviations
- ICAM-4:
-
Intercellular adhesion molecule 4
- MR:
-
Mendelian randomization
- GWAS:
-
Genome-wide association studies
- OR:
-
Odds ratio
- SD:
-
Standard deviation
- CI:
-
Confidence interval
- CES:
-
Cardioembolic stroke
- LAS:
-
Large artery stroke
- SVS:
-
Small vessel stroke
- STROBE-MR:
-
Strengthening the Reporting of Observational Studies in Epidemiology using Mendelian Randomization
- SNPs:
-
Single-nucleotide polymorphisms
- MEGASTROKE:
-
Multi-ancestry Genome-Wide Association Study launched by the International Stroke Genetics Consortium
- LD:
-
Linkage disequilibrium
- IVW:
-
Inverse-variance weighted
- MR-RAPS:
-
Mendelian randomization-Robust Adjusted Profile Score
References
Feigin VL, Stark BA, Johnson CO, et al. Global, regional, and national burden of stroke and its risk factors, 1990–2019: a systematic analysis for the global burden of Disease Study 2019. Lancet Neurol. 2021;20(10):795–820. https://doi.org/10.1016/s1474-4422(21)00252-0.
Feske SK. Ischemic stroke. Am J Med. Dec 2021;134(12):1457–64. https://doi.org/10.1016/j.amjmed.2021.07.027.
Bailly P, Tontti E, Hermand P, Cartron JP, Gahmberg CG. The red cell LW blood group protein is an intercellular adhesion molecule which binds to CD11/CD18 leukocyte integrins. Eur J Immunol Dec. 1995;25(12):3316–20. https://doi.org/10.1002/eji.1830251217.
Spring FA, Parsons SF, Ortlepp S, et al. Intercellular adhesion molecule-4 binds alpha(4)beta(1) and alpha(V)-family integrins through novel integrin-binding mechanisms. Blood Jul. 2001;15(2):458–66. https://doi.org/10.1182/blood.v98.2.458.
Ihanus E, Uotila LM, Toivanen A, Varis M, Gahmberg CG. Red-cell ICAM-4 is a ligand for the monocyte/macrophage integrin CD11c/CD18: characterization of the binding sites on ICAM-4. Blood Jan. 2007;15(2):802–10. https://doi.org/10.1182/blood-2006-04-014878.
Toivanen A, Ihanus E, Mattila M, Lutz HU, Gahmberg CG. Importance of molecular studies on major blood groups–intercellular adhesion molecule-4, a blood group antigen involved in multiple cellular interactions. Biochim Biophys Acta Mar. 2008;1780(3):456–66. https://doi.org/10.1016/j.bbagen.2007.09.003.
Hermand P, Gane P, Huet M, et al. Red cell ICAM-4 is a novel ligand for platelet-activated alpha IIbbeta 3 integrin. J Biol Chem Feb. 2003;14(7):4892–8. https://doi.org/10.1074/jbc.M211282200.
Du VX, Huskens D, Maas C, Al Dieri R, de Groot PG, de Laat B. New insights into the role of erythrocytes in thrombus formation. Semin Thromb Hemost Feb. 2014;40(1):72–80. https://doi.org/10.1055/s-0033-1363470.
Zennadi R, Hines PC, De Castro LM, Cartron JP, Parise LV, Telen MJ. Epinephrine acts through erythroid signaling pathways to activate sickle cell adhesion to endothelium via LW-alphavbeta3 interactions. Blood Dec. 2004;1(12):3774–81. https://doi.org/10.1182/blood-2004-01-0042.
Hu X, De Silva TM, Chen J, Faraci FM. Cerebral vascular Disease and Neurovascular Injury in ischemic stroke. Circ Res Feb. 2017;3(3):449–71. https://doi.org/10.1161/CIRCRESAHA.116.308427.
Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med Apr. 2008;15(8):1133–63. https://doi.org/10.1002/sim.3034.
Davies NM, Holmes MV, Davey Smith G. Reading mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ Jul. 2018;12:362:k601. https://doi.org/10.1136/bmj.k601.
Georgakis MK, Gill D, Rannikmae K, et al. Genetically determined levels of circulating cytokines and risk of stroke. Circulation Jan. 2019;8(2):256–68. https://doi.org/10.1161/CIRCULATIONAHA.118.035905.
Choi Y, Lee SJ, Spiller W, et al. Causal Associations between serum bilirubin levels and decreased stroke risk: a two-sample mendelian randomization study. Arterioscler Thromb Vasc Biol Feb. 2020;40(2):437–45. https://doi.org/10.1161/ATVBAHA.119.313055.
Yuan S, Tang B, Zheng J, Larsson SC. Circulating lipoprotein lipids, apolipoproteins and ischemic stroke. Ann Neurol Dec. 2020;88(6):1229–36. https://doi.org/10.1002/ana.25916.
Zheng J, Baird D, Borges MC, et al. Recent developments in mendelian randomization studies. Curr Epidemiol Rep. 2017;4(4):330–45. https://doi.org/10.1007/s40471-017-0128-6.
Skrivankova VW, Richmond RC, Woolf BAR, et al. Strengthening the reporting of Observational Studies in Epidemiology using mendelian randomization: the STROBE-MR Statement. JAMA Oct. 2021;26(16):1614–21. https://doi.org/10.1001/jama.2021.18236.
Sun BB, Maranville JC, Peters JE, et al. Genomic atlas of the human plasma proteome. Nat Jun. 2018;558(7708):73–9. https://doi.org/10.1038/s41586-018-0175-2.
Malik R, Chauhan G, Traylor M, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet Apr. 2018;50(4):524–37. https://doi.org/10.1038/s41588-018-0058-3.
Di Angelantonio E, Thompson SG, Kaptoge S, et al. Efficiency and safety of varying the frequency of whole blood donation (INTERVAL): a randomised trial of 45 000 donors. The Lancet. 2017;390(10110):2360–71. https://doi.org/10.1016/s0140-6736(17)31928-1.
Adams HP Jr, Bendixen BH, Kappelle LJ, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke Jan. 1993;24(1):35–41. https://doi.org/10.1161/01.str.24.1.35.
Burgess S, Thompson SG, Collaboration CCG. Avoiding bias from weak instruments in mendelian randomization studies. Int J Epidemiol Jun. 2011;40(3):755–64. https://doi.org/10.1093/ije/dyr036.
Burgess S, Thompson SG. Bias in causal estimates from mendelian randomization studies with weak instruments. Stat Med May. 2011;20(11):1312–23. https://doi.org/10.1002/sim.4197.
Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in mendelian randomization studies. Int J Epidemiol Oct. 2013;42(5):1497–501. https://doi.org/10.1093/ije/dyt179.
Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol Nov. 2013;37(7):658–65. https://doi.org/10.1002/gepi.21758.
Borenstein M, Hedges LV, Higgins JPT, Rothstein HR. Introduction to meta-analysis. Int Coach Psychol Rev 2009. https://onlinelibrary.wiley.com/doi/book/10.1002/9780470743386
Burgess S, Thompson SG. Mendelian randomization: methods for causal inference using genetic variants (2nd ed.). Chapman and Hall/CRC. 2021. https://doi.org/10.1201/9780429324352
Greco MF, Minelli C, Sheehan NA, Thompson JR. Detecting pleiotropy in mendelian randomisation studies with summary data and a continuous outcome. Stat Med Sep. 2015;20(21):2926–40. https://doi.org/10.1002/sim.6522.
Engels EA, Schmid CH, Terrin N, Olkin I, Lau J. Heterogeneity and statistical significance in meta-analysis: an empirical study of 125 meta-analyses. Stat Med Jul. 2000;15(13):1707–28. https://doi.org/10.1002/1097-0258(20000715)19:13>1707::aid-sim491<3.0.co;2-p.
Burgess S, Bowden J. Integrating summarized data from multiple genetic variants in Mendelian randomization: bias and coverage properties of inverse-variance weighted methods.arXiv. 2015. https://arxiv.org/abs/1512.04486.
Rees JMB, Wood AM, Dudbridge F, Burgess S. Robust methods in mendelian randomization via penalization of heterogeneous causal estimates. PLoS ONE. 2019;14(9):e0222362. https://doi.org/10.1371/journal.pone.0222362.
Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife May. 2018;30(7). https://doi.org/10.7554/eLife.34408.
Zhao Q, Wang J, Hemani G, Bowden J, Small DS. Statistical inference in two-sample summary-data mendelian randomization using robust adjusted profile score. the Annals of Statistics. arXiv. 2019. https://arxiv.org/abs/1801.09652.
Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol Apr. 2015;44(2):512–25. https://doi.org/10.1093/ije/dyv080.
Zennadi R, Moeller BJ, Whalen EJ, et al. Epinephrine-induced activation of LW-mediated sickle cell adhesion and vaso-occlusion in vivo. Blood Oct. 2007;1(7):2708–17. https://doi.org/10.1182/blood-2006-11-056101.
Delahunty M, Zennadi R, Telen MJ. LW protein: a promiscuous integrin receptor activated by adrenergic signaling. Transfus Clin Biol Mar-Apr. 2006;13(1–2):44–9. https://doi.org/10.1016/j.tracli.2006.02.022.
Traylor M, Farrall M, Holliday EG, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11(11):951–62. https://doi.org/10.1016/s1474-4422(12)70234-x.
Maida CD, Norrito RL, Daidone M, Tuttolomondo A, Pinto A. Neuroinflammatory mechanisms in ischemic stroke: focus on cardioembolic stroke, background, and therapeutic approaches. Int J Mol Sci Sep. 2020;4(18). https://doi.org/10.3390/ijms21186454.
Goebel J, Gaida BJ, Wanke I, et al. Is histologic Thrombus composition in Acute Stroke Linked to Stroke etiology or to interventional parameters? AJNR Am J Neuroradiol Apr. 2020;41(4):650–7. https://doi.org/10.3174/ajnr.A6467.
Acknowledgements
We thank the authors and participants of all GWASs that we have used for making their results publicly available, which provided summary statistics data for making these analyses. Full acknowledgement and funding statements for each of these resources are available via the relevant cited reports. The MEGASTROKE project received funding from sources specified at http://www.megastroke.org/acknowledgments.html.
Funding
This study was supported by the China Postdoctoral Science Foundation (grant: 2021M690115) and National Natural Science Foundation of China (grant: 82103921).
Author information
Authors and Affiliations
Contributions
The study was conceived and designed by LS, ZZ, and JZ. LS, ZZ, and JZ coordinated the study. LS, DG, YJ, MS, PY, YW, FL, ZZ, and JZ contributed to data collection. LS performed the statistical analysis and prepared the first draft of manuscript. ZZ and JZ revised the paper and helped to write the final draft of manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The protocol and data were approved by the ethics committee of the original genome-wide association studies (GWASs), and written informed consent was obtained from each participant prior to data collection.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Sun, L., Guo, D., Jia, Y. et al. Intercellular adhesion molecule 4 and ischemic stroke: a two-sample Mendelian randomization study. Thrombosis J 21, 40 (2023). https://doi.org/10.1186/s12959-023-00485-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12959-023-00485-4